Abstract
Severe combined immunodeficiency (SCID) mice have widely been used as hosts for human tumor cell xenograft study. This animal model, however, is labor intensive. As zebrafish is largely emerging as a promising model system for studying human diseases including cancer, developing efficient immunocompromised strains for tumor xenograft study are also demanded in zebrafish. Here, we have created the Prkdc-null SCID zebrafish model which provides the stable immune-deficient background required for xenotransplantation of tumor cell. In this study, the two transcription activator-like effector nucleases that specifically target the exon3 of the zebrafish Prkdc gene were used to induce a frame shift mutation, causing a complete knockout of the gene function. The SCID zebrafish showed susceptibility to spontaneous infection, a well-known phenotype found in the SCID mutation. Further characterization revealed that the SCID zebrafish contained no functional T and B lymphocytes which reflected the phenotypes identified in the mice SCID model. Intraperitoneal injection of human cancer cells into the adult SCID zebrafish clearly showed tumor cell growth forming into a solid mass. Our present data show the suitability of using the SCID zebrafish strain for xenotransplantation experiments, and in vivo monitoring of the tumor cell growth in the zebrafish demonstrates use of the animal model as a new platform of tumor xenograft study.
Introduction
The DNA-dependent protein kinase catalytic subunit encoded by Prkdc gene functions in DNA nonhomologous end-joining in mammalian cells [1], [2]. This major DNA double-strand break repair process also functions during lymphocyte development because of its fundamental role in V(D)J recombination mediating immunoglobulin and T-cell receptor gene assembly [3], [4]. Consequently, malfunctioning of the DNA nonhomologous end-joining process in animals causes severe combined immunodeficiency (SCID), and this has been usefully applied to animals to develop the tumor study model with immune-deficient background [5]. In fact, the SCID animal models are now widely used for xenograft study and have contributed tremendously to current understanding of various cancers’ initiation and progression, including prostate cancer [6], ovarian cancer [7], melanoma [8], non–small cell lung cancer [9], multiple myeloma [10], colon cancer [11], and gastric cancer [12]. Use of immune-deficient mouse model has been most commonly accepted to study pathophysiological phenotypes of immune disorders. Thus, different types of genetically engineered mouse strains are now available. These include the single-gene mutation strains such as nude (nu) strain, Scid (scid) strain, nonobese (NOD) strain, recombination activating gene (RAG) strains, and NOD/Scid hybrid strain, etc. [13].
Use of zebrafish in immunological studies has also been introduced since early 2000 [14], and the zebrafish has proven to be one of the best vertebrate models for the immunological studies [15], [16]. In these studies, the gene-disruption strategies were effectively used to define the immunological meanings: Hematopoietic cell transplantation in the zebrafish blood mutant was demonstrated to understand the blood-forming system [15]. The Rag1 mutant of zebrafish was generated and characterized to define the lymphocyte population [16]. In addition, a recent study has revealed that rag2 inactivation of zebrafish shows a reduced number of functional T and B cells, allowing tumor cell engraftment [17]. These studies suggest that the zebrafish has also its potential for the use of the animal as an immune-deficient model system.
Recently, transcription activator-like effector nuclease (TALEN) has been used for the complete elimination of gene function in model or organisms [18], [19]. This technique is based on creating the artificial nuclease that will cut the DNA near a predetermined site and thereby provides a knockout mutation of the gene of interest. Chromosome breaks created by the engineered nuclease undergoes nonhomologous end-joining in the absence of a repair template, introducing the short DNA insertions or deletions that create the targeted gene knockouts. In this study, we applied the TALEN which specifically targets and knocks out the Prkdc gene of zebrafish. Molecular analyses revealed that the TALEN introduced a frame mutation of the Prkdc gene, causing a complete knockout of the gene function. Histologic investigations showed that the transgenic zebrafish contained retarded growth of hematologic organs and impaired lymphocytes development, revealing immunodeficiency of the zebrafish. Intraperitoneal injection of human cancer cell lines into the SCID zebrafish successfully demonstrated the real-time monitoring of the tumor cell growth. The aim of our study was to develop an efficient and laboratory-beneficial zebrafish model for human tumor xenograft study.
Material and Methods
Isolation of Zebrafish PRKDC Gene and Establishment of TALEN Construct
Human PRKDC (protein kinase, DNA-activated, catalytic polypeptide) homolog of zebrafish was searched on the genome database (http://www.ncbi.nlm.nih.gov) (accession no: mRNA, XM_009303401:GI688565118; genomic DNA, NC_007118:GI68550835). The TALEN constructs which target exon 3 of the zebrafish PRKDC were designed by using a software program (TAL Effector Nucleotide Targeter 2.0: TALEN Targeter) of the Bogdanove laboratory (https://boglab.plp.iastate.edu/node/add/talen). The TALEN sequences that recognize the exon 3 of the zebrafish Prkdc gene were 5′-TATGAATTTCTTAGGGGCAT-3′ (left arm, RVD sequence: NG NI NG NN NI NI NG NG NG HD NG NG NI NN NN NN NN HD NI NG) and 5′-TCCTCGGACAGTGGCTGACA-3′ (right arm, RVD sequence: NG HD HD NG HD NN NN NI HD NI NN NG NN NN HD NG NN NI HD NI) (Figure 1). The nucleotide sequence of the spacer between two TALEN targets was 5'-TTCTACAGAGAA-3'. Both TALEN constructs were generated by using TALEN Toolbox kit (Addgene) and by following the protocol provided by Feng Zhang laboratory (reference nature protocol). After the sequence verification, the capped mRNA was produced by in vitro transcription of the plasmid individually consisting of left or right arm sequences using mMESSAGE mMACHINE T7 ULTRA kit (Ambion Co.).
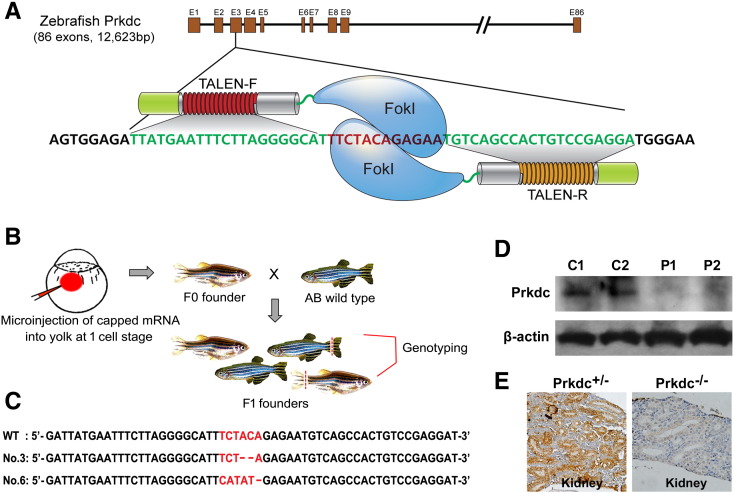
Figure 1.
Schematic illustration of TALEN-mediated knockout of the zebrafish Prkdc gene. (A) Generation of TALEN constructs. Exon 3 of the Prkdc gene was targeted with the 12-bp spacer flanked by left and right TALEN arms. (B) Mutagenesis by microinjection of capped mRNA and strategy for genotyping of the F1 progenies. (C) Sequence confirmation of the mutations with two-nucleotide deletion and six-nucleotide deletion followed by five-nucleotide insertion (mutation no. 3 and 6, respectively). Both mutations cause a shift of the translational frame. (D) Western blot analysis for Prkdc. Kidney and spleen were dissected from several zebrafish, and the pooled samples were processed for the Western blot experiment. C, Prkdc+/−; P, Prkdc−/−. (E) IHC experiment for Prkdc done with the kidney dissections.
Establishment of Prkdc-Null SCID Zebrafish
DNA break at the targeted site was induced by injecting the capped mRNA into the yolk of AB zebrafish embryos using an MMPI-2 microinjector at single cell stage. Injection mixture was prepared by reconstituting the mRNAs (final concentration of each mRNA, 30 ng/ml) in Danieu's buffer mixed with 0.03% phenol red. The F1 progenies were obtained by backcrossing the F0 adult zebrafish to AB wild-type zebrafish. For screening of the germline mutation, the F1 progenies were individually checked by sequence analysis of the polymerase chain reaction (PCR) products amplified from the genomic DNA isolated by tail fin clipping. Primer sequences used to amplify the exon3 sequences were 5′-TTCGCAGGTCTCTGTCTACTGAAAA-3′ (forward) and 5′-CTAGTGCAACAAAGATGACATG-3′ (reverse). The identified heterozygote mutant zebrafish were backcrossed again with the AB zebrafish to produce F2 progenies. The homozygote Prkdc-null zebrafish were screened from the F3 progenies produced by inbreeding of the F2 heterozygote progenies.
Animal Stocks and Embryo Care
The zebrafish used in this study were propagated and raised in a standardized aquaria system (Genomic-Design Co., Daejeon, Korea) (http://zebrafish.co.kr). The system provides continuous water flow, biofiltration tank, constant temperature maintenance at 28.5°C, UV sterilization, and 14-hour light and 10-hour dark cycle. The embryos were maintained in E3 media for 5 days then transferred to a water tank and raised. The embryos to be processed for whole mount analyses were placed in the E3 media with 0.003% phenylthiourea at 24 hpf to inhibit pigmentation. To provide pathogen-free condition for the immune-deficient SCID zebrafish, trimethoprim-sulfomethoxazole was added to water every other week. We strictly followed the Guidelines for the Welfare and Use of Animals in Cancer Research [19].
Ethics Approval
This study got ethics approval from the ethics committee of Department of Laboratory Animal Resources, Yonsei Biomedical Research Institute, Yonsei University College of Medicine (2015-0205).
Statistical Analysis
Statistical analysis was performed by using SPSS 11.0 software. Wilcoxon rank and Kruskal-Wallis rank sum tests were used for the validation of statistical significance.
Histology, Immunohistochemistry (IHC), and In Situ Hybridization (ISH)
Histologic evaluation was done by using 4-μm sections of paraffin-embedded tissue. Hematoxylin and eosin (H&E) staining was performed according to standard protocols [16]. IHC and ISH experiments were carried out as previously described [20], [21]. Antibodies used for IHC were mouse anti-PCNA (ab14370, Abcam), rabbit anti-Ki67 (sc-15,402, Santa Cruz), rabbit anti-RFP (600-401-379, Rockland Inc.), mouse anti-zebrafish Prkdc-C (E7F4J7, Abmart), mouse anti-pancytokeratin (ab961, Abcam), and mouse anti-HLA class1 ABC (ab70328, Abcam). For ISH experiment, partial cDNA (798 bp) sequence of zebrafish Rag1 was PCR-amplified by using F-zRag1 (5′-TTCTGAAGATGCTCCCAGAGC-3′) and R-zRag1 (5′-TGGCACCGTGTGATATTCTTT-3′) and cloned into pCRII vector (Invitrogen, Carlsbad, CA). Riboprobes were generated by using T7 digoxigenin labeling kit (Roche Diagnostics GmbH, Mannheim, Germany). Hybridization was done at 65°C overnight, and a series of stringent wash was done at 68°C. The hybridized riboprobes were then detected by anti-dig antibody binding and visualized by incubating with NBT/BCIP AP substrate solution (Roche Diagnostics GmbH). Counterstaining was done with neutral red. Olympus MVX10 was used for microscopic observation of zebrafish embryos. Photographs from the slide sections were obtained by using Olympus BX51.
Reverse transcriptase (RT)–PCR and Western Blot Analyses
RT-PCR was performed by using the collected sample of the dissected kidney and spleen from three adult zebrafish of each group. RNA sample was extracted by using TRIzol reagent (Invitrogen, Carlsbad, CA). The cDNA was synthesized by using a Maxima First Strand cDNA Synthesis Kit (Thermo Scientific Fermentas, K1641, Glen Burnie, MD). The primer sequences for RT-RCR are shown in Supplementary Table 1. For Western blot assay, whole cell extracts were prepared from the pooled samples of the dissected zebrafish kidney and spleen as described previously [22], [23]. Twenty milligrams of each sample was separated on a 10% SDS–polyacrylamide gel and transferred onto a PVDF membrane (Amersham, GE Health, Sweden). The membrane was incubated for overnight at 4°C with anti–Prkdc-C antibody in a PBS blocking solution (nonfat dry milk). Horseradish peroxidase–conjugated secondary antibody was used for postreaction. Labeled proteins were then detected by ECL reagents and Hyperfilm ECL (Amersham Biosciences).
Cytology
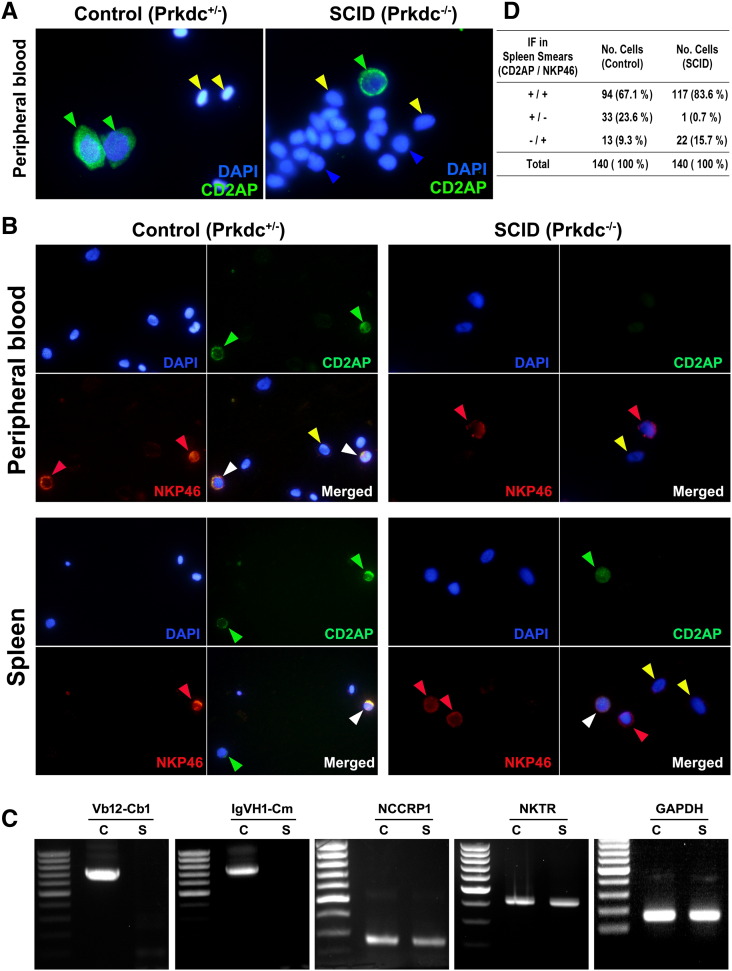
For peripheral blood collection, 3-month-old zebrafish were anesthetized with 0.03% tricaine. Blood was then obtained by cardiac puncture with micropipette tips coated with heparin and was smeared onto glass slides for observation. For kidney and spleen, each organ was dissected under stereomicroscope from 3-month-old zebrafish and directly smeared onto glass slides by using thick smear method. The blood, kidney, and spleen smears were processed through Wright-Giemsa stain as previously described for morphological analyses and differential cell counts [15]. Immunofluorescence (IF) experiments were performed by using the cell smears with the primary antibodies rabbit anti–CD2-associated protein (CD2AP) (sc-9137, Santa Cruz Biotechnology Inc.) and mouse anti-NKP46 (sc-53,599, Santa Cruz Biotechnology Inc.). Cy2-conjugated anti-rabbit and cy3-conjugated anti-mouse antibodies were used as secondary antibodies. Fluorescence images were obtained by using an Olympus BX51.
Flow Cytometry and Cell Sorting
Peripheral blood was collected by cardiac puncture. The hematopoietic cells of kidney and spleen were isolated from the euthanized 3-month-old Prkdc−/+ and Prkdc−/− zebrafish and processed for flow cytometry. For this experiment, the kidney and the spleen from each of the three heterozygote and homozygote mutant zebrafish were dissected under a stereomicroscope and suspended in an ice-cold 0.9× PBS with 5% FBS by repeated pipetting using a P1000 pipettor. The cell suspensions were then passed through a 40-μm Falcon nylon cell strainer (Beckton Dickinson). Propidium iodide (Sigma) was added to a concentration of 1 μm/ml to exclude dead cells. The suspended cells were then kept on ice until analysis. Flow cytometry analysis was done based on propidium iodide exclusion, forward scatter, and side scatter with a FACS flowcytometer (Beckton Dickinson AriaIII). The distinct fractions of the forward- and side-scattered cells were individually sorted. The sorted cells were the smeared onto slide glasses and processed for Wright-Giemsa stain.
Xenograft Experiment
Human cancer cell lines were purchased from American Type Culture Collection (Manassas, VA) and maintained in the American Type Culture Collection–recommended growth media at 37°C, 5% CO2. All of the cancer cells were then transfected with lentiviral particles to transduce pLemiR-nonsilencing plasmid vector containing the TurboRED fluorescent protein (tRFP) sequence (Open Biosystem, Huntsville, AL). A week before the xenograft experiment, the SCID zebrafish were transferred to 35°C aquaria system for adaptation at the desired temperature. For tumor cell xenograft, the tRFP-expressing tumor cells were grown until 70% of confluence is reached. The cells were then resuspended in PBS (2.5 × 106/ml), and 5 × 104 (20 μl) cells were introduced into the SCID zebrafish by intraperitoneal injection using a Hamilton syringe. The injected zebrafish were maintained at 35°C environment, monitoring transcutaneous fluorescence every 2 to 3 weeks. The injected zebrafish were sacrificed at 6 weeks and processed for histologic evaluation.
Results
Generation of Prkdc-Null (SCID) Zebrafish
Human PRKDC homolog of zebrafish is comprised of 86 exons with a cDNA length of 12,623 bp. Among these, we found that the exon 3 with 108 bp was ideal for TALEN targeted mutagenesis (Figure 1A). Thus, two TALEN constructs of left and right arms were prepared and applied to induce a targeted mutation by injecting the mRNAs into the yolk of AB zebrafish at one cell stage. By genotyping eight F1 progenies, two mutations (mutation no. 3 and 6) were identified at the spacer region of the TALENs. Sequence analysis showed that the mutation no. 3 and 6 contained AC dinucleotide deletion and TCTACA deletion followed by CATAT insertion, respectively, in the exon3 of the Prkdc gene, causing a frameshift mutation (Figure 1, B and C). We also checked other sites that show sequence similarity to the targeted region at sequence level. Potential off-target sites were searched using a software program (TAL Effector Nucleotide Targeter 2.0: Paired Target Finder) of the Bogdanove laboratory (https://boglab.plp.iastate.edu/node/add/talen). Among the possible off-targets, the six paired targets that showed a high rate of similarity were selected and analyzed for the mutation possibility by sequence analysis. The sequence analysis revealed that insertion or/and deletion did not occur at any of the off-target sites among the transgenic zebrafish (data not shown), excluding any undesired mutations from the genome of the zebrafish.
F2 progenies of the zebrafish were produced by backcrossing the heterozygote F1 founders to wild-type zebrafish. The adult F2 progenies were then screened for harboring heterozygote mutation and were in-crossed to produce F3 progenies. Homozygote Prkdc-null zebrafish were screened within the F3 progenies and expanded by in-crossing of the homozygotes to secure a stable line of the SCID zebrafish. We also confirmed the complete loss of the Prkdc expression by Western blot and IHC experiments (Figure 1, D and E). For Western blot analysis, the kidney and spleen dissected out from the homozygote zebrafish were used for the total protein isolation. IHC experiment was done with the paraffin-embedded tissue sections obtained from the zebrafish kidney. The two experiments showed that the homozygote zebrafish did not express the Prkdc gene, suggesting that a complete loss of the gene function occurred in the transgenic zebrafish. Taken together, our analyses revealed that the homozygote Prkdc-null SCID zebrafish was successfully generated.
Growth Retardation and Vulnerability to Infection
The SCID zebrafish were viable and showed slightly retarded growth in length compared with their age-matched wild-type zebrafish until 3 months of age (Figure 2, A and B). The growth then became comparable to that of wild-type zebrafish at adult stage. The SCID zebrafish were also fertile and could be maintained as homozygote for Prkdc mutation but showed a reduced reproducibility with short egg-laying period that occurred at 3 to 6 months of age. Both the mutation no. 3 and 6 lines contained virtually identical morphological phenotype. The mutation no. 6 zebrafish, however, was more prolific than the mutation no. 3 zebrafish, and thus, we carried out further detailed experiments by using the mutation no. 6 line.
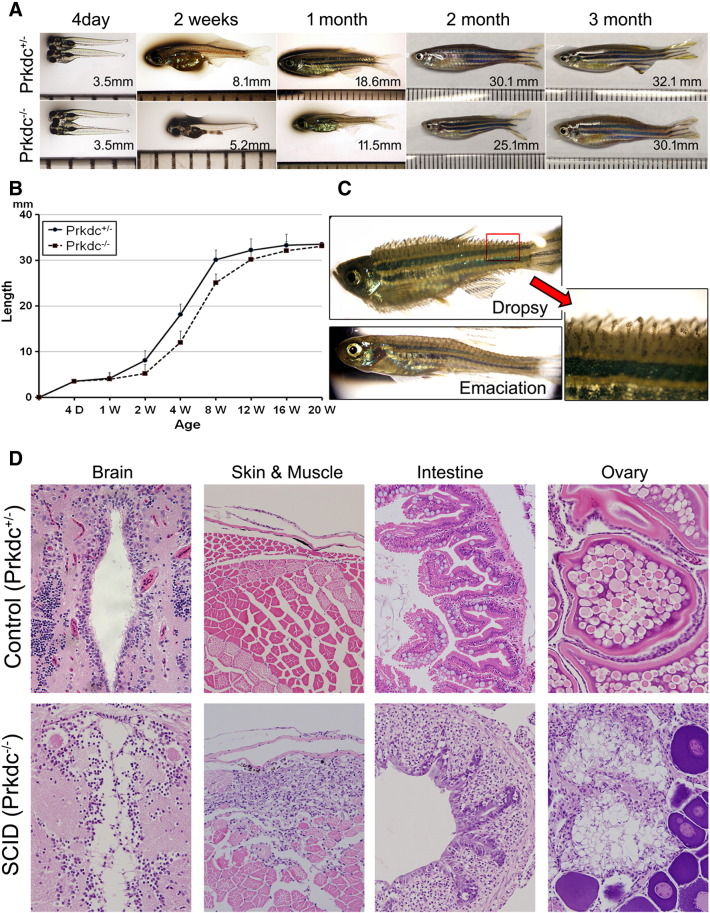
Figure 2.
Growth phenotypes and microscopic observations of the Prkdc-null SCID zebrafish. (A and B) Growth retardation. The SCID zebrafish showed a retarded growth in length compared with the age-matched wild-type controls until 3 months. (C) Infectious manifestation. The immune-deficient SCID zebrafish often succumbed to infection. The emaciated zebrafish died within 7 days, whereas the “dropsy” zebrafish died within a couple of days. Red arrow shows a magnified view of the dropsy zebrafish fin. (D) Microscopic findings of the infection and inflammation occurred in the internal organs. The zebrafish with “dropsy” phenotype revealed that the disseminated infection occurred in the internal organs including brain, intestine, and ovary, suggesting septicemia. The emaciated zebrafish showed a predominant muscle necrosis with inflammation.
As it has been documented in mouse and rat SCID models, the SCID mutation is assumed to cause a virtual loss of both T and B lymphocytes. Susceptibility to spontaneous infection is a well-known phenotype found in the SCID mutation. In fact, the SCID zebrafish were vulnerable to spontaneous infection. We often found that the SCID zebrafish showed the emaciation phenotype caused by muscle necrosis due to local infection (Figure 2C). Once the emaciation developed, the infected hosts usually died within 7 days. The “dropsy” phenotype, a well-known disease caused by septisemia in teleost fish, was also found. Histologic analysis revealed that the dropsy zebrafish contained inflammatory infiltration in the majority of organs including brain, intestine, and ovary (Figure 2D). Predominant muscle necrosis with the inflammation was found in the emaciated zebrafish. The SCID zebrafish with “dropsy” usually died within a couple of days, indicating that the “dropsy” is a more disastrous infection than the emaciation. The results suggest that the Prkdc-null SCID zebrafish are vulnerable to infection possibly because of the reduced immunity.
Analysis of Hematologic Organs
We used Rag1 as an effective marker for the thymus development (Figure 3A). ISH experiment with 4-day-old embryos showed that the SCID zebrafish contained dramatically reduced thymus development compared with the one of our heterozygote control. Histologic analysis of the thymus in the adult zebrafish revealed that the mutant also had a significantly decreased number of lymphoid cells. Because blood production occurs in kidney in adult zebrafish, we investigated the zebrafish kidney under a stereomicroscope (Figure 3B). The microscopic observation with the naked kidney clearly showed morphological differences between the mutant and wild-type zebrafish. Hematopoietic tissue of the mutant zebrafish kidney showed much reduced development with the decreased tissue density compared with that of the heterozygote (Figure 3B, red arrowhead). Further observations with H&E staining of the kidney tissue section revealed that the Prkdc-null SCID zebrafish contained a significantly decreased number of hematopoietic cells (Figure 3B and see the microscopic view). The spleen, a lymphoid organ, was also investigated (Figure 3C). We found that the spleen size was considerably smaller in the SCID zebrafish (1.2 ± 0.2 mm) than in the heterozygote control (2.0 ± 0.3 mm) (P < .05). Histologic observation of the spleen showed that the SCID zebrafish contained a decreased number of lymphoid cells.
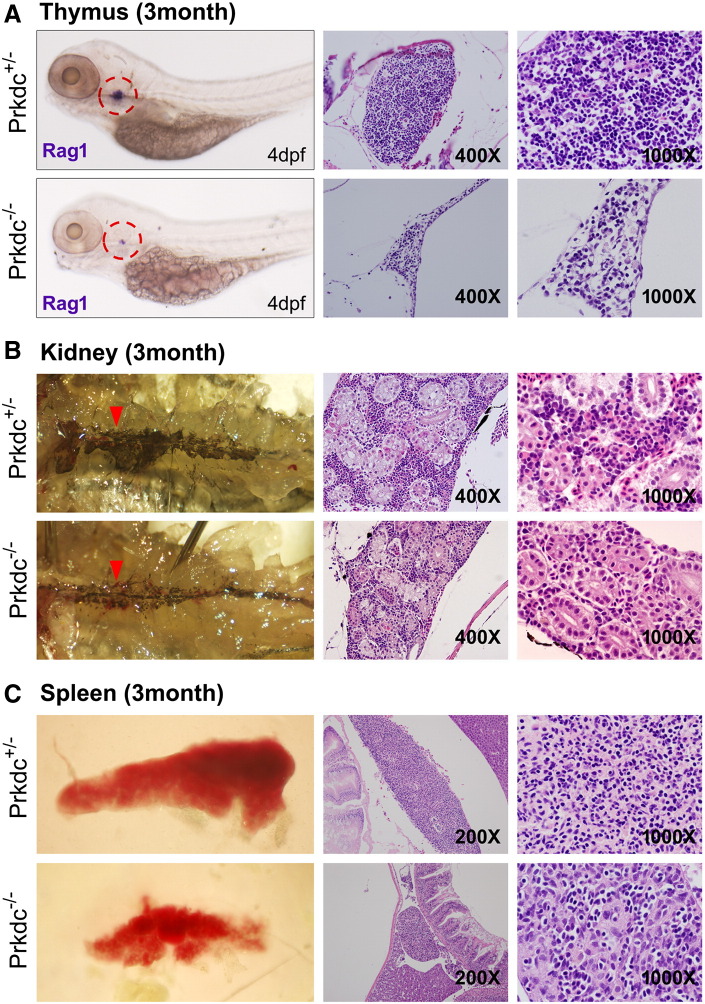
Figure 3.
Microscopic observation of the hematopoietic organs in the Prkdc-null SCID zebrafish. (A) ISH and H&E images of the thymus. The ISH experiment for Rag1 in the SCID zebrafish embryo at 4 dpf showed that the mutant zebrafish contained a very rudimentary thymus development (dotted red circle). H&E staining images at 3 months of age showed that the number of lymphocytes was severely decreased in the SCID zebrafish (prkdc−/−) compared with the heterozygote (prkdc+/−). (B) Microscopic observation of the zebrafish kidney. The retroperitoneal hematopoietic kidney tissue appears as dark linear structure. The tissue appeared as thinner with less density in the SCID zebrafish compared with that of the heterozygote (red arrowhead). H&E staining of the kidney also showed the consistency in which the kidney of the SCID zebrafish was thinner than that of the heterozygote and contained scanty hematopoietic cells between the renal tubules. (C) Gross and H&E images of spleen. The spleen of the SCID zebrafish was much smaller than that of the heterozygote. Compared with the spleen of the heterozygote, small number of lymphocytes was shown to be scattered between splenocytes in the SCID zebrafish.
Analysis of Hematologic Cells
We also evaluated the hematopoietic cells in the separately prepared cell smears from peripheral blood, kidney, and spleen (Figure 4A). Observation of the smears after staining with Wright-Giemsa suggested that the peripheral blood and the internal organs of the SCID zebrafish contained significantly decreased number of lymphoid cells and increased number of myelomonocytes compared with the ones of the heterozygote control (Figure 4A). Our manual cell counting indicated that the lymphocyte fraction in the peripheral blood was present at 63.6% and 27.5% in the control and the SCID zebrafish, respectively (Table 1). The decreased number of lymphocytes in the SCID zebrafish was replaced by monocyte/macrophage lineage cells. The kidney from SCID zebrafish contained much sparse marrow cells and more fat globules compared with control, which reflect the hypoplastic kidney of SCID zebrafish on gross images (Figure 3B). Differential cell counting again revealed decreased lymphocytes fraction in SCID zebrafish. Similar result was also obtained from the spleen (Table 1).
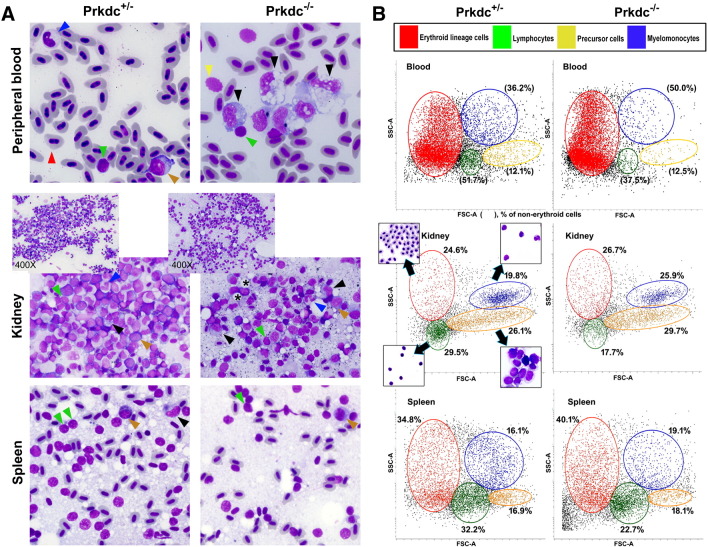
Figure 4.
Hematologic findings of the major blood cell lineages. (A) Wright-Giemsa staining with the smeared cells from the peripheral blood, kidney, and spleen. All images are 1000× view except insets (400×) in middle column. Arrowheads are colored as green for lymphocytes, red for erythroid lineage cells, blue for neutrophils, yellow for thrombocytes, black for monocytes/macrophages, and brown for blasts/precursor cells. Microscopic observation of these organs revealed much decreased number of the lymphocytes in the SCID zebrafish compared with the one in the heterozygote control (detailed counting results are shown in Table 1). The lymphocytes appeared as small cells with a higher nuclear to cytoplasmic ratio, and the monocytes/macrophages appeared as larger cells with a lower nuclear to cytoplasmic ratio and cytoplasmic vacuoles. Direct smear of dissected kidney and spleen also revealed lower lymphocyte fractions in SCID zebrafish. Note the compact cells in the smear of kidney marrow from control and frequent fat globules (*) in the smear from SCID zebrafish. (B) Flow cytometry. Forward scatter (FSC) and side scatter (SSC) are proportional to cell size and cellular granularity, respectively. Peripheral blood and single cell suspensions from the dissected kidney and spleen were processed for flow cytometry. The flow cytometry demonstrated four main cell populations (noted in the box). Note that lymphocyte fractions are markedly decreased in the SCID zebrafish compared with those of the heterozygote control.
Table 1.
Differential Cell Counts by Manual Analyses
| Peripheral Blood (%) |
Kidney (%) |
Spleen (%) |
|||||
|---|---|---|---|---|---|---|---|
| Control | SCID | Control | SCID | Control | SCID | ||
| Number | 8 | 8 | 6 | 6 | 6 | 6 | |
| Myelomonocyte | Neutrophil | 20.6 ± 3.8 | 33.1 ± 4.8 | 8.5 ± 3.3 (11.7 ± 4.9) | 10.6 ± 2.7 (15.3 ± 3.6) | 5.0 ± 1.2 (10.2 ± 2.4) | 5.7 ± 1.5 (13.2 ± 3.2) |
| Eosinophil | 1.1 ± 0.7 | 0.5 ± 0.3 | 0.6 ± 0.7 (0.8 ± 0.9) | 1.1 ± 1.1 (1.6 ± 1.5) | 1.8 ± 1.3 (3.7 ± 2.6) | 2.1 ± 1.1 (4.9 ± 2.4) | |
| Monocyte | 14.2 ± 3.1 | 38.3 ± 7.9* | 5.9 ± 3.9 (8.1 ± 4.3) | 10.6 ± 4.3 (15.3 ± 6.9) | 4.2 ± 1.6 (8.6 ± 3.3) | 8.3 ± 2.6 (19.2 ± 5.5)* | |
| Lymphocyte | 63.6 ± 14.3 | 27.5 ± 9.3* | 33.5 ± 8.3 (46.0 ± 11.1) | 19.3 ± 4.2* (27.8 ± 6.1) | 36.7 ± 6.6 (75.1 ± 13.6) | 25.5 ± 4.1* (59.0 ± 9.1) | |
| Precursor (blast) cell | 0.5 ± 0.3 | 0.6 ± 0.5 | 24.4 ± 4.2 (33.5 ± 5.3) | 27.9 ± 4.3 (7.7 ± 3.2) | 1.2 ± 0.6 (2.5 ± 1.2) | 1.6 ± 0.3 (3.7 ± 0.7) | |
| Erythroid lineage cell | NC | NC | 27.1 ± 7.3 | 30.5 ± 7.7 | 51.1 ± 10.2 | 56.8 ± 10.3 | |
Numbers in parenthesis, % of nonerythroid cells. NC, not counted. Statistical significance was calculated by Kruskal-Wallis rank sum test.
P < .05 compared with control.
Further analysis of the cell suspensions was done by flow cytometry experiment (Figure 4B and Table 2). As revealed in the cell counting, the SCID zebrafish had a considerably decreased number of the lymphocytes in peripheral blood, kidney, and spleen compared with the heterozygote control, whereas other cell scatter populations including erythrocyte and myelomonocyte fractions were similar to each other. The results suggested that the Prkdc-null SCID zebrafish underwent an impaired lymphoid development.
Table 2.
Differential Cell Counts by Flow Cytometry
| Blood (%) |
Kidney (%) |
Spleen (%) |
||||
|---|---|---|---|---|---|---|
| Control | SCID | Control | SCID | Control | SCID | |
| Number | 12 | 12 | 12 | 12 | 12 | 12 |
| Myelomonocyte | 2.1 ± 0.2 (36.2 ± 3.7) | 2.8 ± 0.3 (50.0 ± 6.7)* | 19.8 ± 3.7 (26.3 ± 5.2) | 25.9 ± 4.5 (35.3 ± 7.1) | 16.1 ± 2.2 (24.7 ± 3.7) | 19.1 ± 3.4 (31.8 ± 5.8) |
| Lymphocyte | 3.0 ± 0.4 (51.7 ± 6.7) | 2.1 ± 0.2 (37.5 ± 3.9)* | 29.5 ± 5.7 (39.1 ± 6.5) | 17.7 ± 3.2* (24.1 ± 4.5) | 32.2 ± 3.3 (49.4 ± 5.7) | 22.7 ± 1.7* (37.8 ± 3.1) |
| Precursor (blast) cell | 0.7 ± 0.2 (12.1 ± 3.7) | 0.7. ± 0.4 (12.5 ± 7.7) | 26.1 ± 4.4 (34.6 ± 6.4) | 29.7 ± 6.3 (40.5 ± 7.9) | 16.9 ± 7.3 (25.9 ± 9.9) | 18.1 ± 7.7 (30.2 ± 10.7) |
| Erythroid lineage cell | 94.2 ± 3.0 | 94.4 ± 3.1 | 24.6 ± 2.5 | 26.7 ± 2.6 | 34.8 ± 4.0 | 40.1 ± 4.4 |
Numbers in parenthesis, % of nonerythroid cells. Statistical significance was calculated by Kruskal-Wallis rank sum test.
P < .05 compared with control.
Lymphocytes in the SCID Zebrafish Are Natural Killer (NK) Cells
We performed IF analyses with the cell suspensions from peripheral blood and spleen (Figure 5). Although the CD2AP is used as a marker for the activated T lymphocytes, NK cells are also known to be reactive to the CD2AP, whereas NKP46 is widely used as the specific marker for NK cells. This explains that the CD2AP-reactive lymphocytes can be either activated T lymphocytes or NK cells, whereas the NKP46-positive cells are solely to be the NK cells irrespective of the CD2AP expression. As shown in the Figure 5A, the CD2AP-positive cells were found in the peripheral blood from both the heterozygote control and the SCID zebrafish. When the cell suspensions from the peripheral blood and spleen were subjected to both CD2AP and NKP46 markers, however, no lymphocytes from the SCID zebrafish were solely reactive to the CD2AP marker, suggesting that all of the SCID zebrafish lymphocytes are NK cells (Figure 5B). On the other hand, the heterozygote control zebrafish showed positive signals for CD2AP and/or NKP46 markers. The lymphocyte counting based on the CD2AP or NKP46 positivity was also summarized in Figure 5D. In the heterozygote control, the positive signals were detected by either CD2AP or NKP46 (or by the both markers). In the SCID zebrafish, however, almost all CD2AP-positive cells were also detected by the NKP46, suggesting that they are NK cells. In conclusion, the SCID zebrafish contained no activated T lymphocytes, whereas the control zebrafish contained both activated T lymphocytes and NK cells.
Figure 5.
IF and RT-PCR analyses for activated lymphocytes. (A) IF detected by CD2AP marker. The CD2AP-reactive lymphocytes are present in the peripheral blood from both SCID and control zebrafish, suggesting that the cells are either activated T lymphocytes or NK cells. DAPI staining was used to detect the blood cells. (B) Double IF detected by CD2AP (Cy2) and NKP46 (Cy3) markers. The NKP46 is a specific marker for NK cells, whereas the CD2AP marker is reactive to both T lymphocytes and NK cells. (C) RT-PCR analysis. Glyceraldehyde 3-phosphate dehydrogenase gene detection was used as control. Nonspecific cytotoxic cell receptor protein-1 and NK specific protein (NK cell triggering receptor) gene detections indicate NK cell. TCR V(D)JC (Vb12-Cb1) and immunoglobulin (IgVH1-Cm) gene transcript detections reveal V(D)J recombination and heavy-chain rearrangement. (D) Counted numbers of cells depending on CD2AP or NKP46 positivity. Green arrowhead, positive for CD2AP; red arrowhead, positive for NKP46; white arrowhead, positive for both CD2AP and NKP46; blue arrowhead, lymphocytes negative for both CD2AP and NKP46; yellow arrowhead; red blood cells.
We also performed nested RT-PCR experiments using the mRNAs extracted from the dissected kidney and spleen to determine if the hematopoietic tissues included functional T or B lymphocytes (Figure 5C). The RT-PCR results demonstrated that the SCID zebrafish hematopoietic tissues expressed the mRNAs encoding nonspecific cytotoxic cell receptor protein-1 and NK cell triggering receptor but lacked the TCR V(D)JC (Vb12-Cb1) and the immunoglobulin (IgVH1-Cm) gene transcripts, suggesting that the SCID zebrafish undergoes no V(D)J recombination and heavy-chain rearrangement. The results indicate that the SCID zebrafish contains no functional T and B cells.
Xenograft by Intraperitoneal Injection of Cancer Cell Lines
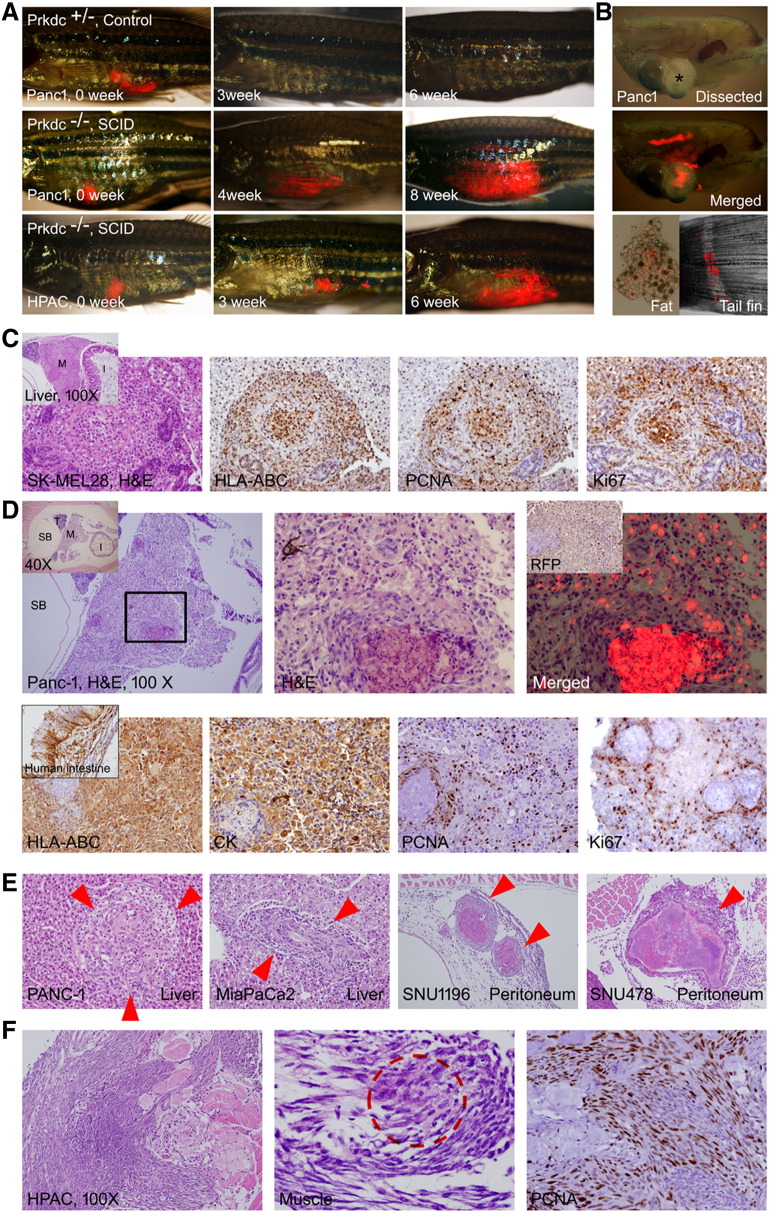
Intraperitoneal injections of the human brain (glioblastoma, U-87MG), melanoma (CloneM3, SK-MEL28, B16F10), leukemia (K562), pancreatic (Panc-1, HPAC, Miapaca-2, BxPC-3), and bile duct (SNU245, SNU478, SNU1196) cancer cell lines were done to examine the xenograft potential with the SCID zebrafish (Figure 6). The experiment showed that adult zebrafish well tolerated the intraperitoneal injection up to 20-μl volume of tumor cell suspension. We found however that approximately 10% of traumatic death occurred after the injection. Another adverse event was occurrence of the “dropsy” phenotype (10% among the injected zebrafish). This adverse event possibly resulted from the procedure-related infection.
Figure 6.
Xenograft of tumor cells. Various tumor cells (5 × 104) expressing TurboRFP were injected into intraperitoneal space by using a Hamilton syringe. If not indicated, all H&E and IHC images are 400×. (A) Gross images. The engraftment allows repeated observation of the growing tumors by visualization of transcutaneous RFP. Tumor engraftment was never formed in Prkdc+/−. (B) Dissected images. Bright field (top, asterisk shows a mass formed by the growing tumor) and merged RFP (middle) images showing an engrafted tumor with RFP expression. Dissected fat tissue (bottom left) showed scattered RFP-expressing tumor cells. Metastatic tumor cells (bottom right) at tail fin are also noted. (C) A mass formed in the liver in SK-MEL28 injected zebrafish. Inset is a low-power view showing mass (M) in the liver. HLA-ABC positivity of the tumor cells indicates that the mass originated from injected human melanoma cells. Many of the tumor cells are positive for PCNA and Ki-67, suggesting active proliferation. (D) A discrete mass formed in the Panc-1–injected zebrafish. Inset in the left is a low-power view indicating testis (T), swim bladder (SB), intestine (I), and mass (M). Enlarged view of the boxed area is shown in the middle. Merged TurboRFP and H&E image is shown on the right. Inset in the right is IHC for RFP. Lower column images are findings of IHC stain. Xenografted tumor cells are positive for HLA-ABC, for pan-cytokeratin (Pan-CK), and also for PCNA and Ki-67 frequently. (E) The tumors formed in liver and peritoneal cavity. (F) The xenografted HPAC cells showing the invasive growth, which results in destruction of muscle (M) and skin. The fibroblast with occasional tumor cells (dotted red circle shown in the middle). The proliferating fibroblasts are highly positive to PCNA.
Lentiviral-induced TurboRFP expression of injected cells allowed a real-time monitoring of the cell growth in the live zebrafish. When the cancer cell lines were intraperitoneally administrated into the Prkdc-null SCID zebrafish, the growing tumors were well visualized by RFP expression (Figure 6, A and B). Besides, repeated observation every 2 to 3 weeks allowed monitoring the growth of engrafted tumor. SCID zebrafish, however, were more vulnerable to tricaine anesthesia that they just tolerated up to 1 minute in tricaine solution, which was 2 minutes in control. Among the various cancer cells, melanoma (SK-MEL28, CloneM3, and B16F10) and brain tumor cells (U-87MG) revealed higher rate of microscopic and macroscopic tumor engraftment compared with the gastrointestinal cancer cells. When each cell line was injected into 12 zebrafish, overall engrafted tumors were found in approximately 50% to 70% of the xenotransplanted SCID zebrafish (Table 3). The engraftment was never formed in heterozygotes for Prkdc mutation. The most common site of engraftment was peritoneum followed by liver. Macroscopic tumor engraftment refers to visible mass on dissection and was observed in minority of the injected zebrafish, of which the size measured up to 2 mm.
Table 3.
Engraftment of Xenotransplated Cancer Cells
| Cancer Cells (Each Cell Line into 12 Zebrafish) |
Microscopic Engraftment | Macroscopic Engraftment | Engrafted Organs |
|---|---|---|---|
| U-87MG | 75% | 25% | Peritoneum, liver |
| CloneM3 | 66.7% | 25% | Peritoneum, liver |
| SK-MEL28 | 75% | 33.3% | Peritoneum, liver |
| B16F10 | 66.7% | 16.7% | Peritoneum, liver |
| K562 | 66.7% | 25% | Peritoneum, intestine |
| Panc-1 | 66.7% | 16.7% | Peritoneum, liver, pancreas, tail fin |
| HPAC | 58.3% | 16.7% | Peritoneum, muscle, liver |
| Miapaca-2 | 50% | 16.7% | Peritoneum, liver, ovary |
| BxPC-3 | 33.3% | 0% | Peritoneum, liver |
| SNU245 | 41.7% | 0% | Peritoneum, liver |
| SNU478 | 58.3% | 16.7% | Peritoneum, liver |
| SNU1196 | 58.3% | 16.7% | Peritoneum, liver |
Tissue section with H&E staining of the zebrafish clearly showed the growing tumor which was specifically localized and overlapped with the RFP expression (Figure 6C). The results indicate that the xenografted tumor has been efficiently formed. IHC experiment further verified that the xenografted tumors originated from injected cells as they expressed RFP, HLA-ABC, and pan-cytokeratin (Figure 6, C and D). The engrafted tumor cells revealed enhanced proliferation as evidenced by increased PCNA and Ki67 expression.
Interestingly, the tumor mass was often found to be composed of the massive fibroblast proliferation with the occasional tumor cells (HPAC cells), reflecting the desmoplastic reaction occurring in pancreatic cancer (Figure 6F). Taken together, the results indicate that our xenograft with the intraperitoneal injection of the cancer cell lines into the Prkdc-null SCID zebrafish has effectively developed tumor generation.
Discussion
SCID mice have widely been used as hosts for human tumor cell xenograft study [24]. The SCID mice include the gene mutation which results in the failure of normal development of both T and B lymphocytes. The PRKDC gene which is responsible for DNA double-strand break repair system has also been used to develop the SCID mouse model [5]. As zebrafish has developed into a powerful tool for immunological study, the zebrafish has also been used as a xenograft tool for human cancer studies [25], [26], [27]. Xenotransplanting human tumor cell into the zebrafish embryos before the maturation of adaptive immune function has successively shown in vivo imaging of early tumor growth and invasion, and tumor angiogenesis [27], [28]. Death of the engrafted embryos, however, does not permit a long-term observation of the tumor behavior. Irradiation approach has recently been done to develop an immune-depleted adult zebrafish [29]. Xenotransplantation of cancer stem cells (CSCs) into the adult zebrafish showed a malignant proliferation of the engrafted tumor cell, suggesting that the zebrafish could be a good model for CSC study in vivo. However, recovering immune system of the irradiated zebrafish after few weeks that may cause tumor rejection remains as a concern.
Gene mutation strategies for generating immune-deficient zebrafish models have been used in zebrafish. The recombination activation gene 1 (rag1) mutation of zebrafish showed lack of T and B lymphocytes, suggesting its potential for the use of the mutant zebrafish as new model system [16]. Later, the first immune-deficient zebrafish model was created by the Rag2E450fs gene knockout strategy [17]. The zebrafish permitted robust and long-term engraftment of multiple tissues and cancer. In our present study, we used TALEN-based knockout of the Prkdc gene to generate the new SCID zebrafish model that becomes the counterpart of mouse and rat SCID models. The TALENs specifically recognized the Prkdc gene sequence and induced mutation of the gene (Figure 1). As expected, the current SCID zebrafish clearly showed that the hematologic organs’ development was severely retarded, causing an impaired lymphoid development (Figure 3). The SCID zebrafish were virtually devoid of functioning T and B cells with relatively increased fraction of the monocytes. Other than T or B cells, lineage analysis of the lymphocytes revealed that almost all of the lymphocytes in the SCID zebrafish were found to be NK cells. These findings recapitulate the hematologic phenotypes observed in the SCID mouse and rat models, which indicate that the Prkdc gene is genetically preserved between teleost fish and mammals.
The NK cells are versatile, especially in the immune reaction against tumor cells without restriction by MHC I or II molecules. The functioning NK cells in the SCID zebrafish may impede the growth of transplanted tumor cells. Thus, the next step for refining the SCID zebrafish model may be the sequential knockout of interleukin-2 receptor γ (duplicated as a and b in zebrafish) to generate the SCID gamma model which improves the xenograft rate. Additionally, pigmentation in adult zebrafish can give imaging limitation for real-time tracing of the tumor cell invasion, intravasation, extravasation, and angiogenesis in the SCID zebrafish due to the opacity. This may be overcome by generating a transparent background obtained from in-crossing of the SCID zebrafish with Casper (mitfa/roy mutant) zebrafish [30], [31].
Several studies have been reported to demonstrate various laboratory benefits of using the zebrafish as a xenograft model. One of the reasons is because the zebrafish model provides the unique tool for visualization of tumor cell behavior. Previously, the zebrafish xenograft model was introduced to track CSC in tumor invasion and metastasis [27], [28]. The transparent zebrafish enabled tracking of the xenografted CSC behavior and monitoring the process of tumor cell invasion in a real-time fashion. Anti-CSC drug evaluation using the zebrafish xenograft model has also been reported, demonstrating the suitability of using this animal model for high-throughput screening of anti-tumor agent [32]. There are also limitations of using the zebrafish especially when the study undergoes for a specific human organ cancer. For instance, the zebrafish does not have organs which are unique to mammals. In a recent study with a zebrafish xenograft model, however, human breast cancer cell lines were injected and studied for invasive and metastatic behavior in vivo [28]. This study showed an application potential of using the zebrafish model for the mechanistic understanding of the human cancer and also for the development of pharmacological inhibition for the treatment of the metastatic cancer. These studies suggest that the zebrafish and their transparent embryos present a beneficial model system over the mouse or rat model not only for studying tumor invasion and metastasis but also for screening anticancer drug.
Another laboratory benefit of using the zebrafish over the mouse or rat model is availability of the large number of offspring which provides cost-effectiveness for cancer studies. Maintaining a large number of the xenografted zebrafish in a small growth chamber allows easy counting to develop statistical data. For this reason, our study demonstrates maintenance of the immune-deficient zebrafish lines in the small growth chamber. Lack of functioning T and B cells, however, rendered the SCID zebrafish vulnerable to infection. Spontaneous infection, especially after tail-fin clipping for genotyping, was a cumbersome problem. Difficulty of maintaining pathogen-free condition (i.e., Specific pathogen-free (SPF) environment in mouse facility) in the aquaria system can also be an issue for the expansion of immune-deficient zebrafish models. We overcame this difficulty by maintaining the SCID zebrafish in the separate system with meticulous care of breeding water and weekly addition of antibiotics, decreasing the infection rate to below 10%. This will minimize the burden of expense which occurs during maintenance of the immune-deficient mouse model in Specific pathogen-free (SPF) environment.
Xenotransplantation experiment was done by utilizing various cancer cells including melanoma and brain, pancreas, and bile duct cancer cells (Figure 6). Many of the previous studies have used leukemia or melanoma cells which are rapid growing and highly tumorigenic because of a higher rate of genetic aberration. Current model also revealed higher engraftment rates by melanoma and brain cancer cells than by gastrointestinal cancer cells. Pancreatic and biliary tumors are notorious for evading early diagnosis, of which the treatment is largely dependent on systemic chemotherapy. Although xenograft rates were not very high in the current model, successful engraftment of these tumor cells can provide a valuable platform for cancer research and personalized treatment.
To summarize, generation of the homozygote Prkdc-null mutation in the zebrafish allowed the immune-deficient background lacking the functional T and B cells (Figure 5), and this provided an efficient environment for intraperitoneal injection of the human cancer cell lines to grow into tumor (Figure 6). Our study presents a complete knockout of Prkdc gene of zebrafish and demonstrates the suitability of using this gene mutation to develop a new xenograft animal model for cancer research. The successful xenotransplantation of the gastrointestinal tumors shown in this study may expand the application of the immunocompromised zebrafish strain to drug sensitivity test. The SCID zebrafish model will provide a new platform of tumor xenograft study for tumor biology and immunology as well as for anticancer drug development.
The following is the supplementary data related to this article.
Primers used for RT-PCR.
Acknowledgement
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C1324), and by an institutional grant of Yonsei University College of Medicine (6-2013-0022).
Footnotes
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C1324), and by an institutional grant of Yonsei University College of Medicine (6-2013-0022).
References
- 1.Jeggo P, Lavin MF. Cellular radiosensitivity: how much better do we understand it? Int J Radiat Biol. 2009;85(12):1061–1081. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- 2.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417(3):639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soulas-Sprauel P, Rivera-Munoz P, Malivert L, Le Guyader G, Abramowski V, Revy P, de Villartay JP. V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene. 2007;26(56):7780–7791. doi: 10.1038/sj.onc.1210875. [DOI] [PubMed] [Google Scholar]
- 4.Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst) 2006;5:1030–1041. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 5.Mashimo T, Takizawa A, Kobayashi J, Kunihiro Y, Yoshimi K, Ishida S, Tanabe K, Yanagi A, Tachibana A, Hirose J. Generation and characterization of severe combined immunodeficiency rats. Cell Rep. 2012;2:685–694. doi: 10.1016/j.celrep.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Bastide C, Bagnis C, Mannoni P, Hassoun J, Bladou F. A nod scid mouse model to study human prostate cancer. Prostate Cancer Prostatic Dis. 2002;5:311–315. doi: 10.1038/sj.pcan.4500606. [DOI] [PubMed] [Google Scholar]
- 7.Lee CH, Xue H, Sutcliffe M, Gout PW, Huntsman DG, Miller DM, Gilks CB, Wang YZ. Establishment of subrenal capsule xenografts of primary human ovarian tumors in SCID mice: potential models. Gynecol Oncol. 2005;96:48–55. doi: 10.1016/j.ygyno.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 8.Lozupone F, Pende D, Burgio VL, Castelli C, Spada M, Venditti M, Luciani F, Lugini L, Federici C, Ramoni C. Effect of human natural killer and γδ T cells on the growth of human autologous melanoma xenografts in SCID mice. Cancer Res. 2004;64:378–385. doi: 10.1158/0008-5472.can-03-1501. [DOI] [PubMed] [Google Scholar]
- 9.Forest V, Peoc'h M, Campos L, Guvotat D, Vergnon JM. Effects of cryotherapy or chemotherapy on apoptosis in a non–small-cell lung cancer xenografted into SCID mice. Cryobiology. 2005;50:29–37. doi: 10.1016/j.cryobiol.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Rebouissou C, Wijdenes J, Autissier P, Tarte K, Costes V, Liautard J, Rossi JF, Brochier J, Klein B. A gp130 interleukin-6 transducer dependent SCID model of human multiple myeloma. Blood. 1998;91:4727–4737. [PubMed] [Google Scholar]
- 11.Ochiai M, Ubagai T, Kawamori T, Imai H, Sugimura T, Nakagama H. High susceptibility of SCID mice to colon carcinogenesis induced by azoxymethane indicates a possible caretaker role for DNA-dependent protein kinase. Carcinogenesis. 2001;22:1551–1555. doi: 10.1093/carcin/22.9.1551. [DOI] [PubMed] [Google Scholar]
- 12.Wacheck V, Heere-Ress E, Halaschek-Wiener J, Lucas T, Meyer H, Eichler HG, Jansen B. Bcl-2 antisense oligonucleotides chemosensitize human gastric cancer in a SCID mouse xenotransplantation model. J Mol Med. 2001;79:587–593. doi: 10.1007/s001090100251. [DOI] [PubMed] [Google Scholar]
- 13.Workman P, Aboagye EO, Balkwill F. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand Immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- 15.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multileneage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 16.Petrie-Hanson L, Hohn C, Hanson L. Characterization of rag1 mutant zebrafish leukocytes. BMC Immunol. 2009;10:1–8. doi: 10.1186/1471-2172-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Q, Abdelfattah NS, Blackburn JS, Moore JC, Martinez SA, Moore FE, Lobbardi R, Tenente IM, Ignatius MS, Berman JN. Optimized cell ransplantation using adult rag2 mutant zebrafish. Nat Methods. 2014;11(8):821–824. doi: 10.1038/nmeth.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA doublestrand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung IH, Jung DE, Park YN, Song SY, Park SW. Aberrant Hedgehog ligand induce progressive pancreatic fibrosis by paracrine activation of myofibrolasts and ductular cells in transgenic zebrafish. PLoS One. 2011;6:1–15. doi: 10.1371/journal.pone.0027941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park SW, Davison JM, Rhee J, Hruban RH, Maitra A, Leach SD. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134:2080–2090. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Airaksinen S, Rabergh CMI, Sistonen L, Nikinmaa M. Effects of heat shock and hypoxia on protein synthesis in rainbow trout (Oncorhynchus mykiss) cells. J Exp Biol. 1998;201:2543–2551. doi: 10.1242/jeb.201.17.2543. [DOI] [PubMed] [Google Scholar]
- 23.Jung IH, Choi JH, Chung YY, Lim GL, Park YN, Park SW. Predominant activation of JAK/STAT3 pathway by interleukin-6 is implicated in hepatocarcinogenesis. Neoplasia. 2015;17(7):586–597. doi: 10.1016/j.neo.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belizario JE. Immunodeficient mouse model: an overview. Open Immuno J. 2009;2:79–85. [Google Scholar]
- 25.Stoletov K, Klemke R. Catch of the day: zebrafish as a human cancer model. Oncogene. 2008:1–12. doi: 10.1038/onc.2008.95. [DOI] [PubMed] [Google Scholar]
- 26.Konantz M, Balci TB, Hartwig UF, Dellaire G, André MC, Berman JN, Lengerke C. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 2012;1266:124–137. doi: 10.1111/j.1749-6632.2012.06575.x. [DOI] [PubMed] [Google Scholar]
- 27.Yang XJ, Cui W, Gu A, Xu C, Yu SC, Li TT, Cui YH, Zhang X, Bian XW. A novel zebrafish xenotransplantation model for study of glioma stem cell invasion. PLoS One. 2013;8(4):e61801. doi: 10.1371/journal.pone.0061801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drabsch Y, He S, Zhang L, Snaar-Jagalska BE, ten Dijke P. Transforming growth factor-β signalling controls human breast cancer metastasis in a zebrafish xenograft model. Breast Cancer Res. 2013;15(6):R106. doi: 10.1186/bcr3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Shimada Y, Kuroyanagi J, Nishimura Y, Umemoto N, Nomoto T, Shintou T, Miyazaki T, Tanaka T. Zebrafish xenotransplantation model for cancer stem-like cell study and high-throughput screening of inhibitors. Tumor Biol. 2014;35:11861–11869. doi: 10.1007/s13277-014-2417-8. [DOI] [PubMed] [Google Scholar]
- 30.White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2(2):183–189. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heilmann S, Ratnakumar K, Langdon EM, Kansler ER, Kim IS, Campbell NR, Perry EB, McMahon AJ, Kaufman CK, van Rooijen E. A quantitative system for studying metastasis using transparent zebrafish. Cancer Res. 2015;75(20):4272–4282. doi: 10.1158/0008-5472.CAN-14-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang XJ, Cui W, Gu A, Xu C, Yu SC, Li TT, Cui YH, Zhang X, Bian XW. A synthetic dl-nordihydroguaiaretic acid (Nordy), inhibits angiogenesis, invasion and proliferation of glioma stem cells within a zebrafish xenotransplantation model. PLoS One. 2014;9(1):e85759. doi: 10.1371/journal.pone.0085759. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for RT-PCR.