Abstract Abstract
Background
We present data from a DNA taxonomy register of the abyssal Cnidaria collected as part of the Abyssal Baseline (ABYSSLINE) environmental survey cruise ‘AB01’ to the UK Seabed Resources Ltd (UKSRL) polymetallic-nodule exploration area ‘UK-1’ in the eastern Clarion-Clipperton Zone (CCZ), central Pacific Ocean abyssal plain. This is the second paper in a series to provide regional taxonomic data for a region that is undergoing intense deep-sea mineral exploration for high-grade polymetallic nodules. Data were collected from the UK-1 exploration area following the methods described in Glover et al. (2015b).
New information
Morphological and genetic data are presented for 10 species and 18 records identified by a combination of morphological and genetic data, including molecular phylogenetic analyses. These included 2 primnoid octocorals, 2 isidid octocorals, 1 anemone, 4 hydroids (including 2 pelagic siphonophores accidentally caught) and a scyphozoan jellyfish (in the benthic stage of the life cycle). Two taxa matched previously published genetic sequences (pelagic siphonophores), two taxa matched published morphological descriptions (abyssal primnoids described from the same locality in 2015) and the remaining 6 taxa are potentially new species, for which we make the raw data, imagery and vouchers available for future taxonomic study. We have used a precautionary approach in taxon assignments to avoid over-estimating species ranges. The Clarion-Clipperton Zone is a region undergoing intense exploration for potential deep-sea mineral extraction. We present these data to facilitate future taxonomic and environmental impact study by making both data and voucher materials available through curated and accessible biological collections. For some of the specimens we also provide image data collected at the seabed by ROV, wich may facilitate more accurate taxon designation in coming ROV or AUV surveys.
Keywords: DNA barcode, deep sea, coral, sea anemony, sea jelly, baseline, survey, mining, CCZ, megafauna
Introduction
We present data from a DNA taxonomy register of the abyssal Cnidaria collected as part of the Abyssal Baseline (ABYSSLINE) environmental survey cruise ‘AB01’ to the UK Seabed Resources Ltd (UKSRL) polymetallic-nodule exploration area ‘UK-1’ in the eastern Clarion-Clipperton Zone (CCZ), central Pacific Ocean (Smith et al. 2013).
This is the second paper in a series to provide taxonomic data for a region that is undergoing intense deep-sea mineral exploration for high-grade polymetallic nodules regulated by Sponsoring States (here the United Kingdom Government) and the International Seabed Authority (ISA 2014, Glover and Smith 2003, Glover et al. 2015b, Wedding et al. 2015). As with the previous Echinodermata paper (Glover et al. 2016) this Cnidaria contribution is not yet a comprehensive faunal guide to the region, but a data paper that may be updated with new additions following future collections and analyses. New versions will contain all the data contained in the previous version, plus additional descriptions and records from future research cruises.
The abyssal zone of the world’s oceans has been defined as the seafloor between depths of 3000m and 6000m, a bathymetric zone that encompasses 54% of the geographic surface of the planet (Smith et al. 2008). Cnidarians form a morphologically and functionally diverse group in this region. Current online data sources list 616 cnidarian species recorded at abyssal depths from between 3000m and 6000m (OBIS 2016) out of a total of 1,922 cnidarian species recorded from depths greater than 500m (Glover et al. 2015a).
The Clarion-Clipperton Zone (hereafter, CCZ) is so called as it lies mainly between the Clarion and Clipperton Fracture Zones, topographical highs that extend longitudinally across almost the entire eastern Pacific. There is no strict definition of the region, but it has come to be regarded as the area between these fracture zones that lies within international waters and encompasses the main areas of commercial interest for polymetallic-nodule mining. Areas licensed for mining by the International Seabed Authority (ISA), as well as mining reserve areas and areas protected from mining by the ISA (ISA 2014, Wedding et al. 2013) extend from 115°W (the easternmost extent of the UK-1 exploration area) to approximately 158°W, and from 22°N to 2.5°S (Fig. 1). This is an area of 6 million sq km, approximately 1.7% of the ocean’s surface.
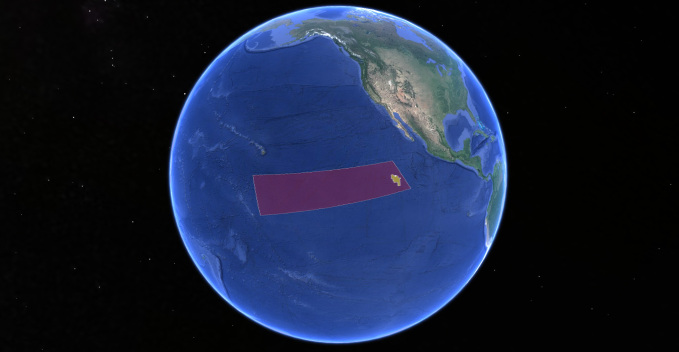
Figure 1.
The Clarion-Clipperton Zone, central Pacific Ocean (purple box) is a 6 milllion sq km region at the time of writing containing only 8 online databased records of cnidarian species (OBIS 2016). The UK Seabed Resources Ltd ‘UK-1’ polymetallic nodule exploration area is highlighted (yellow box). Image via Google Earth, IBCAO, Landsat, Data from SIO, NOAA, U.S Navy, NGA, GEBCO.
Within the entire 6 million sq km CCZ, as defined above, current online data sources prior to this publication list three known benthic species of cnidarians from 8 records (OBIS 2016). This is obviously the result of lack of sampling and/or taxonomy given that an abundant and diverse cnidarian fauna is suspected in the region from photographic and video survey (e.g. Amon et al. 2016, Foell and Pawson 1986). The goal of the DNA taxonomy part of the ABYSSLINE program is to begin filling these gaps in our knowledge and to make data publically available, which will eventually allow for a complete taxonomic synthesis of the CCZ supported by openly-available molecular and morphological data. Here we provide version 1.0 of the Cnidaria taxonomic synthesis from the ABYSSLINE program, consisting of taxon records, high-resolution imagery, genetic data from multiple markers and phylogenetic analysis from the first research cruise (AB01) aboard the RV Melville in October 2013. In this paper we report seven species of benthic cnidarians from the CCZ with the mention of an additional three pelagic ones, and a total of 18 records. Benthic surveys such as ABYSSLINE often recover miscellaneous pelagic taxa (e.g recovered on wires or equipment) and one of the goals of the DNA taxonomy part of this project was to ensure that these useful data records are made available, particularly given the lack of data on pelagic species in the open ocean. This open data publication is the second of equivalent similar data publications including also the Annelida, Mollusca, Bryozoa, Echinodermata, Porifera and other taxa forming a suite of taxonomic syntheses of biodiversity in the region, supported by a contract between the company UK Seabed Resources Ltd and the Natural History Museum, London, Uni Research, Bergen, and the University of Hawaii at Manoa (Glover et al. 2015b, Glover et al. 2016).
Materials and methods
It is widely accepted that knowledge of baseline biodiversity and biogeography in the CCZ is severely hampered by a lack of modern DNA-supported taxonomic studies (International Seabed Authority 2014). With this in mind, four fundamental principles underpin our methodological pipeline: (1) A sampling design pipeline with consideration to the spatial scale of the required data, the differing biases in sampling gear and the requirement for at-sea taxonomic study, (2) A field pipeline with consideration to the successful collection of high-quality specimens using live-sorting in a 'cold-chain' from depths of 4000-5000m in the central tropical Pacific, (3) A laboratory pipeline with consideration to the needs to collect both DNA sequences and morphological data in a timely and cost-effective manner suited to the immediate needs of the science community and, (4) A data and sample management pipeline that includes the publication of results with consideration to the accessibility of data and materials. Our complete methodology for DNA taxonomy in the CCZ, including deployment protocols for the various sampling gears, methods for live-sorting and microscope photography at sea and details of sample and data curation are provided in a separate open-access publication (Glover et al. 2015b). Details of the taxonomical methods, including primers used for DNA taxonomy, are also given in the CCZ Echinodermata paper (Glover et al. 2016).
Field Pipeline
The ABYSSLINE environmental baseline survey includes three 30x30 km survey boxes (strata), distributed across the UK-1 area, and an additional reference sites outside of the UK-1 area (Glover et al. 2015b, Smith et al. 2013). Within each survey stratum, sample sites for a variety of benthic sampling gears are selected randomly – a randomized, stratified sampling design that assumes no a priori knowledge of the benthic environment (Smith et al. 2013). The UK-1 strata are being sampled in a series of oceanographic cruises during the course of the project, which commenced in July 2013, with the first cruise (AB01) taking place in October 2013 aboard the RV Melville. During this cruise, the first stratum was comprehensively mapped with multibeam bathymetry and sampled for a range of biological, environmental and geophysical parameters (Fig. 2, Smith et al. 2013).
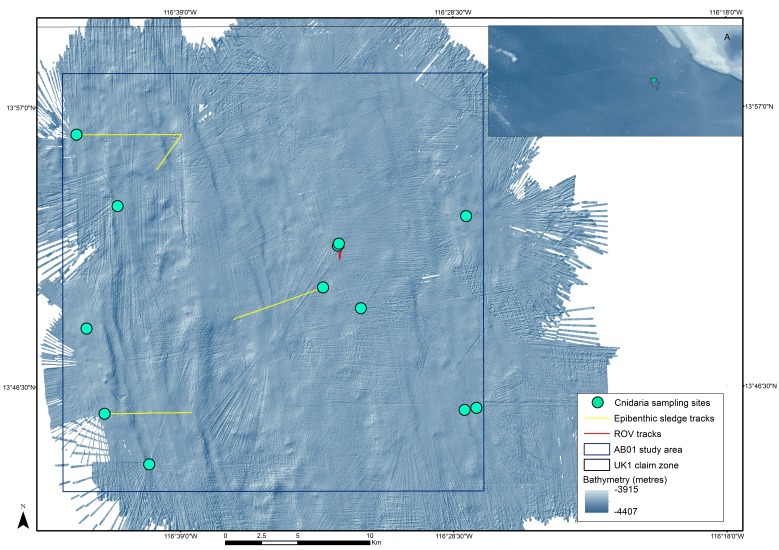
Figure 2.
'UK-1 Stratum A' ABYSSLINE biological baseline survey box sited within the UK-1 polymetallic nodule exploration area. Stratum A is a 30x30km survey box in the northern sector of the 58,000 km2 exploration area. Cnidaria sample localities are indicated by green circles from the AB01 RV Melville survey cruise, October 2013. Inset map A: the site location within the central Pacific, with GEBCO 2014 bathymetry (global 30 arc-second interval grid data set). Seafloor bathymetry from the RV Melville ABYSSLINE cruise is shown in the main map.
A comprehensive description of our DNA taxonomy pipeline is provided in Glover et al. 2015b. In summary, deep-sea benthic specimens from the UK-1 Stratum A were collected using a range of oceanographic sampling gears including box core (BC), epibenthic sledge (EBS), remotely operated vehicle (ROV) and megacore (MC) (Figs 2, 3). Geographic data from sampling activities were recorded on a central GIS database. Live-sorting of sediment and specimen samples was carried out aboard the RV Melville under the ‘cold-chain’ pipeline, in which material was immediately transferred and maintained in chilled, filtered seawater held at 2-4°C (Fig. 3). Specimens were preliminarily identified at sea and imaged live using stereomicroscopes with attached digital cameras and strobe lighting. The specimens were then transferred to individual microtube vials containing an aqueous solution of 80% non-denatured ethanol, numbered and barcoded into a database and kept chilled until return to the Natural History Museum (NHM), London. Larger, megafaunal-sized, animals were sub-sampled for DNA (with the tissue and DNA sample being taken to NHM, London) with the remaining intact specimen preserved in 10% formalin solution and taken to the University of Hawaii, Honolulu, USA for further study.
Figure 3.
ABYSSLINE UK-1 polymetallic nodule exploration area field pipeline for DNA taxonomy. ABYSSLINE AB01 cruise sampling aboard RV Melville in October 2013. (a) Preparing Box Core (BC) for deployment, (b) BC entering the water, (c) Megacore entering the water, (d-f) Epibenthic Sledge shown on recovery in water and cod-end where samples are taken, (g) controlling BC deployment on seafloor, (h) echosounder trace showing BC approaching seabed reflection, (i) successful BC surface after recovery, 50cm x 50cm, (j) carefully sifting mud in chilled filtered seawater (approx. temp 5-7°C) to remove live animals in undamaged state, (k) live-sorting aboard ship, taking samples for DNA and photomicrographs of specimens down to <1mm in size. All images by Glover, Dahlgren & Wiklund. A more comprehensive description of our methods is provided in Glover et al. 2015b.
Laboratory Pipeline
Extraction of DNA was done with DNeasy Blood and Tissue Kit (Qiagen) using a Hamilton Microlab STAR Robotic Workstation. About 1800 bp of 18S, 450 bp of 16S, and 650 bp of cytochrome c oxidase subunit I (COI) were amplified using primers listed in Table 1. PCR mixtures contained 1 μl of each primer (10μM), 2 μl template DNA and 21 μl of Red Taq DNA Polymerase 1.1X MasterMix (VWR) in a mixture of total 25 μl. The PCR amplification profile consisted of initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 45 s, extension at 72°C for 2 min, and a final extension at 72°C for 10 min. PCR products were purified using Millipore Multiscreen 96-well PCR Purification System, and sequencing was performed on an ABI 3730XL DNA Analyser (Applied Biosystems) at The Natural History Museum Sequencing Facility, using the same primers as in the PCR reactions plus two internal primers for 18S (Table 1).
Table 1.
Primers used for PCR and sequencing of 18S, 16S and COI.
| Primer | Sequence 5'-3' | Reference |
|---|---|---|
| 18S | ||
| 18SA | AYCTGGTTGATCCTGCCAGT | Medlin et al. 1988 |
| 18SB | ACCTTGTTACGACTTTTACTTCCTC | Nygren and Sundberg 2003 |
| 620F | TAAAGYTGYTGCAGTTAAA | Nygren and Sundberg 2003 |
| 1324R | CGGCCATGCACCACC | Cohen et al. 1998 |
| CO1 | ||
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | Folmer et al. 1994 |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | Folmer et al. 1994 |
| polyLCO | GAYTATWTTCAACAAATCATAAAGATATTGG | Carr et al. 2011 |
| polyHCO | TAMACTTCWGGGTGACCAAARAATCA | Carr et al. 2011 |
| 16S | ||
| 16SbrH | CCGGTCTGAACTCAGATCACGT | Palumbi 1996 |
| Ann16SF | GCGGTATCCTGACCGTRCWAAGGTA | Sjölin et al. 2005 |
Overlapping sequence fragments were merged into consensus sequences using Geneious R6 (www.geneious.com, Kearse et al. (2012) and aligned using MAFFT (Katoh 2002) for 18S and 16S, and MUSCLE (Edgar 2004) for COI, both programs used as plugins in Geneious, with default settings. Bayesian phylogenetic analyses (BA) were conducted with MrBayes 3.1.2 (Ronquist and Huelsenbeck 2003). Analyses were run for 10-20 million generations, of which 2.5-5 million generations were discarded as burn-in.
Data Pipeline
The field and laboratory pipelines created a series of databases and sample sets that were then integrated into a data-management pipeline (Fig. 4). This includes the transfer and management of data and samples between a central collections database, a molecular collections database, an online scratchpad (website for faunal data) and external repositories (e.g GenBank, WoRMS, OBIS, GBIF) through a DarwinCore archive. This provides a robust data framework to support DNA taxonomy, in which openly-available data and voucher material is key to quality data standards. A further elaboration of the data pipeline is published in Glover et al. (2015b)
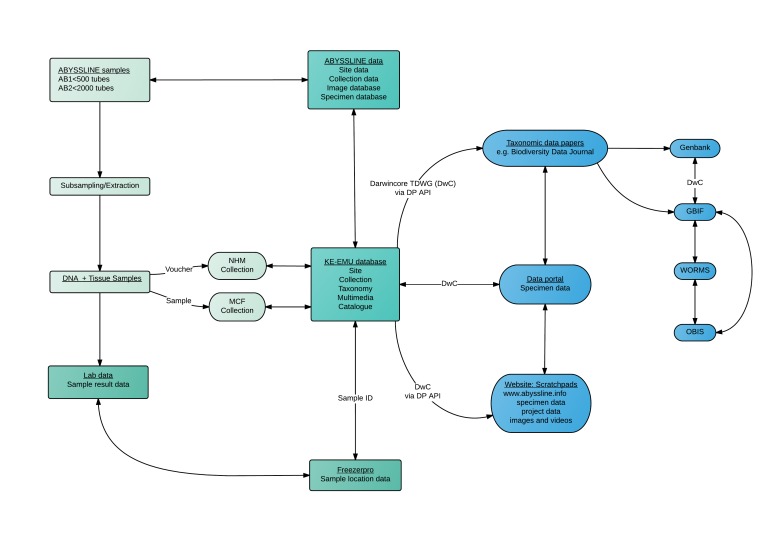
Figure 4.
Data and sample management workflow on the ABYSSLINE DNA taxonomy project. Processes relating to a) physical samples are shown in grey, b) institution level data in dark green and c) externally-available data in blue.
Taxonomic Assignments
All future studies of biogeographic and bathymetric ranges, gene-flow, extinction risks, natural history, reproductive ecology, functional ecology and geochemical interactions of CCZ species are dependent on accurate identifications faciliated by taxonomy. This taxonomy, presented here, is itself dependent on a sound theoretical underpinning – a species concept - coupled with the availability of both raw data and voucher samples. Here we use a phylogenetic species concept, sensu Donoghue 1985 with species determined by DNA-based phylogenetic analysis and the recognition of distinct monophyletic groups as species. For those taxa where the typical morphological data that allows determination of species are missing, we provide the lowest-level taxonomic name possible, but determination to species with genetic data. For species similar to a morphologically well-defined species name where we lack comparable genetic data from type material or from the type locality, or when genetic data previously published in Genbank is incompatible with ours, we use the open nomenclature expression ”cf.".
All DNA vouchers (including archived frozen tissue) and genetic data are accessible through the Natural History Museum, London, together with the morphological data presented in this paper. Additional morphological material is held at both NHM London and the University of Hawaii (detailed in Table 2). As such our species hypotheses are easily open to further evaluation and iterative improvement, e.g. full descriptions for new taxa with improved data from future cruises. A localized identification field guide to the CCZ fauna will be the subject of a future publication as more species are described, but for the present we recommend DNA-based identification (barcoding) of our species coupled with morphological comparisons made possible through this publication.
Table 2.
Taxon treatments presented in this paper. Includes Class, DNA Taxonomy ID (a species-level identification based on combined DNA and morphological evidence), GUID (Global Unique Identifier link to data record on http://data.nhm.ac.uk), ABYSSLINE Record number, NHM Accession number, NHM Molecular Collection Facility (MCF) sample ID number (NHMUK_MCF#) and NCBI GenBank accession number (Genbank#) for successfully sequenced genetic markers. Record numbers include both material archived at NHM (all DNA tissue samples held in the NHM MCF, plus morphological material including whole specimens for some taxa), University of Hawaii (AB01_CSXX) and National Museum of Natural History (USNM). GenBank numbers for data downloaded from GenBank for phylogenetic analysis are presented in Suppl. material 1.
| Class | DNA Taxonomy ID | GUID# | ABYSSLINE Record# | NHMUK_Acc# | NHMUK_MCF# | Genbank# |
|---|---|---|---|---|---|---|
| Anthozoa | Abyssoprimnoa gemina | 6e976c27-c70c-434a-8be3-f6155e36567f | NHM_341 USNM 1268848 AB01_CS09 |
NHMUK 2016.1 | 0185546837 0185546838 0185546851 |
KX384618
KX384626 |
| Anthozoa | Calyptrophora persephone | dc143dd0-a366-479a-9e23-ca28fe79761b | NHM_462 USNM 1268754 AB01_CS16 |
NHMUK 2016.2 | 0185546835 0185546836 0185546847 |
KX384619
KX384627 |
| Anthozoa | Mopseinae sp. (NHM_330) | 52fc66a5-d396-49be-a210-4ac58a164aec | NHM_002 | NHMUK 2016.4 | 0185546839 0185546840 0185546862 |
KX384629 |
| Anthozoa | Mopseinae sp. (NHM_330) | c1c12d85-274b-4ae5-bf60-72c82b70dc16 | NHM_019 | NHMUK 2016.5 | 0185546841 0185546863 |
KX384610
KX384621 KX384630 |
| Anthozoa | Mopseinae sp. (NHM_330) | 39fb53c7-b5f4-4e9a-a8da-756129250937 | NHM_035 | NHMUK 2016.6 | 0185546842 0185546864 |
KX384611 |
| Anthozoa | Mopseinae sp. (NHM_330) | fb7d366b-dcd6-448e-8fe0-7625d811483d | NHM_136 | NHMUK 2016.7 | 0185541733 0185546843 0185546865 |
KX384631 |
| Anthozoa | Mopseinae sp. (NHM_330) | 6102fb1a-c452-47a8-9a52-ca1159c06cb0 | NHM_154 | NHMUK 2016.8 | 0185546830 0185546866 |
KX384632 |
| Anthozoa | Mopseinae sp. (NHM_330) | d73e2414-e2ad-4c07-9e12-0864ce6ef3c1 | NHM_203 | NHMUK 2016.9 | 0185546867 |
KX384612
KX384633 |
| Anthozoa | Mopseinae sp. (NHM_330) | d5fdef19-8efd-4865-bc53-725568bac8df | NHM_330 AB01_CS07 |
NHMUK 2016.10 | 0185546828 0185546829 0185546854 |
KX384613
KX384634 |
| Anthozoa | Mopseinae sp. (NHM_330) | d24a6561-7c54-4607-89cd-6ddaaa557ca9 | NHM_333 | NHMUK 2016.11 | 0185541760 0185546826 0185546827 |
KX384622
KX384635 |
| Anthozoa | Mopseinae sp. (NHM_330) | e54932dc-d64b-431f-8e55-fa93504e2bb6 | NHM_376 | NHMUK 2016.12 | 0185546824 0185546825 0185546849 |
KX384636 |
| Anthozoa | Keratoisidinae sp. (NHM_417) | d260bc48-6a67-4fb4-b504-863b6d174da0 | NHM_417 AB01_CS14 |
NHMUK 2016.13 | 0185546822 0185546823 0185546848 |
KX384623
KX384637 |
| Anthozoa | Hormathiidae sp. (NHM_416) | bad617b4-5dff-4c9d-8ea8-c04ef29b0313 | NHM_416 AB01_CS13 |
NHMUK 2016.3 | 0185541761 0185546820 0185546821 |
KX384620 |
| Hydrozoa | Abylopsis eschscholtzii | 8cab45b5-4868-4d7a-a2a1-92f8b9ab3649 | NHM_477 | NHMUK 2016.14 | 0185546846 |
KX384609
KX384617 |
| Hydrozoa |
Corymorphidae sp. (NHM_398) |
dc50e246-ae77-401d-9d57-8f0c8df2b07d | NHM_398 | NHMUK 2016.15 | 0185546845 | KX384628 |
| Hydrozoa | Siphonophorae sp. (NHM_339) | 42ffb986-0c1c-45ed-be56-7b9512ed746f | NHM_339 | NHMUK 2016.16 | 0185546808 0185546809 0185546852 |
KX384615
KX384625 |
| Hydrozoa | Stephanomia amphytridis | a1573f3a-6acf-46b9-b43e-db3a9ac90d2b | NHM_249 | NHMUK 2016.17 | 0185546811 0185546812 0185546832 |
KX384616 |
| Scyphozoa | Nausithoe sp. (NHM_353) | 86032849-ddd2-4690-b5e3-a3d1a7d09d3d | NHM_353 | NHMUK 2016.18 | 0185546810 0185546850 |
KX384614
KX384624 |
Data resources
The following sections detail the phylogenetic analysis and data resources that underpins the species hypotheses presented in the taxon treatments. A full list of all taxa including Natural History Museum Accession Numbers, NHM Molecular Collection Facility (NHM-MCF) FreezerPro numbers and NCBI GenBank Accession numbers is provided in Table 2.
Phylogenetic analyses
All specimens in Table 2 with the exception of NHM_083 and NHM_321 were sequenced, searched via Basic Local Alignment Search Tool (BLAST) for comparison on NCBI Genbank and subsequently analysed in MrBayes for phylogenetic relationship studies.
Although phylogenetic trees were produced for all species, we only present one of the trees here, the octocorals (Fig. 5). In all other analyses, the specimens were too distant from each other to incorporate in one tree per group, and their affiliations are instead explained in the text below.
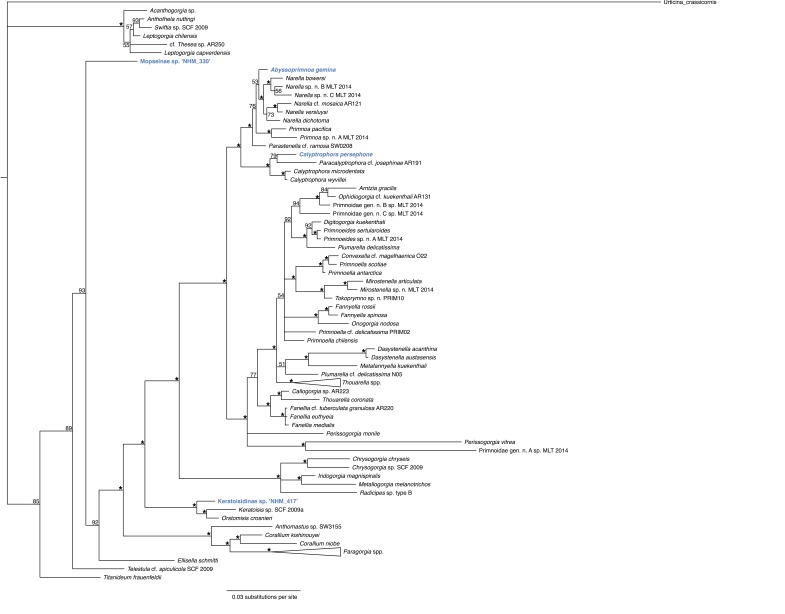
Figure 5.
Phylogenetic analysis of the octocorals. 50% majority rule consensus tree from the Bayesian analyses, combining the two genes 18S and 16S, and using in total 83 taxa (Suppl. material 1). Some of the clades are collapsed in order to make the tree smaller and easier to read.
Phylogenetic analysis of the octocorals (Fig. 5) reveals the presence of four distinct lineages of ABYSSLINE specimens which we interpret as the four species described below based on their genetic data. Among Hydrozoa, two of the sequenced species had a match on Genbank; Abylopsis eschscholtzii NHM_477 (matching with accession number GQ119937) and Stephanomia amphytridis NHM_249 (matching with GQ120047), and subsequent alignment view and tree building (trees not showed) confirmed the identifications. For some of the other taxa, trees were constructed in order to narrow down the taxonomic identity, and thus the hydrozoan NHM_398 was shown to fall within the family Corymorphidae, while the scyphozoan NHM_353 fell close to Nausithoe atlantica. As only COI from one species of Nausithoe was available, it is impossible to prove that NHM_353 actually belongs to that genus. However, with an uncorrected ‘p’ and K2P value of 0.04 between Nausithoe atlantica and NHM_353, which is comparable to values of 0.06 between Atolla vanhoeffeni and Atolla wyvillei (also Coronatae, as is Nausithoe), we suggest Nausithoe sp. for NHM_353 for now. The siphonophore NHM_339 was impossible to identify to any taxonomic level below order. The hexacoral NHM_416 was identified by morphology to the family Hormathiidae, and while the molecular result was inconclusive it was not contradictory, and future studies might resolve the generic affiliation.
Taxon treatments
Abyssoprimnoa gemina
Cairns, 2015
Materials
Type status: Other material. Occurrence: catalogNumber: 6e976c27-c70c-434a-8be3-f6155e36567f; recordNumber: NHM_341; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594618; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384618 | KX384626; Taxon: taxonConceptID: Abyssoprimnoa gemina; scientificName: Abyssoprimnoa gemina; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Primnoidae; genus: Abyssoprimnoa; specificEpithet: gemina; scientificNameAuthorship: Cairns, 2015; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4111; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.761666666667; decimalLongitude: -116.46033333333; geodeticDatum: WGS84; Identification: identifiedBy: Stephen Cairns, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Bowers & Connelly Megacore; eventDate: 2013-10-18; eventTime: 15:54; habitat: Abyssal plain; fieldNumber: MC08; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
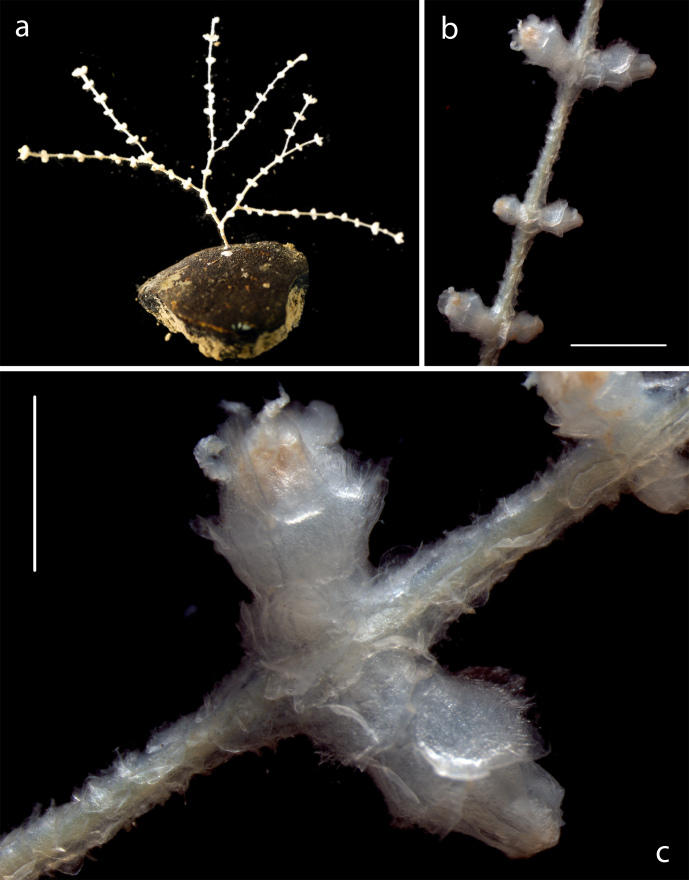
Small uniplanar dichotomously branched colonies, having paired globose polyps (Fig. 6).
Figure 6.
Abyssoprimnoa gemina Cairns, 2015. (a) Specimen NHM_341 attached to nodule. (b) Detail from same specimen. (c) Polyp from same specimen. Scale bars (b) 2 mm, (c) 1 mm. Image attribution Glover, Dahlgren & Wiklund 2016.
Genetic data for this taxon with new GenBank accession numbers are provided in Table 2.
Diagnosis
Species described by Stephen Cairns based on this specimen (NHM_341) from UK-1 claim area and additional material collected in the German claim area (Cairns 2015). No genetic matches at Genbank but forms a cluster with Narella and Primnoa sequences published.
Calyptrophora persephone
Cairns, 2015
Materials
Type status: Other material. Occurrence: catalogNumber: dc143dd0-a366-479a-9e23-ca28fe79761b; recordNumber: NHM_462; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594705; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384619 | KX384627; Taxon: taxonConceptID: Calyptrophora persephone; scientificName: Calyptrophora persephone; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Primnoidae; genus: Calyptrophora; specificEpithet: persephone; scientificNameAuthorship: Cairns, 2015; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4160; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.726616666667; decimalLongitude: -116.67; geodeticDatum: WGS84; Identification: identifiedBy: Stephen Cairns, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: USNEL Box Core; eventDate: 2013-10-22; eventTime: 07:25; habitat: Abyssal plain; fieldNumber: BC14; fieldNotes: Collected from 0-2 cm layer of box core using a 300 micron sieve; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
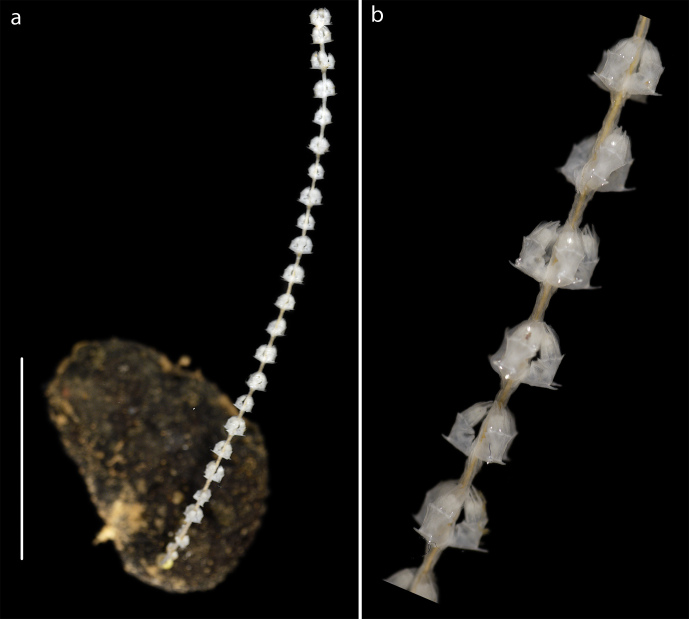
Unbranched upright colonies with worls of three to four polyps facing upward on stem Fig. 7.
Figure 7.
Calyptrophora persephone Cairns, 2015. (a) Specimen NHM_462 attached to nodule, (b) Detail, same specimen. Scale bar 30 mm. Image attribution Glover, Dahlgren & Wiklund 2016.
Genetic data for this taxa with new Genbank accession numbers are provided in Table 2.
Diagnosis
Species described by Stephen Cairns based on this specimen (NHM_462) and additional material collected in the German claim area (Cairns 2015). No genetic matches at Genbank but forms a cluster with other Calyptrophora sequences published
Mopseinae sp. 'NHM_330'
Materials
Type status: Other material. Occurrence: catalogNumber: 52fc66a5-d396-49be-a210-4ac58a164aec; recordNumber: NHM_002; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5595039; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384629; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4171; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.881666666667; decimalLongitude: -116.46666666667; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: USNEL Box Core; eventDate: 2013-10-08; eventTime: 17:15; habitat: Abyssal plain; fieldNumber: BC03; fieldNotes: Collected from 0-2 cm layer of box core using a 300 micron sieve; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: c1c12d85-274b-4ae5-bf60-72c82b70dc16; recordNumber: NHM_019; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594370; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384610 | KX384621 | KX384630; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4336; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.8372; decimalLongitude: -116.55843333333; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Brenke Epibenthic Sledge; eventDate: 2013-10-09; eventTime: 10:26; habitat: Abyssal plain; fieldNumber: EB01; fieldNotes: Collected from epi net (on the epibenthic sledge); Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: 39fb53c7-b5f4-4e9a-a8da-756129250937; recordNumber: NHM_035; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594384; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384611; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4336; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.8372; decimalLongitude: -116.55843333333; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Brenke Epibenthic Sledge; eventDate: 2013-10-09; eventTime: 10:26; habitat: Abyssal plain; fieldNumber: EB01; fieldNotes: Collected from epi net (on the epibenthic sledge); Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: fb7d366b-dcd6-448e-8fe0-7625d811483d; recordNumber: NHM_136; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594458; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384631; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4080; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.758333333333; decimalLongitude: -116.69851666667; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Brenke Epibenthic Sledge; eventDate: 2013-10-11; eventTime: 10:32; habitat: Abyssal plain; fieldNumber: EB02; fieldNotes: Collected from epi net (on the epibenthic sledge); Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: 6102fb1a-c452-47a8-9a52-ca1159c06cb0; recordNumber: NHM_154; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594475; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384632; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4080; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.758333333333; decimalLongitude: -116.69851666667; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Brenke Epibenthic Sledge; eventDate: 2013-10-11; eventTime: 10:32; habitat: Abyssal plain; fieldNumber: EB02; fieldNotes: Collected from epi net (on the epibenthic sledge); Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: d73e2414-e2ad-4c07-9e12-0864ce6ef3c1; recordNumber: NHM_203; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594513; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384612 | KX384633; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4054; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.824116666667; decimalLongitude: -116.53425; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: USNEL Box Core; eventDate: 2013-10-14; eventTime: 21:37; habitat: Abyssal plain; fieldNumber: BC07; fieldNotes: Collected from 0-2 cm layer of box core using a 300 micron sieve; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: d5fdef19-8efd-4865-bc53-725568bac8df; recordNumber: NHM_330; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594610; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384613 | KX384634; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4043; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.7603833333333; decimalLongitude: -116.467933333333; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Remotely Operated Vehicle; eventDate: 2013-10-17; eventTime: 19:46; habitat: Abyssal plain; fieldNumber: RV05; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: d24a6561-7c54-4607-89cd-6ddaaa557ca9; recordNumber: NHM_333; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594613; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384622 | KX384635; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4075; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.76085; decimalLongitude: -116.4653; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Remotely Operated Vehicle; eventDate: 2013-10-17; eventTime: 19:06; habitat: Abyssal plain; fieldNumber: RV05; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Type status: Other material. Occurrence: catalogNumber: e54932dc-d64b-431f-8e55-fa93504e2bb6; recordNumber: NHM_376; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594645; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384636; Taxon: taxonConceptID: Mopseinae sp. (NHM_330); scientificName: Mopseinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4182; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.933066666667; decimalLongitude: -116.71628333333; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Brenke Epibenthic Sledge; eventDate: 2013-10-19; eventTime: 12:16; habitat: Abyssal plain; fieldNumber: EB05; fieldNotes: Collected from epi net (on the epibenthic sledge); Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
Branched or unbranched colonies attached to nodules. Identified to family based on general morphology (Fig. 8).
Figure 8.
Mopseinae sp. 'NHM_330'. (a) Specimen NHM_330 attached to nodule imaged by Remotely Operated Vehicle in situ. (b) Specimen NHM_002. (c) Specimen NHM_330 imaged alive on ship. (d) Detail of NHM_330. (e) Detail of polyp, specimen NHM_330. Scale bars (c) 1 cm, (e) 500 µm. Image attribution (a) Diva Amon and Craig R Smith, (b-e) Glover, Dahlgren & Wiklund.
Genetic data for this taxon with new GenBank accession numbers are provided in Table 2.
Diagnosis
No genetic matches on Genbank and does not cluster with other Isididae species available at Genbank but occupy a position ancestral to most other sequenced taxa in suborder Calcaxonia (order Alcyonacea) (Fig. 5). Based on morphology this taxon is here referred to the subfamily Mopseinae Gray, 1870. The genus is indeterminate at this time but morphologically resembles Primnoisis (Phil Alderslade pers com).
Keratoisidinae sp. 'NHM_417'
Materials
Type status: Other material. Occurrence: catalogNumber: d260bc48-6a67-4fb4-b504-863b6d174da0; recordNumber: NHM_417; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594676; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384623 | KX384637; Taxon: taxonConceptID: Keratoisidinae sp. (NHM_417); scientificName: Keratoisidinae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Alcyonacea; family: Isididae; scientificNameAuthorship: Lamouroux, 1812; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4026; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.8645666666667; decimalLongitude: -116.548233333333; geodeticDatum: WGS84; Identification: identifiedBy: Les Watling, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Remotely Operated Vehicle; eventDate: 2013-10-20; eventTime: 11:05; habitat: Abyssal plain; fieldNumber: RV06; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
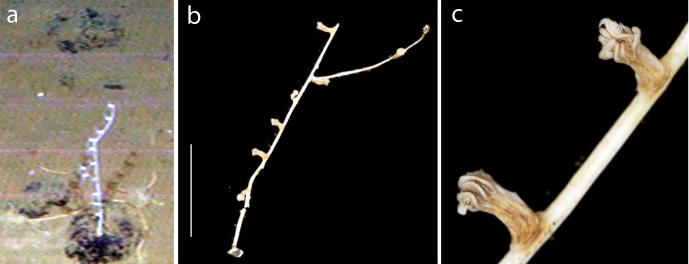
Branched colony attached to nodule. Identified to family based on general morphology (Fig. 9).
Figure 9.
Keratoisidinae sp. 'NHM_417'. (a) Specimen imaged in situ by Remotely Operated Vehicle. Entire specimen. (b) Specimen after recovery. (c) Detail of polyps. Scale bar 3 cm. Image attribution (a) Diva Amon and Craig R Smith, (b-c) Glover, Dahlgren & Wiklund.
Genetic data for this taxon with new GenBank accession numbers are provided in Table 2.
Diagnosis
The specimen superficially resembles Bathygorgia profunda Wright (1885) first described in the Narrative (Part 2) of the HMS Challenger voyage and more fully described in Wright and Studer (1889) where a type locality from station 241 between Yokohama and Hawaii (depth 4206m) is indicated. Morphologically-similar specimens based on AUV or ROV image data is also referred to this species in the CCZ megafauna atlas (International Seabed Authority 2016). However, this specimen is distinguished by the presence of large needle sclerites (Fig. 9) that extend between the bases of the tentacles (which are not present in Bathygorgia), and the body sclerites being predominantly small needles rather than the disorganised rods that typify Bathygorgia. The present specimen belongs to a new genus and species (Watling et al. in prep). For a recent discussion on Keratoisid corals, see Dueñas et al. (2014) , Lapointe and Watling (2015) and Moore et al. 2016.
No genetic matches at Genbank but forms a cluster with Keratoisis and Orstomisis sequences published (Fig. 5).
Hormathiidae sp. 'NHM_416'
Materials
Type status: Other material. Occurrence: catalogNumber: bad617b4-5dff-4c9d-8ea8-c04ef29b0313; recordNumber: NHM_416; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594675; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384620; Taxon: taxonConceptID: Hormathiidae sp. (NHM_416); scientificName: Hormathiidae; kingdom: Animalia; phylum: Cnidaria; class: Anthozoa; order: Actiniaria; family: Hormathiidae; scientificNameAuthorship: Carlgren, 1932; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4025; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.8645283333333; decimalLongitude: -116.548211666667; geodeticDatum: WGS84; Identification: identifiedBy: Estefania Rodriguez, Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Remotely Operated Vehicle; eventDate: 2013-10-20; eventTime: 22:54; habitat: Abyssal plain; fieldNumber: RV06; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
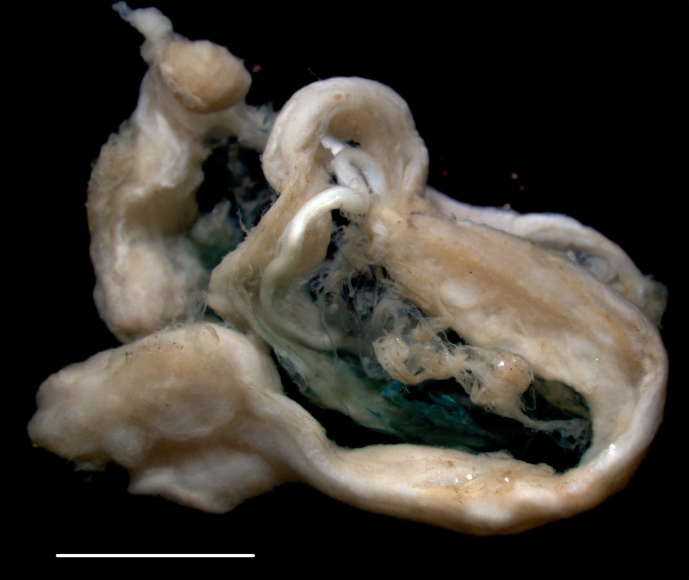
Description
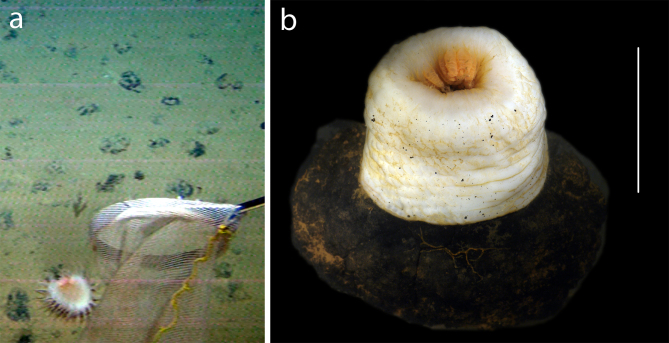
Relatively large anemone attached to nodule. Column pale, white, with verrucae arranged in rows. No pronounced limbus. Morphology and colour of tentacles not observed (Fig. 10).
Figure 10.
Hormathiidae sp. NHM_416. Scale bar 5 cm. Image attribution (a) Diva Amon & Craig R Smith, (b) Glover, Dahlgren & Wiklund.
Genetic data for this taxon with new GenBank accession numbers are provided in Table 2.
Diagnosis
Assigned here to the family Hormathiidae Carlgren (1932) based on morphology, and most similar to species in the genus Phelliactis Simon (1892) (Estefania Rodriguez pers. com.). Attempts to collect a full suite of genetic marker data was unsuccessful, and limited to the 18S gene only. A phylogenetic analysis of the 18S gene including hexacoral taxa available at Genbank placed this specimen with low support as sister to a sequence assigned to Capnea georgiana (Carlgren 1927). The result is most probably an artefact due to the restricted dataset and homoplasy in the 18S gene. Previous analyses has suggested limitation of this gene alone for species barcoding and genus level phylogenetic reconstruction in hexacorals (Rodríguez et al. 2014).
Abylopsis eschscholtzii
(Huxley, 1859)
Materials
Type status: Other material. Occurrence: catalogNumber: 8cab45b5-4868-4d7a-a2a1-92f8b9ab3649; recordNumber: NHM_477; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5595047; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384609 | KX384617; Taxon: taxonConceptID: Abylopsis eschscholtzii; scientificName: Abylopsis eschscholtzii; kingdom: Animalia; phylum: Cnidaria; class: Hydrozoa; order: Siphonophorae; family: Abylidae; genus: Abylopsis; specificEpithet: eschscholtzii; scientificNameAuthorship: Huxley, 1859; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4160; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.726616666667; decimalLongitude: -116.67; geodeticDatum: WGS84; Identification: identifiedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: USNEL Box Core; eventDate: 2013-10-22; eventTime: 07:25; habitat: Abyssal plain; fieldNumber: BC14; fieldNotes: Collected from 0-2 cm layer of box core using a 300 micron sieve; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
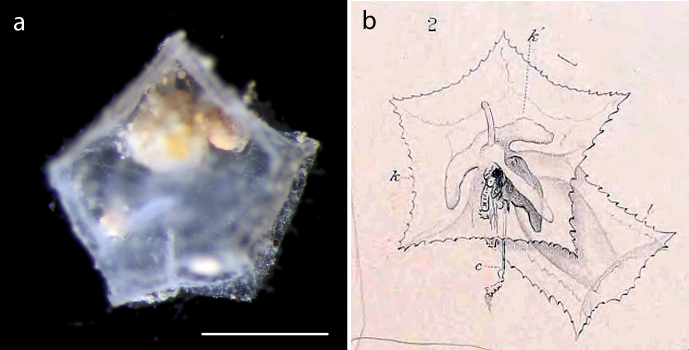
Pentagonal, transparent disk found in a boxcore sample, possibly from the water column (Fig. 11). The animal cavity contained sediment.
Figure 11.
Abylopsis eschscholtzii (Huxley, 1859). (a) Specimen NHM_477. (b) Drawing from original description by Huxley 1859. Scale bar 1 mm. Image attribution (a) Glover, Dahlgren & Wiklund 2016.
Genetic data for this taxon with new GenBank accession numbers are provided in Table 2.
Diagnosis
The CO1 data suggest that the specimen is identical to Abylopsis eschscholtzii (Huxley, 1859) (Genbank Accession # GQ119937). No type locality was assigned to the name in the species description. The original description state that the species occurred in 'all the seas that was traversed' (Huxley 1859).
Corymorphidae sp. 'NHM_398'
Materials
Type status: Other material. Occurrence: catalogNumber: dc50e246-ae77-401d-9d57-8f0c8df2b07d; recordNumber: NHM_398; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594663; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384628; Taxon: taxonConceptID: Corymorphidae sp. (NHM_398); scientificName: Corymorphidae; kingdom: Animalia; phylum: Cnidaria; class: Hydrozoa ; order: Anthoathecata; family: Corymorphidae; scientificNameAuthorship: Allman,1872; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4500; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.863283333333; decimalLongitude: -116.54885; geodeticDatum: WGS84; Identification: identifiedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Diva Amon; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: USNEL Box Core; eventDate: 2013-10-20; eventTime: 03:39; habitat: Abyssal plain; fieldNumber: BC12; fieldNotes: Collected from 0-2 cm layer of box core using a 300 micron sieve; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
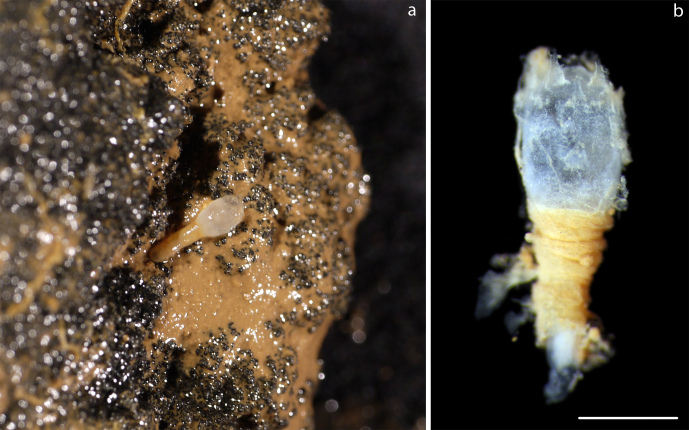
Small bulb-like corymorph hydrozoan found attached to a nodule with the crown apparently retracted (Fig. 12).
Figure 12.
Corymorphidae sp. NHM_398. (a) Specimen NHM_398 attached to nodule, (b) Detail, same specimen. Scale bar 1,5 mm. Image attribution Glover, Dahlgren & Wiklund 2016.
Genetic data for this taxa with new Genbank accession numbers are provided in Table 2.
Diagnosis
Phylogenetic analyses based on gene data suggest that this hydrozoan fall within the family Corymorphidae (tree not showed).
Siphonophorae sp. 'NHM_339'
Materials
Type status: Other material. Occurrence: catalogNumber: 42ffb986-0c1c-45ed-be56-7b9512ed746f; recordNumber: NHM_339; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594616; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384615 | KX384625; Taxon: taxonConceptID: Siphonophorae sp. (NHM_339); scientificName: Siphonophorae; kingdom: Animalia; phylum: Cnidaria; class: Hydrozoa ; order: Siphonophorae; scientificNameAuthorship: Eschscholtz, 1829; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4111; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.761666666667; decimalLongitude: -116.46033333333; geodeticDatum: WGS84; Identification: identifiedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Bowers & Connelly Megacore; eventDate: 2013-10-18; eventTime: 15:54; habitat: pelagic; fieldNumber: MC08; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
Tentacle from pelagic jellyfish found on pinger (Fig. 13).
Figure 13.
Siphonophorae sp. NHM_339. Detached tentacle found on pinger. Scale bar 5 mm. Image attribution Glover, Dahlgren & Wiklund 2016.
Genetic data for this taxon with new GenBank accession numbers are provided in Table 2.
Diagnosis
No genetic matches at Genbank but in a COI tree (not shown) it is sister to Nectopyramis natans although with quite low support, while the 18S sequence BLAST search is nearest to Rhizophysa eysenhardti.
Stephanomia amphytridis
Lesueur & Petit, 1807
Materials
Type status: Other material. Occurrence: catalogNumber: a1573f3a-6acf-46b9-b43e-db3a9ac90d2b; recordNumber: NHM_249; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 1; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5594546; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384616; Taxon: taxonConceptID: Stephanomia amphytridis; scientificName: Stephanomia amphytridis; kingdom: Animalia; phylum: Cnidaria; class: Hydrozoa; order: Siphonophorae; family: Stephanomiidae; genus: Stephanomia; specificEpithet: amphytridis; scientificNameAuthorship: Lesueur & Petit, 1807; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4076; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.811683333333; decimalLongitude: -116.71001666667; geodeticDatum: WGS84; Identification: identifiedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: Bowers & Connelly Megacore; eventDate: 2013-10-16; eventTime: 19:50; habitat: pelagic; fieldNumber: MC06; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
Fragment of specimen stuck on megacore at retrieval. No image.
Genetic data for this taxon with new GenBank accession numbers are provided in Table 2.
Diagnosis
The CO1 data suggest that the specimen is identical to Stephanomia amphytridis Lesueur and Petit (1807) (Genbank Accession # GQ120047).
Nausithoe sp. 'NHM_353'
Materials
Type status: Other material. Occurrence: catalogNumber: 86032849-ddd2-4690-b5e3-a3d1a7d09d3d; recordNumber: NHM_353; recordedBy: Adrian Glover, Helena Wiklund, Thomas Dahlgren, Maggie Georgieva; individualCount: 2; preparations: tissue voucher stored in 80% non-denatured ethanol aqueous solution and DNA voucher stored in elution buffer; otherCatalogNumbers: 5595043; associatedSequences: http://ncbi.nlm.nih.gov/nucleotide/KX384614 | KX384624; Taxon: taxonConceptID: Nausithoe sp. (NHM_353); scientificName: Nausithoe; kingdom: Animalia; phylum: Cnidaria; class: Scyphozoa; order: Coronatae; family: Nausithoidae; genus: Nausithoe; scientificNameAuthorship: Kölliker, 1853; Location: waterBody: Pacific; stateProvince: Clarion Clipperton Zone; locality: UK Seabed Resources Ltd exploration claim UK-1; verbatimLocality: UK-1 Stratum A; maximumDepthInMeters: 4150; locationRemarks: RV Melville Cruise MV1313; decimalLatitude: 13.888333333333; decimalLongitude: -116.68998333333; geodeticDatum: WGS84; Identification: identifiedBy: Andy Gooday, Adrian Glover, Helena Wiklund, Thomas Dahlgren; dateIdentified: 2016-03-01; identificationRemarks: identified by DNA and morphology; Event: samplingProtocol: USNEL Box Core; eventDate: 2013-10-19; eventTime: 02:25; habitat: Abyssal plain; fieldNumber: BC11; fieldNotes: Collected from 0-2 cm layer of box core using a 300 micron sieve; Record Level: language: en; institutionCode: NHMUK; collectionCode: ZOO; datasetName: ABYSSLINE; basisOfRecord: PreservedSpecimen
Description
Specimens attached to nodule. Seemingly empty tubes with transverse rings and longitudinal striations (Fig. 14).
Figure 14.
Nausithoe sp. NHM_353. (a) Specimen NHM_321 attached to nodule (arrow). Image attribution Glover, Dahlgren & Wiklund 2016.
Genetic data for this taxon (NHM_353) with new GenBank accession numbers are provided in Table 2.
Diagnosis
ID based on Genbank BLAST place the material close to Nausithoe atlantica Broch (1914). The tubes are morphologically similar to Atorella sibogae (Leloup 1937) (top fig. p 119 in Morandini and Jarms (2005). The species was previously assigned to Stephanoscyphus based on general polyp morphology (Morandini and Jarms 2005). Type locality of Atorella sibogae is Malayan archipelago at 794 m depth. The specimens NHM_083 and NHM_321 failed to produce any DNA data and the identification is therefor provisional.
Discussion
After more than three decades of explorations focused on the CCZ, faunal records from the area (e.g. in the OBIS or GBIF databases) remain scarce. For the entire 6 million sq km area, there are no more than 220 records belonging to 52 taxa. Limiting the search for the same area to benthic records of Cnidaria yielded seven records of five taxa. In this study we report 18 records for 10 taxa, most of which benthic, that is more than doubling the known data for the region from our relatively small sample set. There were only two matches in Genbank for any of our cnidarian sequences. They were for the two pelagic hydrozoans Abylopsis eschscholtzii and Stephanomia amphytridis, two species with supposedly cosmopolitan distributions but previously not reported from the CCZ. A third pelagic species (Siphonophorae sp.) did not match any previously sequenced taxon available on Genbank.
Deep-sea corals are, in general, considered as vulnerable to anthropogenic stressors such as ocean acidification (Orr et al. 2005) and increased turbidity from activities such as trawling or oil and gas extraction (Roberts 2002). Changes in species composition and abundance of the diverse, and to a considerable extent, novel coral fauna of the CCZ, may become an important indicator of environmental impact from future mining activities. Our data show that DNA sampling of this fauna in conjuction with detailed morphological examination is required to properly assess CCZ coral diversity. We hope that the rapid publication of this and similar taxonomic data from other groups (Glover et al. 2016) will help ongoing and future efforts to build a taxonomic baseline of the CCZ suited to future impact assessments and environmental monitoring.
Supplementary Material
Taxa table
Dahlgren et al.
Data type: Genbank accession numbers
Brief description: List of taxa downloaded from GenBank that are included in the phylogenetic analysis of Octocorallia, with their Genbank accession numbers. Accession numbers for taxa sequenced in this study are found in Table 2.
File: oo_86279.xlsx
Acknowledgements
The ABYSSLINE (ABYSSal baseLINE) environmental survey of the UK-1 exploration area is supported by a collaborative partnership between six non-profit global academic research institutes (University of Hawaii at Manoa, Natural History Museum, Uni Research, National Oceanography Centre, Senckenberg Institute, IRIS Norway) and through an arrangement with UK Seabed Resources Ltd. Additional support was provided by the Swedish research council FORMAS (TGD). We would like to acknowledge the support of Magdalena Georgieva from the Natural History Museum team in sampling on board ship, and Estefania Rodriguez from the American Museum of Natural History for insights into anemone systematics and the identification of the hormathiid anemone in this study. We would also like to acknowledge the expert support from the Senckenberg Institute team in the deployment and recovery of successful Brenke Epibenthic Sledge samples (Nils Brenke, Pedro Martinez and Inga Morhbeck). This study was made possible only by the dedicated help from the entire scientific party and the Masters and Crew of the Research Vessel Melville during the first cruise of the ABYSSLINE project in October 2013. Thanks also to Jackie Mackenzie-Dodds (Molecular Collection Facility, NHM).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Data were provided by all the authors. The manuscript was first drafted by TGD and AGG and subsequently edited by all the authors.
References
- Amon D. J., Ziegler A. F., Dahlgren T. G., Glover A. G., Goineau A., Gooday A. J., Wiklund H., Smith C. R. First insights into the abundance and diversity of abyssal megafauna in a polymetallic-nodule region in the eastern Clarion-Clipperton Zone. Scientific Reports. 2016 doi: 10.1038/srep30492. [DOI] [PMC free article] [PubMed]
- Broch Hjalmar. Scyphomedusae from the Michael Sars North Atlantic Deep-Sea Expedition 1910. Rep. 'Sars' N. Atl. Deep Sea Exped. 1914;3(1):1–20. [Google Scholar]
- Cairns Stephen D. New abyssal Primnoidae (Anthozoa: Octocorallia) from the Clarion-Clipperton Fracture Zone, equatorial northeastern Pacific. http://dx.doi.org/10.1007/s12526-015-0340-x. Marine Biodiversity. 2015;l:1. doi: 10.1007/s12526-015-0340-x. [DOI] [Google Scholar]
- Carlgren O. Actinaria and Zoantharia. Odhner T., editor. 102Further Zoological Results from the Swedish Antarctic Expedition 1901-1903. 1927;2
- Carlgren O. Die Ceriantharien, Zoantharien und Actiniarien des arktischen Gebietes. In: Römer F., Schaudinn F., editors. Eine Zusammenstellung der arktischen Tierfor- men mit besonderer Berücksichtigung des Spitzbergen-Gebietes auf Grund der Ergebnisse der Deutschen Expedition in das Nördliche Eismeer im Jahre 1898. Vol. 6. Gustav Fischer; Jena: 1932. 266 [Google Scholar]
- Carr Christina M., Hardy Sarah M., Brown Tanya M., Macdonald Tara A., Hebert Paul D. N. A tri-oceanic perspective: DNA barcoding reveals geographic structure and cryptic diversity in canadian polychaetes. http://dx.doi.org/10.1371/journal.pone.0022232. PLoS ONE. 2011;6(7):e22232. doi: 10.1371/journal.pone.0022232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B. L., Gawthrop A., Cavalier-Smith T. Molecular phylogeny of brachiopods and phoronids based on nuclear-encoded small subunit ribosomal RNA gene sequences. http://dx.doi.org/10.1098/rstb.1998.0351. Philosophical Transactions of the Royal Society B: Biological Sciences. 1998;353(1378):2039–2061. doi: 10.1098/rstb.1998.0351. [DOI] [Google Scholar]
- Dueñas Luisa F., Alderslade Phil, Sánchez Juan A. Molecular systematics of the deep-sea bamboo corals (Octocorallia: Isididae: Keratoisidinae) from New Zealand with descriptions of two new species of Keratoisis. http://dx.doi.org/10.1016/j.ympev.2014.01.031. Molecular Phylogenetics and Evolution. 2014;74:15–28. doi: 10.1016/j.ympev.2014.01.031. [DOI] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. http://dx.doi.org/10.1093/nar/gkh340. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foell E. J., Pawson D. L. Photographs of Invertebrate Megafauna from Abyssal Depths of the North-Eastern Equatorial Pacific Ocean. The Ohio Journal of Science. 1986;86(3):61–68. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular marine biology and biotechnology. 1994;3(5):294–9. [PubMed] [Google Scholar]
- Glover A. G., Smith C. R. The deep-sea floor ecosystem: current status and prospects of anthropogenic change by the year 2025. http://dx.doi.org/10.1017/s0376892903000225. Environmental Conservation. 2003;30(3):219–241. doi: 10.1017/s0376892903000225. [DOI] [Google Scholar]
- Glover A. G., Higgs N. H., Horton T. World Register of Deep-Sea Species. http:// www.marinespecies.org/deepsea
- Glover A. G., Dahlgren T. G., Wiklund H., Mohrbeck I., Smith C. R. An End-to-End DNA Taxonomy Methodology for Benthic Biodiversity Survey in the Clarion-Clipperton Zone, Central Pacific Abyss. http://dx.doi.org/10.3390/jmse4010002. Journal of Marine Science and Engineering. 2015;4(1):2. doi: 10.3390/jmse4010002. [DOI] [Google Scholar]
- Glover Adrian G, Wiklund Helena, Rabone Muriel, Amon Diva J, Smith Craig R, O'Hara Tim, Mah Christopher L, Dahlgren Thomas G. Abyssal fauna of the UK-1 polymetallic nodule exploration claim, Clarion-Clipperton Zone, central Pacific Ocean: Echinodermata. http://dx.doi.org/10.3897/BDJ.4.e7251. Biodiversity data journal. 2016;4:e7251. doi: 10.3897/BDJ.4.e7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley Thomas Henry. The oceanic Hydrozoa : a description of the Calycophoridæ and Physophoridæ observed during the voyage of H.M.S. "Rattlesnake," in the years 1846-1850 / by Thomas Henry Huxley. Ray Society; London: 1859. 143. [DOI] [Google Scholar]
- Authority International Seabed. Workshop on taxonomic methods and standardization of macrofauna in the CCZ. https://www.isa.org.jm/workshop/workshop-taxonomic-methods-and-standardization-macrofauna-clarion-clipperton-fracture-zone. [2016-05-16T00:00:00+03:00];
- Authority International Seabed. Atlas of Abyssal Megafauna Morphotypes of the Clarion Clipperton Fracture Zone. http://ccfzatlas.com/wiki/index.php?title=Main_Page. [2016-03-23T00:00:00+02:00];
- ISA International Seabed Authority. http://www.isa.org.jm
- Katoh K. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. http://dx.doi.org/10.1093/nar/gkf436. Nucleic Acids Research. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. http://dx.doi.org/10.1093/bioinformatics/bts199. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe Abby, Watling Les. Bamboo corals from the abyssal Pacific: Bathygorgia. http://dx.doi.org/10.2988/0006-324x-128.2.125. Proceedings of the Biological Society of Washington. 2015;128(2):125–136. doi: 10.2988/0006-324x-128.2.125. [DOI] [Google Scholar]
- Leloup E. Hydropolypes et scyphopolypes recueillis par C. Dawydoff sur les côtes de l'Indochine française. Mém. Mus. R. d'hist. nat. Belgique, Sér. 2. 1937;12:1–73. [Google Scholar]
- Lesueur C. A., Petit N. M. Voyage de ´ couvertes aus terres australes. Atlas. Imprimerie Impériale; Paris: 1807. . [Google Scholar]
- Medlin Linda, Elwood Hille J., Stickel Shawn, Sogin Mitchell L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. http://dx.doi.org/10.1016/0378-1119(88)90066-2. Gene. 1988;71(2):491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- Moore K., Alderslade P., Miller K. A taxonomic revision of the genus Primnoisis Studer [& Wright], 1887 (Coelenterata: Octocorallia: Isididae) using morphological and molecular data. http://dx.doi.org/10.11646/zootaxa.4075.1.1. Zootaxa. 2016;4075(1):1–141. doi: 10.11646/zootaxa.4075.1.1. [DOI] [PubMed] [Google Scholar]
- Morandini André Carrara, Jarms Gerhard. New combinations for two coronate polyp species (Atorellidae and Nausithoidae, Coronatae, Scyphozoa, Cnidaria) Contributions to Zoology. 2005;74:117–123. [Google Scholar]
- Nygren Arne, Sundberg Per. Phylogeny and evolution of reproductive modes in Autolytinae (Syllidae, Annelida) http://dx.doi.org/10.1016/s1055-7903(03)00095-2. Molecular Phylogenetics and Evolution. 2003;29(2):235–249. doi: 10.1016/s1055-7903(03)00095-2. [DOI] [PubMed] [Google Scholar]
- OBIS Ocean Biogeographic Information System. http://www.iobis.org
- Orr James C., Fabry Victoria J., Aumont Olivier, Bopp Laurent, Doney Scott C., Feely Richard A., Gnanadesikan Anand, Gruber Nicolas, Ishida Akio, Joos Fortunat, Key Robert M., Lindsay Keith, Maier-Reimer Ernst, Matear Richard, Monfray Patrick, Mouchet Anne, Najjar Raymond G., Plattner Gian-Kasper, Rodgers Keith B., Sabine Christopher L., Sarmiento Jorge L., Schlitzer Reiner, Slater Richard D., Totterdell Ian J., Weirig Marie-France, Yamanaka Yasuhiro, Yool Andrew. Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. http://dx.doi.org/10.1038/nature04095. Nature. 2005;437(7059):681–686. doi: 10.1038/nature04095. [DOI] [PubMed] [Google Scholar]
- Palumbi Steve. Nucleic acid II: the polymerase chain reaction. In: Hillis DM, Moritz G, Mable BK, editors. Molecular Systematics. Sinauer Associates; Sunderland, Massachusetts: 1996. [Google Scholar]
- Roberts C. Deep impact: the rising toll of fishing in the deep sea. http://dx.doi.org/10.1016/s0169-5347(02)02492-8. Trends in Ecology & Evolution. 2002;17(5):242–245. doi: 10.1016/s0169-5347(02)02492-8. [DOI] [Google Scholar]
- Rodríguez Estefanía, Barbeitos Marcos S., Brugler Mercer R., Crowley Louise M., Grajales Alejandro, Gusmão Luciana, Häussermann Verena, Reft Abigail, Daly Marymegan. Hidden among sea anemones: The first comprehensive phylogenetic reconstruction of the order Actiniaria (Cnidaria, Anthozoa, Hexacorallia) reveals a novel group of hexacorals. http://dx.doi.org/10.1371/journal.pone.0096998. PLoS ONE. 2014;9(5):e96998. doi: 10.1371/journal.pone.0096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. http://dx.doi.org/10.1093/bioinformatics/btg180. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Simon J. A. Ein Beitrag zur Anatomie und Systematik der Hexactinien. Druck von Val. Höfling; München: 1892. 102 [Google Scholar]
- Sjölin Erica, Erséus Christer, Källersjö Mari. Phylogeny of Tubificidae (Annelida, Clitellata) based on mitochondrial and nuclear sequence data. http://dx.doi.org/10.1016/j.ympev.2004.12.018. Molecular Phylogenetics and Evolution. 2005;35(2):431–441. doi: 10.1016/j.ympev.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Smith C. R., De Leo F, Bernardino A, Sweetman A. K., Arbizu P. Abyssal food limitation, ecosystem structure and climate change. http://dx.doi.org/10.1016/j.tree.2008.05.002. Trends in Ecology & Evolution. 2008;23(9):518–528. doi: 10.1016/j.tree.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Smith C. R., Dahlgren T. G., Drazen J., Goday A., Glover A. G., Kurras G., Martinez-Arbizu P., Shulse C., Spickermann R., Sweetman A. K., Vetter E. Abyssal Baseline Study (ABYSSLINE) Cruise Report: Abyssal Baseline (AB01) – SRD UK-1 Site Oct 3 – 27, 2013 R/V Melville 13.80N 116.60W. Seafloor Investigations Report. 2013;2013-1304-051J- SRDL-AB01:1–160. [Google Scholar]
- Wedding L. M., Friedlander A. M., Kittinger J. N., Watling L., Gaines S. D., Bennett M., Hardy S. M., Smith C. R. From principles to practice: a spatial approach to systematic conservation planning in the deep sea. http://dx.doi.org/10.1098/rspb.2013.1684. Proceedings of the Royal Society B: Biological Sciences. 2013;280(1773):20131684–20131684. doi: 10.1098/rspb.2013.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedding L. M, Reiter S. M, Smith C. R, Gjerde K. M, Kittinger J. N, Friedlander A. M, Gaines S. D, Clark M. R, Thurnherr A. M, Hardy S. M. Managing mining of the deep seabed. Science. 2015;349:144–145. doi: 10.1126/science.aac6647. [DOI] [PubMed] [Google Scholar]
- Wright E. P. Narrative of the Cruise of the HMS Challenger, with a general account of the scientific results of the expedition. Narrative v.1, pt.2. HMSO; 1885. 1107. [Google Scholar]
- Wright E. P, Studer T. Report on the Alcyonaria collected by H.M.S. Challenger during the Years 1873-76. Vol. 31. HMSO; 1889. 314 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxa table
Dahlgren et al.
Data type: Genbank accession numbers
Brief description: List of taxa downloaded from GenBank that are included in the phylogenetic analysis of Octocorallia, with their Genbank accession numbers. Accession numbers for taxa sequenced in this study are found in Table 2.
File: oo_86279.xlsx