Abstract
The last 6 years have witnessed an explosion of discoveries at the interface of glycobiology and immunology. Binding of clustered oligosaccharides has turned out to be a very frequent mode by which human antibodies have developed broadly neutralizing activity against HIV. This mini-review will cover many recent developments in the HIV antibody field, as well as emerging data about Dengue broadly neutralizing antibodies.
Keywords: Dengue broadly neutralizing antibodies, glycoproteins, HIV broadly neutralizing antibodies, multivalency, protein–carbohydrate interactions
Introduction
The binding of carbohydrates at biological surfaces is an important mechanism of recognition and adhesion, either between cells of the same species, or between host and pathogen. Important examples of such interactions include recognition of egg by sperm during fertilization (Sinowatz et al. 2001), adhesion of cancer cells to vasculature during metastasis (Kannagi et al. 2004), signaling of stem cell differentiation (Lanctot et al. 2007), adhesion of viruses to antigen-presenting cells to facilitate infection (Zhang et al. 2014), and binding of antibodies to viruses to facilitate protection. This review will focus primarily on carbohydrate-mediated antibody-virus interactions, about which a great deal has been discovered in the last 5–6 years.
Multivalency
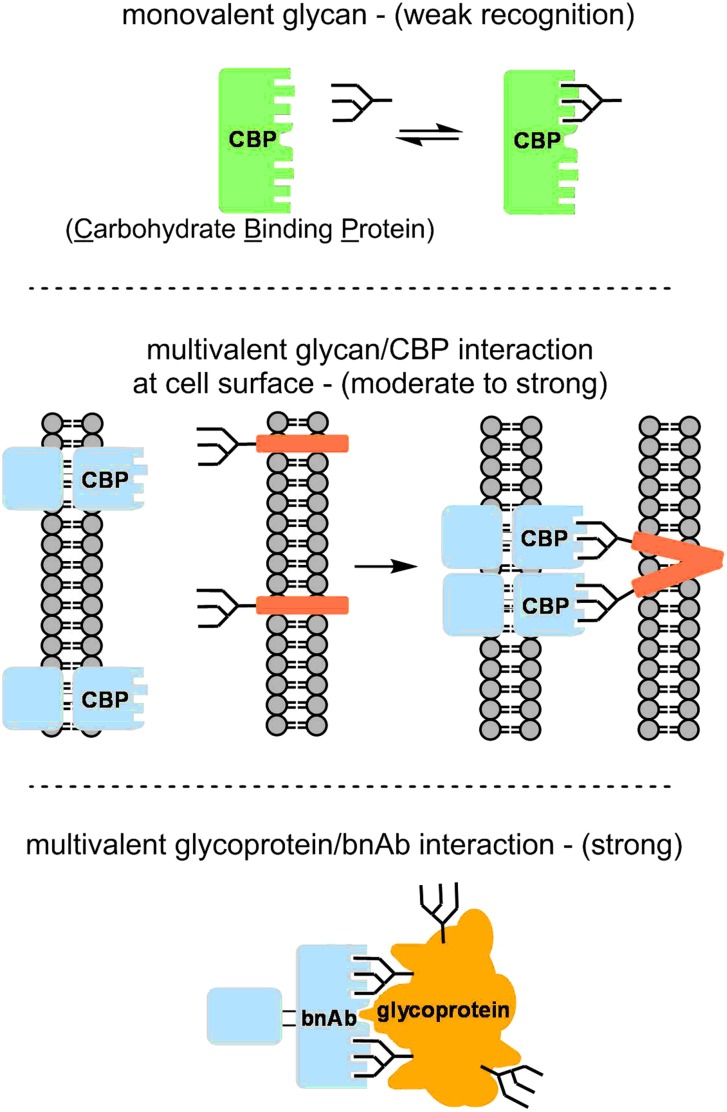
In general, protein–carbohydrate interactions are weak, with Kd's in the millimolar to micromolar range. However, much higher affinities, with Kd's in the nanomolar or picomolar range, can be achieved by multivalent interactions, in which clusters of mono- or oligosaccharides interact with proteins (or protein clusters) containing multiple carbohydrate-binding pockets (Figure 1) (Fasting et al. 2012). This enhanced affinity due to multivalency is sometimes referred to as “avidity”. In many cases, carbohydrate clusters are formed dynamically on membranes, where lateral diffusion of glycolipids or glycoproteins may bring carbohydrates into proximity. Alternatively, several glycans in fixed proximity on a single glycoprotein molecule may comprise a multivalent recognition motif, and polypeptide surfaces may be recognized together with carbohydrates.
Fig. 1.
Multivalent binding enhancement. This figure is available in black and white in print and in color at Glycobiology online.
Many HIV broadly neutralizing antibodies bind to carbohydrates
Antibodies which bind multivalent carbohydrate structures are of high relevance in the design of vaccines against HIV (Horiya et al. 2014). To appreciate such antibodies, one must consider the other types of antibodies which more frequently arise during HIV infection (Burton and Mascola, 2015). The sole targets of HIV neutralizing antibodies are the “Env” proteins gp41 and gp120. These are biosynthesized as gp160, which trimerizes and is then cleaved to gp41 and gp120, which remain associated as an unstable trimer of heterodimers, with gp41 serving as the membrane anchor. Many HIV antibodies bind only to dysfunctional Env monomers that have disassembled from each other or from the virus, but fail to bind trimeric Env or block viral entry (non-neutralizing Abs). Some antibodies do bind to intact trimeric Env, and thus neutralize the virus; however, most of these bind to polypeptide regions of high-sequence variability, and thus neutralize just a narrow subset of viral strains (strain-specific Abs). In contrast, broadly neutralizing antibodies (bnAbs), which develop in 20% of HIV-positive individuals, bind to conserved epitopes on mature trimeric Env, with some neutralizing up to 90% of viral strains. The discovery of bnAbs, starting in the 1990s and accelerating since 2009, is significant because it shows that the human immune system is capable of producing a useful antibody response against HIV. Moreover, by studying bnAbs, we can identify which epitopes on the Env proteins gp120 or gp41 must be targeted to achieve broad neutralization.
Some bnAbs bind to conserved polypeptide epitopes of HIV Env, such as the CD4-binding site, which are essential for viral function, but also sterically recessed, masked by glycans and variable polypeptide sequence at the protein surface (Saphire et al. 2001; Zhou et al. 2010; Jardine et al. 2013). The other major class of bnAbs, of interest in this review, are those which bind to some of the ∼25 N-linked glycans decorating gp120 and gp41. The first such bnAb to be discovered was 2G12, isolated (Buchacher et al. 1994) as a hybridoma from patient serum in the 1990s.
2G12: the first known carbohydrate-directed bnAb
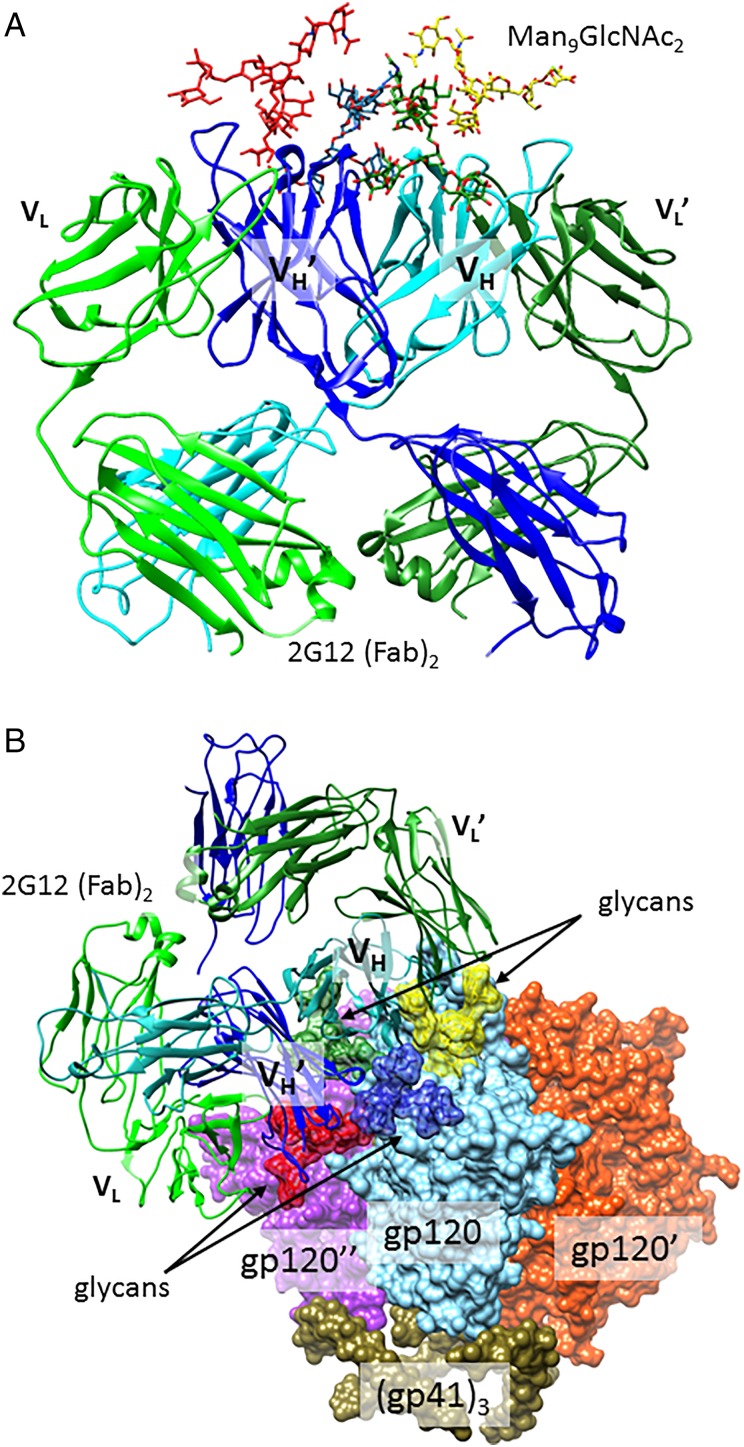
At the time of its discovery (Buchacher et al. 1994), 2G12 was remarkable in neutralizing ∼30% of HIV strains tested, although breadth was largely restricted to Clade B viruses (Binley et al. 2004). 2G12 was soon recognized to bind N-linked glycans, particularly those containing mannose, based on mutation and glycosidase-digestion studies (Trkola et al. 1996; Sanders et al. 2002; Scanlan et al. 2002). In 2003, X-ray crystallography showed in atomic detail that 2G12 cocrystallizes with Man9GlcNAc2 high-mannose glycans with one glycan bound to each of four antibody sites (Figure 2A) (Calarese et al. 2003). From these data, it can be seen that 2G12 interacts only with the Man(α1–2Man) motifs in the non-reducing D1 and D3 termini of the glycans. This crystal structure did not contain gp120 protein, and thus did not directly indicate which sites on gp120 bear the glycans involved in the 2G12 interaction. However, a recent 17-Å cryoEM structure of the 2G12 in complex with trimeric Env, modeled together with several gp120 crystal structures, and neutralization data for glycan deletion mutants, support a model (Figure 2B) in which the four glycans bound are at positions N332, N295, N392 and N339, although the interaction with N339 is less necessary for neutralization, and may be less extensive than the others (Murin et al. 2014). Although isolated Man9 oligosaccharide binds to 2G12 with modest affinity (Kd ∼180 µM) (Wang et al. 2008), gp120 is recognized with very tight affinity (Kd ∼5 nM), as the result of multivalent binding enhancement (Hoorelbeke et al. 2013).
Fig. 2.
2G12 contacts to gp120 glycans. (A) The co-crystal structure of 2G12 Fab dimer in complex with four Man9GlcNAc2 glycans (Calarese et al. 2003; PDB 1OP5). (B) A model of the 2G12 Fab dimer interaction with Man9GlcNAc2 glycans on the surface of trimeric HIV envelope. Glycans at N295, 339, 392 and 332 are depicted in red, blue, yellow and forest green, respectively. All other glycans are deleted for clarity. Although one 2G12 Fab dimer binds to each of the three gp120 protomers, only one 2G12 is shown, for clarity. This model was constructed by aligning the 2G12 (PDB 1OP5) and Env (PDB 4NCO) structures as in Murin et al. (2014). This figure is available in black and white in print and in color at Glycobiology online.
The ability of 2G12 to recognize several glycans simultaneously is facilitated by an extremely unusual domain-exchanged antibody architecture (Figure 2A), in which each heavy chain variable (VH) domain is paired with the light chain variable domain of the opposite Fab , rather than its own VL (Calarese et al. 2003, 2005). The resulting cross-linked Fab dimer contains two “unconventional” mannose-binding sites at the central interface, in addition to “conventional” binding sites at the and interfaces. Numerous laboratories have prepared oligomannose glycan clusters which are recognized by 2G12, but none of these constructs has yet proved useful for eliciting 2G12-like antibodies in vivo (Horiya et al. 2014). The uniqueness of 2G12's domain-exchanged structure among antibodies so far characterized raises the questions of whether domain-exchanged antibodies can be produced in all individuals, and whether they can be elicited by a vaccine. Like most bnAbs, 2G12 is the product of extensive affinity maturation, with numerous (38) mutations from its putative germline antibody sequence (Doores, Fulton, et al. 2010; Huber et al. 2010). Although just five of these mutations are necessary to initiate significant domain exchange in the context of this antibody, it is still questionable whether domain exchange will be found in other antibodies. More important, recently discovered bnAbs exhibit much broader and more potent neutralization than 2G12, without domain-exchanged structure, and therefore much of the interest in the field has shifted from 2G12 to the newer bnAbs.
Advances in discovery and characterization of HIV bnAbs
With improved high-throughput neutralization assays (Binley et al. 2004; Seaman et al. 2010), methods of B-cell sorting (Scheid et al. 2009), sequencing and antibody cloning (Tiller et al. 2008), numerous new carbohydrate-binding bnAbs have now been isolated from cohorts of HIV-positive patients who exhibit highly neutralizing sera (Simek et al. 2009). Although X-ray crystallography has played a major role in their characterization, our understanding of these antibodies and the glycoproteins they bind to has been enhanced by advances in chemical glycobiology, such as carbohydrate microarray technologies (Muthana and Gildersleeve, 2014), chemoenzymatic synthesis (Amin et al. 2013) and improved mass spectrometry of glycopeptides (Desaire, 2013). These new bnAbs lack the unusual domain-exchanged architecture found in 2G12, most exhibit much greater breadth (70–85% of viruses tested), and many exhibit extremely potent neutralization (IC50 ∼0.01–0.1 µg/mL). In all cases, antibody binding and neutralization is dependent not only on additional glycans but also on conserved regions of gp120 polypeptide sequence. Although a great deal of multivalency research has focused on presentations of multiple copies of a single ligand, these examples highlight the importance in nature of “heteromultivalency” (i.e. clustering of several different ligands) (Blanco et al. 2013).
Prior to discussing crystal structures of these antibodies in complex with HIV Env proteins, it is important to point out that the glycans seen in a structure may not be those present in native HIV Env. Viral glycoproteins can be difficult to isolate in quantity from native virions, contain heterogeneous glycosylation and are difficult to crystallize. To facilitate crystallography, glycoproteins are normally produced recombinantly, in the presence of inhibitors of glycan processing pathways, or in GnT I-deficient cells, which results exclusively in high-mannose glycoforms at all sites. Next, glycosidase treatment of a complex is often used to remove conformationally flexible glycans prior to crystallization. Finally, portions of a glycan which are not conformationally restricted by an interaction with the bnAb may be disordered and thus not modeled in the crystal structure.
Although the artificially elevated high-mannose content of gp120 constructs derived from GnT I−/− cell lines may be prone to misinterpretations, it is likely only a moderate deviation from the true glycosylation of native trimeric HIV Env, which is less heterogeneous than that of typical glycoproteins. In fact, mass spectrometric analysis of “native” gp120 (isolated from HIV viral samples produced in infected human lymphocytes) reveals an unusually high percentage (60–80%) of high-mannose glycoforms (Doores, Bonomelli, et al. 2010; Bonomelli et al. 2011). This likely results from the high spatial density of glycosylation sites—∼25 per gp120 monomer, compared with 2–3 in typical mammalian glycoproteins (Apweiler et al. 1999)—which sterically obstructs glycosidase processing. This crowding is greatly enhanced by the trimeric structure of the envelope protein, and this trimer is rendered even more compact upon furin-cleavage (Pritchard, Vasiljevic, et al. 2015). Although many glycan analysis studies have detected a prevalence of complex-type glycans in recombinant gp120-related constructs (Leonard et al. 1990; Go et al. 2011), these analyses have not utilized fully native samples, i.e. trimeric, furin-cleaved, full-length HIV Env, isolated from virions produced in human lymphocytes.
bnAbs which bind to the N332 glycan
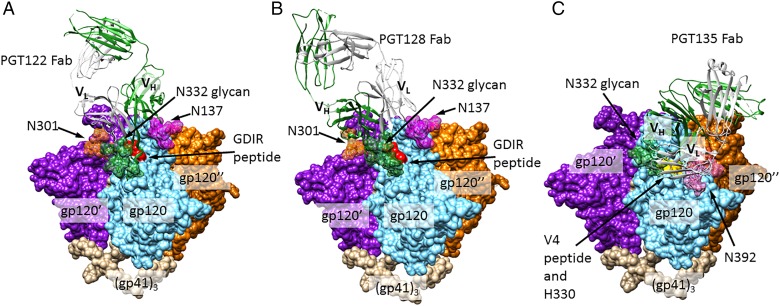
Among the more recently discovered bnAbs, a major class, exemplified by PGT128, PGT122 and PGT135 bnAb families (Figure 3A–C, respectively) (Walker et al. 2011), is similar to 2G12 in binding to two or more glycans, including a Man8/9GlcNAc2 glycan at N332. It is noteworthy that the glycan at N332 is shifted to N334 in some HIV strains, and yet is still recognized by PGT128 and 122, exemplifying a promiscuity of recognition which occurs with some PGT-family antibodies (Sok et al. 2014). PGT128 and −122 potently neutralize ∼70% of HIV strains tested and PGT122 has been shown to protect macaques from infection at very low doses (Moldt et al. 2012). Crystal structure data of a gp120 outer domain protein in complex with PGT128 show that, in addition to the N332 glycan, this bnAb recognizes a glycan at N301 as well as backbone atoms of the 324GDIR327 peptide residues (a highly conserved motif; Garces et al. 2014) at the base of the V3 loop, and potentially other glycans (Pejchal et al. 2011). Extensive antibody contacts are observed with the non-reducing terminal mannose residues and core of the N332 Man8/9GlcNAc2, consistent with glycan analysis data indicating the high prevalence of this glycoform at this position (Pritchard, Spencer, et al. 2015). Interactions with the N301 glycan are focused on the second GlcNAc and three core mannose residues, compatible with recognition of either high-mannose or complex glycan at this position, although PGT128 recognizes only high mannose and not complex-type glycans on a glycan array.
Fig. 3.
N332-dependent bnAbs. (A) Crystal structure of PGT122 Fab bound to BG505 SOSIP.664 gp140 trimer, a native-like, cleaved trimeric HIV Env construct (PDB ID: 4NCO). N301, 322 and 137 glycans are shown in forest green, orange and magenta, respectively, with all other glycans removed for clarity. Identically bound PGT122 Fabs on the other two gp120 protomers are also not shown. (B) A model of the PGT128-HIV Env interaction, created by aligning a gp120 chain from the BG505 Env structure (PDB ID: 4NCO) with the gp120 chain of a PGT128-eODmV3 gp120 co-crystal structure (PDB ID: 3TYG). N332 and 301 glycans depicted (forest green and orange, respectively) are from the PGT128 structure and N137 glycan (magenta) is from the BG505 structure. (C) A model of the PGT135-HIV Env interaction, created in a manner analogous to (B). PGT135 and the N332 and 392 glycans (forest green and pink, respectively) are from a structure of the quaternary complex PGT135 + gp120 core + CD4 + 17b Fab (PDB ID: 4JM2). All glycans outside the bnAb epitopes are omitted for clarity, as are the two Fabs binding other gp120 protomers. This figure is available in black and white in print and in color at Glycobiology online.
PGT122-family antibodies, which are derived from a different human donor than PGT128, bind to a closely related epitope. A crystal structure of PGT122 in complex with a trimeric form of HIV Env (Figure 3B) shows that, in addition to the major interaction with the mannose D3 arm and reducing core of N332 high-mannose glycan, another major interaction is made with the reducing core of the N137 glycan, and a minor interaction is made with N301. For the clonally related PGT121, binding to sialylated biantennary glycan is observed on arrays (Mouquet et al. 2012; Julien, Sok, et al. 2013) and PGT121 has crystalized with this glycan in its N137-glycan-binding site, though it is possible that high-mannose binds this position in PGT122. PGT122-family antibodies, like PGT128, also bind to Env V3 loop residues 324GDIR327, but unlike PGT128, interact with sidechain atoms rather than the backbone, and therefore binding is quite sensitive to mutation of these residues. Antibody paratope residues which recognize the N332 glycan and 324GDIR327 sequence are present in the putative germline ancestor of all PGT122-family antibodies, suggesting that these structures were originally recognized by the germline antibody, and may be important in a vaccine to prime a PGT122-like response (Garces et al. 2014).
PGT135-family antibodies are a third major class of bnAbs which bind to a high-mannose glycan at N332, although PGT135 exhibits substantially less neutralizing breadth (∼33% of strains tested) than PGT122 and PGT128. PGT135 (Figure 3C) can be seen making extensive contacts to a GlcNAc and several Man residues of N332 glycan and also with terminal D1 and D3 mannose residues of a Man8GlcNAc2 glycan at N392. In contrast to PGT122 and PGT128 antibodies, PGT135 does not contact the 324GDIR327 sequence, but instead is critically dependent on 330H, and makes additional contacts to residues of the V4 loop.
Bnabs to Env quaternary interfaces
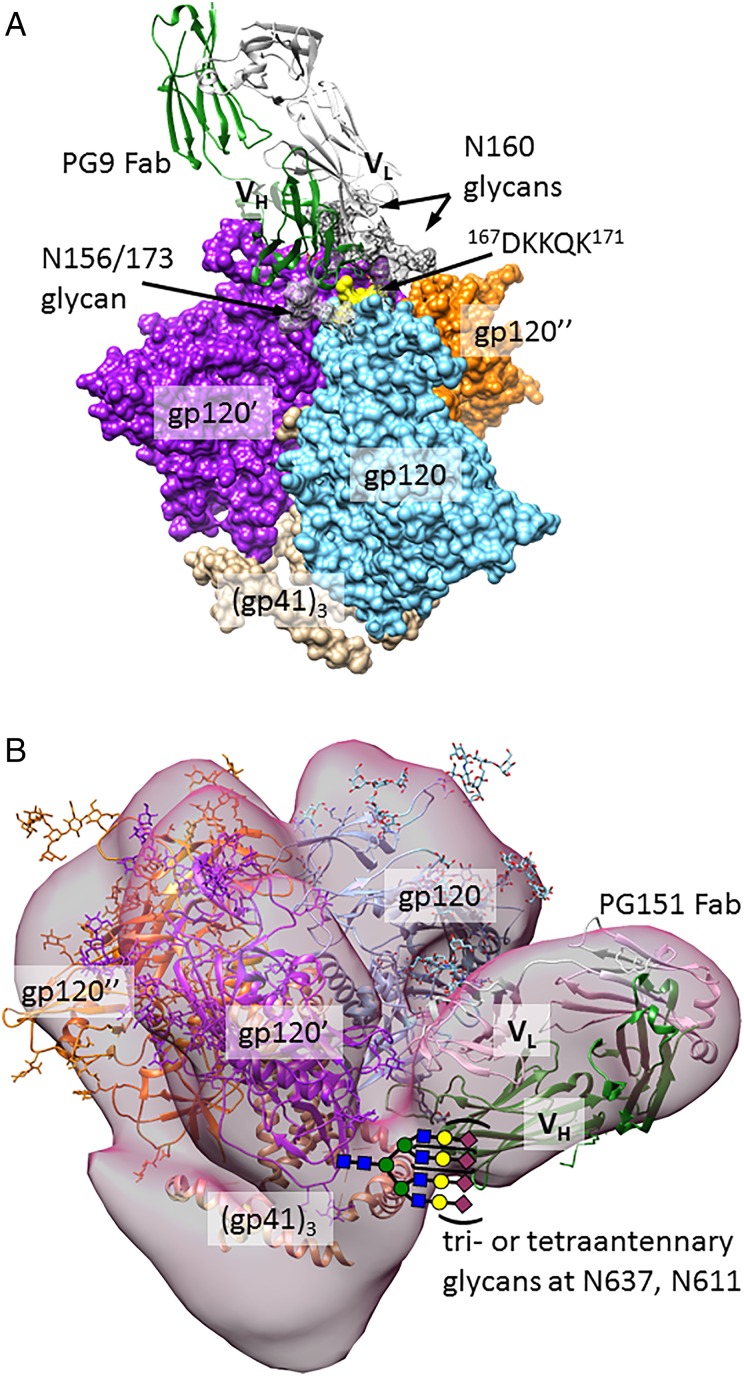
Additional classes of HIV bnAbs bind to glycans at interfaces between protein subunits of HIV Env. PG9 and PG16, reported in 2009, were the first glycan-dependent bnAbs discovered after 2G12 (Walker et al. 2009). Among glycan-dependent antibodies, PG9 exhibits the greatest neutralizing breadth (77% of strains tested). PG9 and PG16 bind only to trimeric forms of Env, making contact near the trimer apex (Julien, Lee, et al. 2013), likely with two Man5GlcNAc glycans from N160 of neighboring gp120 monomers, as well as with a sialylated complex or hybrid glycan (Pancera et al. 2013) at N173 or the spatially close position N156 (Figure 4A) (McLellan et al. 2011). Consistent with this, synthetic glycopeptides exhibit enhanced recognition by PG9 when they contain both sialylated biantennary- and high-mannose glycans (Amin et al. 2013), or two glycans attached to N160-like peptide positions (Alam et al. 2013). In addition to making glycan contacts, PG9 interacts with a lysine-rich section of the gp120 V1/V2 loop, using a CDRH3 loop which contains sulfated tyrosines. As was seen for the N332-glycan-related epitope, this trimer apex epitope is the target of several clonally unrelated antibody families isolated from different patients (PGT141–145 (Walker et al. 2011) and CH01–04 (Bonsignori et al. 2011)).
Fig. 4.
Quaternary structure-dependent bnAbs. (A) A model of the PG9-HIV Env interaction, created by aligning V1/V2 loops of the BG505 trimeric Env structure (PDB ID: 4NCO) with the V1/V2 loop of PG9 Fab-CAP45-V1/V2 structure (PDB ID: 3U4E). Glycans (dark gray for N160 and light grey for N156/173) are from the PG9 structure. All glycans outside the PG9 epitope are omitted for clarity. Only one PG9 Fab binds to the trimer. (B) A model of the PGT151-HIV Env interaction, created as in Blattner et al. (2014), by alignment of an unliganded PGT151 Fab crystal structure (PDB ID: 4NUG) and the trimeric Env crystal structure (PDB ID: 4NCO) with a cryo-EM reconstruction of trimeric Env (EMD 5919). The Env crystal structure was obtained with EndoH-treated protein, and thus lacks most glycans. This figure is available in black and white in print and in color at Glycobiology online.
A very recently discovered class of antibodies binds glycans and peptide at the interface of gp120 and gp41. For the PGT151–158 family, glycan array studies show that these antibodies bind to tri- and tetraantennary complex-type glycans, and similar array signals are obtained for fucosylated and non-fucosylated versions of these glycans (Blattner et al. 2014; Falkowska et al. 2014). For the tetraantennary glycan, non-sialylated and α3-sialylated versions are both recognized, but only a weak signal is detected for the α6-sialylated glycan. Although no crystal structure is available for PGT151 antibodies in complex with HIV Env, a cryoEM structure and neutralization data of glycan deletion mutants are consistent with binding to glycans at positions N611, N637, to peptide residues at the interface between gp41 and gp120, and to additional glycans in a strain-dependent manner. Gp41 glycosylation is highly heterogeneous, which leads to incomplete neutralization by antibodies dependent on gp41 glycans. In the case of PGT151–158, neutralization plateaus at <80% among 26% of viral strains tested; this may complicate efforts to design vaccines based on mimicry of this epitope. Another gp41–gp120 interface antibody with distinct specificity, 35O22, was recently described (Huang et al. 2014); this antibody binds to the N88, N230 and N241 glycans.
bnAbs to dengue viruses
Dengue Virus (DENV) is a mosquito-borne pathogen which infects 50–100 million people annually, causing 500,000 cases of Dengue Hemorrhagic Fever (DHF). This severe form of the disease results in 22,000 deaths annually, mostly in children. There are four distinct serotypes of the virus (DENV1–4), and antibodies against each serotype typically do not neutralize the other three. Although a first infection tends to be mild, antibodies against the first serotype fail to protect against the other serotypes, and actually enhance subsequent infections, leading to DHF. As in the case of HIV, it would be desirable to design vaccines which could elicit broadly neutralizing Dengue antibodies, and, as in the case of HIV, such antibodies have been isolated from DHF patients and were recently structurally characterized (Dejnirattisai et al. 2015; Rouvinski et al. 2015).
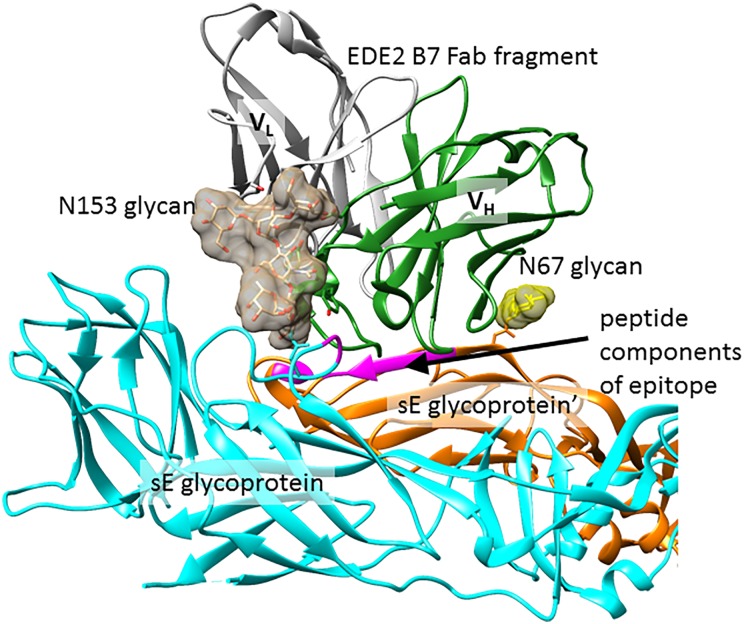
As is the case with the HIV bnAbs described in this review, Dengue bnAbs bind to epitopes comprising one or more glycans and conserved polypeptide residues in the Dengue envelope (E glycoprotein). These bnAbs can be divided into two classes: the EDE2 family, which neutralizes only viruses which are glycosylated at N153, and the EDE1 family, which can neutralize virus without N153. Both types of antibody can be seen binding to N67 and N153 glycans in co-crystal structures with a soluble form of E glycoprotein (Figure 5). Antibody interactions with glycans are focused on the core Man3GlcNAc2 residues. Although the reducing terminal GlcNAc residue is fucosylated in these structures, the fucose makes no contact with the antibody. Crystallographic data contain electron density for further glycan residues at the non-reducing ends, but disorder and/or heterogeneity prevents assignment, and there is no evidence that these residues make further contacts with the antibodies. Insight about the glycoforms present on the virus comes from mass spectrometric analysis of virus-derived glycoprotein E; this shows that the glycans are heterogeneous, including both high-mannose and non-sialylated hybrid- and complex-type glycans, and this is consistent with binding of virus to DC-SIGN (Tassaneetrithep et al. 2003) and galactose-binding lectins (Lei et al. 2015). In addition to contacts with glycans, antibody contacts to the polypeptide surface of the E glycoprotein are extensive. For both EDE1 and EDE2 antibodies, this includes two discontinuous strips of 5–8 peptide residues, one of which is the fusion loop necessary for viral fusion to the host membrane, and further peptide regions on both sides of the protein dimer interface are recognized by individual members of the EDE1 and EDE2 antibody families.
Fig. 5.
Dengue glycoprotein recognition by bnAb EDE2 B7. A crystal structure of dimeric sE glycoprotein in complex with a Fab fragment of bnAb EDE2 B7 (PDB ID: 4UT6). N153 and N67 glycans are shown as charcoal and yellow surfaces, respectively, and peptide components of the epitope are colored purple in both sE glycoprotein chains. The other half of this complex (not shown) is related by C2 symmetry and contains a second Fab fragment making identical contacts to the glycoprotein. This figure is available in black and white in print and in color at Glycobiology online.
Conclusion and outlook
The frequent targeting of carbohydrate or glycopeptide epitopes by broadly neutralizing antibodies is now clear. Multivalent recognition of clustered glycans, or the recognition of a peptide motif together with a glycan, may be the key to achieving specificity despite the possible presence of the glycan in other contexts on “self” proteins. The importance of glycopeptide recognition is unlikely to be limited to HIV and Dengue antibodies; rather it is likely that studies to date have only scratched the surface of this phenomenon. Other important pathogens such as Hepatitis C and Ebola viruses have glycosylated envelope proteins, and recent improvements in technologies for analysis of the human antibody repertoire are likely to uncover many additional epitopes of use in vaccine design.
Funding
The support of the US National Institutes of Health (R01 AI090745 and R01 AI113737) is gratefully acknowledged. Molecular graphics and analyses were performed with the UCSF Chimera package (http://www.cgl.ucsf.edu/chimera). Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Conflict of interest statement
None declared.
Abbreviations
bnAbs, broadly neutralizing antibodies; DENV, dengue virus; DHF, dengue hemorrhagic fever.
References
- Alam SM, Dennison SM, Aussedat B, Vohra Y, Park PK, Fernández-Tejada A, Stewart S, Jaeger FH, Anasti K, Blinn JH et al. . 2013. Recognition of synthetic glycopeptides by HIV-1 broadly neutralizing antibodies and their unmutated ancestors. Proc Natl Acad Sci U S A. 110:18214–18219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MN, McLellan JS, Huang W, Orwenyo J, Burton DR, Koff WC, Kwong PD, Wang L-X. 2013. Synthetic glycopeptides reveal the glycan specificity of HIV-neutralizing antibodies. Nat Chem Biol. 9:521–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1473:4–8. [DOI] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H et al. . 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 78:13232–13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco JLJ, Mellet CO, Fernandez JMG. 2013. Multivalency in heterogeneous glycoenvironments: Hetero-glycoclusters, -glycopolymers and -glycoassemblies. Chem Soc Rev. 42:4518–4531. [DOI] [PubMed] [Google Scholar]
- Blattner C, Lee Jeong H, Sliepen K, Derking R, Falkowska E, de la Peña Alba T, Cupo A, Julien J-P, van Gils M, Lee Peter S et al. . 2014. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 40:669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2011. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS ONE. 6:e23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Hwang K-K, Chen X, Tsao C-Y, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE et al. . 2011. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 85:9998–10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, JungBauer A et al. . 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. Aids Res Hum Retrovir. 10:359–369. [DOI] [PubMed] [Google Scholar]
- Burton DR, Mascola JR. 2015. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 16:571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Lee H-K, Huang C-Y, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong C-H, Wilson IA. 2005. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc Natl Acad Sci U S A. 102:13372–13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, Mimura Y, Kunert R, Zhu P, Wormald MR, Stanfield RL, Roux KH et al. . 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 300:2065–2071. [DOI] [PubMed] [Google Scholar]
- Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T et al. . 2015. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol. 16:170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaire H. 2013. Glycopeptide analysis, recent developments and applications. Mol Cell Proteomics. 12:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. 2010. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A. 107:13800–13805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Fulton Z, Huber M, Wilson IA, Burton DR. 2010. Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged. J Virol. 84:10690–10699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le Khoa M, Ramos A, Doores Katie J, Lee Jeong H, Blattner C, Ramirez A, Derking R, van Gils Marit J, Liang C-H et al. . 2014. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 40:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasting C, Schalley CA, Weber M, Seitz O, Hecht S, Koksch B, Dernedde J, Graf C, Knapp EW, Haag R. 2012. Multivalency as a chemical organization and action principle. Angew Chem Int Ed. 51:10472–10498. [DOI] [PubMed] [Google Scholar]
- Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien JP, Hua YZ, Cupo A, Moore JP et al. . 2014. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 159:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go EP, Hewawasam G, Liao H-X, Chen H, Ping L-H, Anderson JA, Hua DC, Haynes BF, Desaire H. 2011. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. J Virol. 85:8270–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoorelbeke B, van Montfort T, Xue J, LiWang PJ, Tanaka H, Igarashi Y, Van Dammef EJM, Sanders RW, Balzarini J. 2013. HIV-1 envelope trimer has similar binding characteristics for carbohydrate-binding agents as monomeric gp120. FEBS Lett. 587:860–866. [DOI] [PubMed] [Google Scholar]
- Horiya S, MacPherson IS, Krauss IJ. 2014. Recent strategies targeting HIV glycans in vaccine design. Nat Chem Biol. 10:990–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang G-Y, Druz A et al. . 2014. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 515:138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M, Le KM, Doores KJ, Fulton Z, Stanfield RL, Wilson IA, Burton DR. 2010. Very few substitutions in a germ line antibody are required to initiate significant domain exchange. J Virol. 84:10700–10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien J-P, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang P-S, MacPherson S, Jones M et al. . 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science. 340:711–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ et al. . 2013. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. 110:213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A et al. . 2013. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 9:e1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannagi R, Izawa M, Koike T, Miyazaki K, Kimura N. 2004. Carbohydrate-mediated cell adhesion in cancer metastasis and angiogenesis. Cancer Sci. 95:377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot PM, Gage FH, Varki AP. 2007. The glycans of stem cells. Curr Opin Chem Biol. 11:373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Yu H, Dong Y, Yang J, Ye W, Wang Y, Chen W, Jia Z, Xu Z, Li Z et al. . 2015. Characterization of N-glycan structures on the surface of mature dengue 2 virus derived from insect cells. PLoS ONE. 10:e0132122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 265:10373–10382. [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai KF et al. . 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 480:336–U386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B, Rakasz EG, Schultz N, Chan-Hui P-Y, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P et al. . 2012. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 109:18921–18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PNP, Spencer DIR, Seaman MS et al. . 2012. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 109:E3268–E3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murin CD, Julien J-P, Sok D, Stanfield RL, Khayat R, Cupo A, Moore JP, Burton DR, Wilson IA, Ward AB. 2014. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J Virol. 88:10177–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthana SM, Gildersleeve JC. 2014. Glycan microarrays: Powerful tools for biomarker discovery. In IOS Press, 29–41. [DOI] [PubMed]
- Pancera M, Shahzad-ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y et al. . 2013. Structural basis for diverse N-glycan recognition by HIV-1–neutralizing V1–V2–directed antibody PG16. Nat Struct Mol Biol. 20:804–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, Wang S-K, Stanfield RL, Julien J-P, Ramos A, Crispin M et al. . 2011. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 334:1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard LK, Spencer DIR, Royle L, Vasiljevic S, Krumm SA, Doores KJ, Crispin M. 2015. Glycan microheterogeneity at the PGT135 antibody recognition site on HIV-1 gp120 reveals a molecular mechanism for neutralization resistance. J Virol. 89:6952–6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard LK, Vasiljevic S, Ozorowski G, Seabright GE, Cupo A, Ringe R, Kim HJ, Sanders RW, Doores KJ, Burton DR et al. . 2015. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell Reports. 11:1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouvinski A, Guardado-Calvo P, Barba-Spaeth G, Duquerroy S, Vaney M-C, Kikuti CM, Navarro Sanchez ME, Dejnirattisai W, Wongwiwat W, Haouz A et al. . 2015. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 520:109–113. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 76:7293–7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saphire EO, Parren PWHI, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. 2001. Crystal structure of a neutralizing human IgG against HIV-1: A template for vaccine design. Science. 293:1155–1159. [DOI] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann Saphire E, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of α1->2 mannose residues on the outer face of gp120. J Virol. 76:7306–7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H et al. . 2009. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 343:65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG et al. . 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 84:1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G et al. . 2009. Human immunodeficiency virus type 1 elite neutralizers: Individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol. 83:7337–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinowatz F, Töpfer-Petersen E, Kölle S, Palma G. 2001. Functional morphology of the Zona Pellucida. Anatomia, Histologia, Embryologia. 30:257–263. [DOI] [PubMed] [Google Scholar]
- Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien J-P, Menis S, Wickramasinghe L et al. . 2014. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med. 6:236ra263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL et al. . 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 197:823–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 329:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol. 70:1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M et al. . 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 477:466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL et al. . 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 326:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SK, Liang PH, Astronomo RD, Hsu TL, Hsieh SL, Burton DR, Wong CH. 2008. Targeting the carbohydrates on HIV-1: Interaction of oligomannose dendrons with human monoclonal antibody 2G12 and DC-SIGN. Proc Natl Acad Sci U S A. 105:3690–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ren SY, Zuo YF. 2014. DC-SIGN, DC-SIGNR and LSECtin: C-type lectins for infection. Int Rev Immunol. 33:54–66. [DOI] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, Do Kwon Y, Scheid JF, Shi W, Xu L et al. . 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 329:811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]