Abstract
Bacteria have remarkably robust cell shape control mechanisms. For example, cell diameter only varies by a few percent across a given population. The bacterial actin homolog, MreB, is necessary for establishment and maintenance of rod shape although the detailed properties of MreB that are important for shape control remained unknown. In this study, we perturb MreB in two ways: by treating cells with the polymerization-inhibiting drug A22 and by creating point mutants in mreB. These perturbations modify the steady-state diameter of cells over a wide range, from 790 ± 30 nm to 1700 ± 20 nm. To determine which properties of MreB are important for diameter control, we correlated structural characteristics of fluorescently tagged MreB polymers with cell diameter by simultaneously analyzing three-dimensional images of MreB and cell shape. Our results indicate that the helical pitch angle of MreB inversely correlates with the cell diameter of Escherichia coli. Other correlations between MreB and cell diameter are not found to be significant. These results demonstrate that the physical properties of MreB filaments are important for shape control and support a model in which MreB organizes the cell wall growth machinery to produce a chiral cell wall structure and dictate cell diameter.
Introduction
Bacteria come in a large variety of shapes and sizes. Their diameters can range from 200 nm in the case of Mycoplasma to 750 μm or more for Thiomargarita namibiensis, which can be seen with the naked eye (1). Cells also come in a variety of shapes, from spheres and rods to spirals and squares. These shapes can be important for many aspects of bacterial life such as motility, growth, predation, and packing within biofilms (2).
Cell shape in the vast majority of Gram-positive and Gram-negative bacteria is defined by the rigid, exoskeletal peptidoglycan (PG) cell wall (3). Bacterial cells modify their cell wall by cutting existing PG and inserting new glycan strands during growth and division to generate cells of a specific size and shape (4). Despite a tremendous variation in cell morphology, the chemical building blocks of the cell wall and the enzymes that produce and modify it are largely the same across all bacteria. This has led to the hypothesis that cell morphology is dictated by additional proteins that regulate cell wall assembly in space and time.
In most rod-shaped bacteria, such as Escherichia coli and Bacillus subtilis, cell wall insertion is spatially guided by the localization of the bacterial actin-homolog MreB (5, 6, 7). The deletion of MreB, or the inhibition of MreB polymerization using the drug A22, leads to spherical cells that are prone to lysis and have an altered cell wall structure (5, 8, 9). During elongation, we observed that MreB has a preference to localize to regions of small or even negative local Gaussian curvature. This results in growth at the sidewalls of the cell cylinder in addition to straightening of curved cells (10). We also observed curvature-based localization in wall-less, L-form cells where localized MreB is responsible for the de novo growth of rod-shaped cell walls (11).
MreB is also required for maintaining the overall arrangement of glycan strands in the cell wall. Our biophysical experiments using AFM and osmotic shock demonstrate that the E. coli cell wall is an anisotropic material with the stiff glycan strands oriented helically relative to the cell’s long axis (12, 13). To generate this long-range order, MreB-guided growth causes cells to twist as they elongate. Inhibition of MreB leads to the loss of both this twisting behavior and the chiral organization of the cell wall (13).
Why does MreB-guided insertion lead to rod-shape cells and how does MreB sense the local curvature of the cell? Our previous work combining three-dimensional (3D) imaging and computer simulations of cell growth indicated that the ability of MreB to form polymers might hold the answers to these questions. Although random insertion throughout the cell wall inevitably leads to malformed rods, the use of chirally arranged polymers that lie along the cell cylinder evens out insertion and generates smooth rods with chirally ordered glycan strands (13). Another model for rod-shaped growth hypothesizes that MreB polymers apply an inward mechanical force on the cell wall that constrains the growth from being unstable (9). Although this model is able to predict conditions for rod-shaped growth, it fails to predict the MreB-dependent chiral organization of the glycan strands. Other recent attempts to model cell growth do not use chiral polymers, but instead hypothesize that MreB’s role is to colocalize cell wall growth factors randomly on the cell surface (14). This work also fails to predict the cell wall chirality and invokes a global geometric sensing of the cell’s long-axis direction, either by the MreB or the cell wall synthesizing enzymes, without an explanation of how a nonpolymeric molecule might achieve this.
Due to their elongated nature, polymers can sense extended geometric properties of the cell and coordinate enzymatic activity over distances substantially larger than the few-nanometer size of a single globular protein. Our simulations showed that even short polymers, shorter than 200 nm, are sufficient to coordinate cell growth into a uniform rod as long as the polymers remain oriented relative to the cell’s long axis (13). It has been hypothesized that orientation inside the cell comes about due to binding of MreB polymers to the inner membrane and the energetics of polymer and membrane deformation (15). The length of MreB polymers has been the subject of considerable debate due to fluorescent-labeling artifacts, although the most recent published data and data presented in this study indicate that MreB forms short polymers about a micron in length (16, 17).
In our study we address how MreB influences cell diameter by generating an improved fluorescent fusion and using high-resolution 3D imaging to quantitatively correlate MreB’s physical properties with cell diameter across a range of MreB perturbations that alter cell shape. We show that the only property of MreB that significantly correlates with cell diameter is the helical pitch angle of MreB filaments within the cell. These results provide the first evidence, to our knowledge, that the structure and organization of MreB filaments is important for defining cell shape. Our findings support a model for cell shape determination where the helical conformation of MreB polymers gives rise to helical cell wall insertion, which in turn leads to different cell diameters due to changes in the organization of the cell wall.
Materials and Methods
Construction of MreBmsfGFP
The construction of MreBmsfGFP was previously described (10). The specific MG1655 strain differs from that previous work as we found that there are physiological and metabolic differences between MG1655 strains in different labs, presumably from accumulation of genomic mutations over time. For this reason, we chose to use MG1655 that could be traced to back to the Yale Coli Genetic Stock Center. We moved the csrD-kanR-mreBmsfGFP-mreCD operon from our previous MG1655 to MG1655 (CGSC #7740) using the lambda red method followed by selection for kanamycin resistance (18). Colonies were picked and screened using fluorescence microscopy and then sequenced.
Media conditions
Multiple media compositions were used for comparison of cell shape between fluorescently labeled and unlabeled MreB strains. Three medias were used: M media, Lysogeny broth (LB) with 5g NaCl per liter, and M63 with glucose and casamino acids (19, 20). All measurements of MreB polymers and cell shape were conducted in M63 media. Kanamycin sulfate (sigma) at 20 μg/mL was used in overnight cultures and plating but was not used in subcultures used for imaging.
Selection of mutants
Individual colonies of MreBmsfGFP were grown overnight. Cultures were spread the following day on LB plates containing 30 μg/mL kanamycin, 1.5 μg/mL cephalexin, and 10–35 μg A22. We find that 1.5 μg/mL cephalexin aids in the selection of A22 suppressors because cells that lose MreB function are hypersensitive to cephalexin. Individual colonies that grow on the plates were grown in liquid for imaging and mreB was sequenced to identify mutations. We then moved the csrD-kanR-mreBmsfGFP section of the operon containing the A22 resistance mreB mutations into the parental MG1655 strain (CGSC #7740) using the lambda red method followed by selection for kanamycin resistance (18). Fluorescence colonies were picked and sequenced to confirm the transfer of the mutations.
A22 resistance quantification
Each strain was grown in a 96 well plate in LB containing serial dilution of A22 ranging from 0 to 100 μg A22. Growth measurements were made on a BioTek (Winooski, VT) microplate reader to record the OD600 over 16 h of growth at 37°C. The A22 resistance of each MreB mutant was calculated by its IC50.
Imaging of mreB mutants
Strains were grown over night at 37°C in M63 media in the presence of kanamycin to prevent contamination. The next morning, cells were subcultured between 1:10000 and 1:30000 and grown until they reached exponential phase. When the OD600 of the culture was between 0.15 and 0.3, cells were imaged on 1% UltraPure Agarose (Life Technologies, Carlsbad, CA) pads made of M63 media supplemented with glucose and casamino acids. Imaging was conducted in a 20°C temperature-controlled room on a custom-built inverted wide-field fluorescent microscope with a 1.49NA 100× objective (Nikon, Tokyo, Japan). Images for 3D volumes were taken at 100 nm increments in stage position.
A22 treatment imaging
MreBmsfGFP strain was grown over night at 37°C in the presence of kanamycin to prevent contamination. The next morning, cells were subcultured between 1:10000 and 1:30000 in the presence of A22 at concentrations of 0 μg/mL, 0.125 μg/mL, 0.25 μg/mL, 0.5 μg/mL, 0.75 μg/mL, and 1 μg/mL A22. When OD600 of the culture was between 0.15 and 0.3, cells were imaged on 1% UltraPure Agarose (Life Technologies) pads made of M63 media containing the same concentration of A22 as the liquid media. Imaging was then conducted in the same manner as with the mreB mutants.
3D cell shape reconstruction
Cell shapes were measured by fitting 3D images of cells stained with FM 4-64 with an active mesh in MATLAB (The MathWorks, Natick, MA). An initial surface was found by fitting a series of active contours to axial slices of the cell (21). Convolving the surface with the 3D point-spread function (PSF) of our microscope creates a test image that can be compared directly with the image stack from the microscope. The surface was then iteratively deformed to minimize the square difference between the simulated image and the real image (22). The PSF was measured by averaging image stacks of multiple individual 0.1 μm TetraSpeck microspheres (1000–4000) imaged at 100 nm steps in the axial direction. The surface of the cell was used to calculate measurements of cell shape such as length, volume, and diameter. Radius was measured as the average distance from the surface to the centerline after the removal of the pole regions.
MreB polymer measurements
Polymers are measured by fitting the 3D MreB images to a set of polymers confined to the membrane. Using the surface determined by the membrane fitting, a two-dimensional (2D) unwrapped image of the MreB polymers is created. The 2D unwrap is used for intensity-based segmentation to determine the initial location, length, and orientation of the polymers on the surface. We do not distinguish between structures that are due to MreB polymers and those that may be MreB aggregates. Each polymer is modeled by a stiff active contour confined to the membrane and deformed to fit the 3D MreB image. The polymer positions are convolved with the PSF to create a simulated 3D MreB image. Once again, the polymers deform to minimize the square difference between the simulated image and the 3D image from the microscope. The 2D unwrap image is not used for fitting of the polymers. The length of the polymer is measured in 3D and the polymer angle is measured as the angle away from the longitudinal axis of the cell, with angles greater than 90° corresponding to a right-handed helical wrapping on the cell surface and angles less than 90° corresponding to a left-handed helix. This helical pitch angle is a measure of how the polymer is oriented relative to the cell’s long axis and is not the helical pitch between monomers in the filament structure.
The fraction of MreB bound to the membrane is estimated by comparing the raw MreB image of a cell with the FM4-64 image, where the fluorescence is uniformly distributed over the cell membrane, and a simulated image of the cell filled uniformly with fluorescence. A plot of average fluorescence intensity as a function of distance from the centerline is made for each of the MreB, membrane, and simulated filled fluorescent images. Poles are excluded from this analysis. The MreB plot is fit with a linear combination of the normalized membrane and filled plots. The fraction of MreB on the membrane is approximated as the percentage of MreB signal that can be attributed to the membrane distribution of fluorescence.
Correlation significance testing
We used Pearson correlation coefficients to analyze relationships between different data sets. To determine if correlations between two parameters were significant, we compared our data with a noise model in which both data sets were shuffled 10,000 times to create a distribution of correlation values. Correlations were significant if the values were within the 99.67th percentile of the noise distribution, which is equivalent to p < 0.05 after accounting for multiple hypothesis testing using the Bonferroni Correction with 15 tests. These 15 tests allow us to compare the data in one of the six parameters with the data in one of the five remaining parameters. The number is further reduced to 15 due to the symmetric nature of calculating the correlation coefficient.
Results
MreBmsfGFP is minimally perturbative
Previous live-cell fluorescence microscopy studies of MreB localization used fluorescent fusions of yellow fluorescent protein (YFP) to the N-terminus of MreB or of mCherry inserted internally at a nonconserved surface-exposed loop in the protein (23). Both of these strains suffered from physiological defects. The N-terminal YFP fusion, which manifests as a single large helical filament, does not complement a deletion of the MreB protein. On the other hand, the mCherry internal “sandwich” fusion yields multiple small fluorescence structures. Although this fusion rescues the viability of MreB deletion strains, it frequently results in significant cell shape defects suggesting that it too disrupts MreB function (Fig. S1 in the Supporting Material).
Because mCherry has recently been shown to stimulate aggregation when fused to proteins, we sought to find a better probe for live-cell studies by replacing the mCherry in the sandwich fusion with nine different fluorescent proteins, six of which have been shown to cause the least amount of aggregation (24). The majority of the fluorescent proteins tested were not able to restore rod shape in place of the sandwich fusion (Fig. S1). The fusion that generated the most native cell shape was MreBmsfGFP (monomeric-super-folder-GFP). This fluorescent fusion was then encoded on the chromosome at the native mreB locus under native regulation.
We used two criteria to determine the level of perturbation that tagging MreB with msfGFP would cause. First, the tagged strain should have the same growth rate as the untagged version when grown in different media. We grew the fluorescently tagged and untagged strains in three kinds of media: high-sucrose media (M-media), rich LB, and minimal media (M63) supplemented with casamino acids. MreBmsfGFP shows unperturbed growth rate in all media when compared with the unlabeled wild-type (Fig. S2). Both strains exhibited identical exponential growth doubling times in all media, with doubling times of 19 min in M-media, 17 min in LB, and 28 min in M63. Since we wished to study how MreB influences bacterial rod diameter, our second criterion was that the fluorescent fusion should minimally alter cell width. Using our custom cell-shape analysis software (see Materials and Methods), we found that MreBmsfGFP is ∼5% wider than the unlabeled parental strain and equally as rod-shaped (Fig. S3). The average diameters of the unlabeled and labeled cells are 893 ± 3 nm and 934 ± 6 nm, respectively (all values are reported as mean ± 80% confidence interval). Using these two criteria, we conclude that the MreBmsfGFP fluorescent fusion is minimally perturbative.
MreBmsfGFP forms ∼500 nm long polymers along the cell cylinder
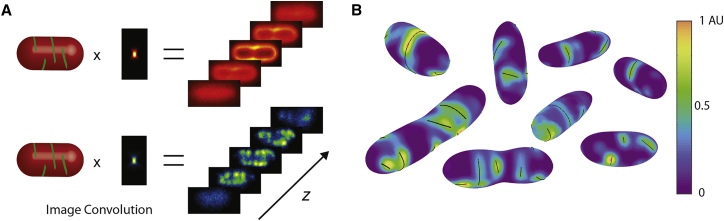
We next examined MreBmsfGFP and measured its polymeric properties. Combining 3D imaging with polymer detection and segmentation software developed in our lab, we were able to calculate the size and orientation of MreB structures with respect to the 3D cell surface. We used a previously developed forward convolution fitting method to estimate the 3D cell shape of cells stained with FM4-64 (10). Polymer detection was performed by fitting the 3D fluorescent images of MreBmsfGFP with semi-rigid segments confined to lie on the measured 3D surface of the cell (Fig. 1 A; Materials and Methods). This analysis revealed that MreB forms extended structures that are larger than the diffraction limit. In M63 media, we measured the average polymer length of MreBmsfGFP to be 500 ± 10 nm (Fig. 2 B). Examples of fits to both MreBmsfGFP structure and cell shape are shown in Fig. 1 B, with the surface color showing the fluorescence intensity of MreBmsfGFP at each point on the surface, and the detected polymers shown in black. We measured an average of seven polymers per cell and these polymers were mostly found in the cylindrical portion of the cell and excluded from the cell poles (Fig. S4). Although not explored here, the MreB polymers we observe are dynamic and move circumferentially over long-time scales as reported previously (7, 25, 26).
Figure 1.
Cell shape and polymer fitting method. (A) Diagram outlines the cell shape and polymer fitting algorithm. Cells expressing MreBmsfGFP under native regulation are membrane-stained with FM4-64 and imaged using 3D fluorescent microscopy. The imaging process can be written as a convolution between the point-spread function (PSF) of our microscope and the spatial distribution of fluorescent molecules, in this case the membrane. To estimate the shape of the surface, a model cell is convolved with the PSF to create a 3D simulated image. The surface is relaxed so that the simulated image best matches the experimental image. A similar process is used to fit the MreB polymers. Each polymer is modeled as a stiff elastic rod confined to the surface of the membrane. Again, a simulated 3D MreB image is created and model filaments relax to best match the experimental image. (B) Representative surface fits of cells expressing MreBmsfGFP are shown. The color of the surface is determined by interpolating the intensity of the 3D MreB image at the points of the surface. The detected polymers are shown in black. To see this figure in color, go online.
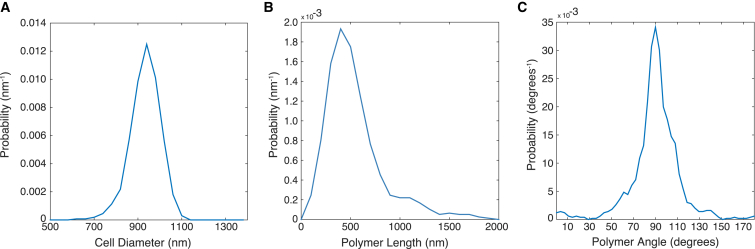
Figure 2.
Probability density functions of (A) cell diameter, (B) MreB polymer lengths, and (C) MreB monomer angles in E. coli expressing MreBmsfGFP. Data is collected from 459 cells, with an average of 7.3 polymers detected per cell. The distribution in (A) shows an average diameter of 934 ± 6 nm. The average MreB polymer length was measured as 500 ± 10 nm. The angle distribution of monomers in (C) is made by weighting the angle distribution of polymers by the length of each polymer, and had a mean angle of 91 ± 1°. All values are shown as mean ± 80% confidence intervals. To see this figure in color, go online.
Our analysis also enabled us to determine the helical pitch angle of MreB polymers relative to the cell’s long axis. Due to the effects of blurring by the microscope, pitch angle was only calculated for polymers with a length greater than 300 nm. On average, MreBmsfGFP polymers had a right-handed helical pitch angle of 91 ± 1°. The angle is measured relative to the long axis of the cell (Fig. 1 C), with angles above 90° indicating a right-handed pitch (p = 0.08). Interestingly, the handedness reported is opposite of the left-handed polymers we previously measured for E. coli using the noncomplementing N-terminal YFP fusion. Based on the functionality of this fusion, we have a higher confidence that this measurement more faithfully represents the normally unlabeled state of MreB and can be used as a basis for further study of MreB function.
Generating different cell shapes by perturbing MreB
Armed with tools for the 3D quantification of cell shape and MreB polymer conformation in the wild-type MreBmsfGFP strain, we sought to probe changes in bacterial cell shape due to the perturbation of MreB. Amino acid substitutions and the application of the MreB polymerization-inhibiting drug A22 at sublethal concentrations both result in cell shape changes that are directly linked to MreB. Although these treatments have been previously shown to cause changes in cell shape, no studies, to our knowledge, have examined the biophysical properties of MreB polymers in these conditions to determine the cause of the observed cell shape modification.
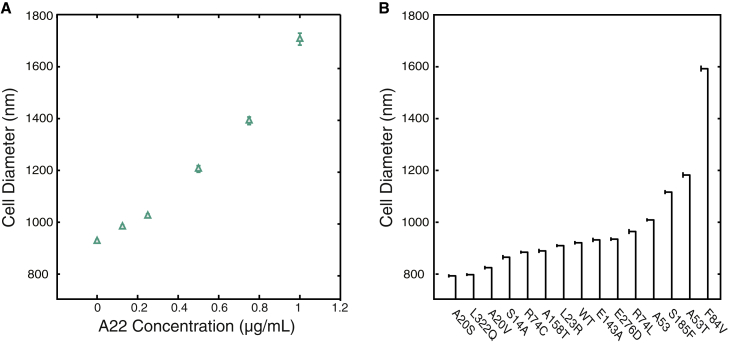
A22 has been shown to inhibit rod shape through a specific interaction with MreB (27, 28). Growing E. coli in the presence of different sublethal concentrations of A22 resulted in the growth of cells with varying steady-state cell diameters (Fig. 3 A). We used concentrations of A22 up to 1 μg/mL because higher concentrations lead to high lethality and potential disruption of nonspecific targets (29). Using this range of A22 concentrations, we reproducibly generated cell populations with stable steady-state diameters ranging from 934 ± 6 nm without the drug to 1700 ± 20 nm at 1 μg/mL. Each of these treatment conditions contain measurements ranging from 42 to 474 cells, with an average of 297 cells.
Figure 3.
Two independent methods to perturb cell diameter. (A) Cells expressing MreBmsfGFP were grown to steady state in different concentrations of the polymerization-inhibiting drug A22. All treatments are below the lethal concentrations of A22. As A22 concentration increases, cells significantly increase their diameter. (B) Cell diameter can also be changed with single-point mutants in the mreBmsfGFP. Fourteen mutants were generated with diameters both larger and smaller than the unmutated form (WT). Mutants are arranged in order of increasing diameter. Error bars represent 80% confidence intervals. To see this figure in color, go online.
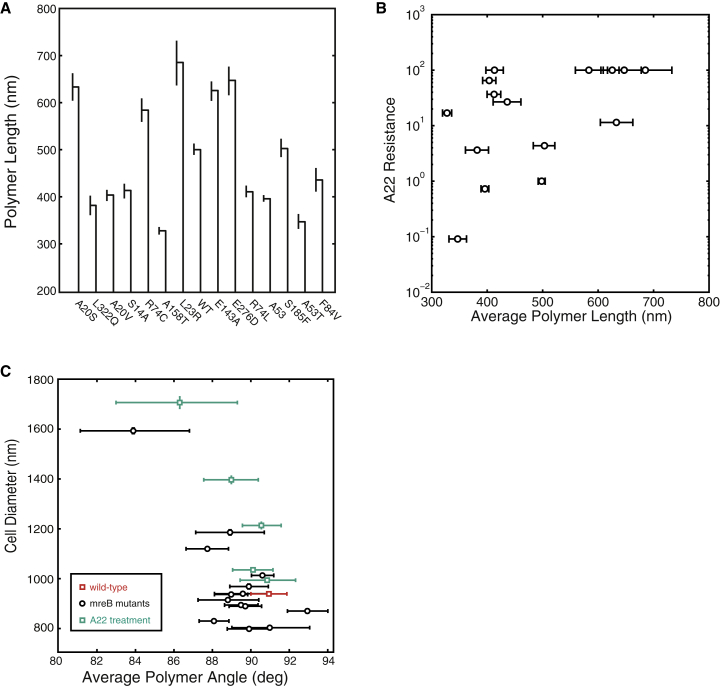
To generate MreB-dependent changes in cell shape genetically, we created a collection of mutations in mreBmsfGFP. The simplest way to generate such a collection of mutations is by selecting for A22-resistant suppressor mutations. Using this method, we isolated 12 different MreB amino-acid substitutions in MreBmsfGFP that showed varying levels of A22 resistance. The mutations were then moved to a fresh parental background to reduce the chances of second-site mutation effects. Importantly, the mutations were distributed across three of MreB’s four subdomains (IA, IB, and IIA; Fig. S5), indicating that the mutations likely perturb a range of MreB properties. We also used site-directed mutagenesis to generate A53T and the deletion of the 53rd amino acid (ΔA53) as this residue was previously shown to increase cell diameter depending on the amino acid substitution (30). We grew all strains in M63 media supplemented with casamino acids and measured between 64 and 1228 cells (478 on average) for each of the 14 different mutations. These mutations altered the average cell diameter compared with the wild-type MreBmsfGFP during steady-state growth (Fig. 3 B) generating cells ranging in diameter from 790 ± 30 nm, which is thinner than wild-type, to 1590 ± 60 nm. Depending on the specific amino acid substitution, the average MreB polymer length ranged between 328 ± 7 nm to 690 ± 50 nm (Fig. 4 A).
Figure 4.
MreB polymer measurements. (A) MreB mutations can alter the MreB polymer length. In some cases, the polymer length is increased with respect of wild-type and in other cases the polymer length is decreased. Mutations are arranged as in Fig. 3B, in order of increasing average cell diameter. Error bars represent 80% confidence intervals. (B) MreB polymer length appears to correlate with the level of A22 resistance of the MreB mutants. Mutations that have longer polymers have high levels of A22 resistance whereas increased sensitivity to A22 is seen in mutations that have shorter polymers. A22 resistance levels are shown as a fold changes from wild-type MreBmsfGFP, with levels capped at 100× that of wild-type since higher levels of A22 can effect proteins other than MreB. (C) The average polymer angle inversely correlates with the cell diameter in both mreB mutant data (black) and A22 treatment data (green). The wild-type mreB is shown in red. The cell diameter increases as the average helical pitch angle of MreB decreases. The handedness of the helical pitch changes in both data sets as 90° is crossed. To see this figure in color, go online.
MreB polymers in A22-treated cells and the effect of A22 on suppressor mutants
Although high concentrations of A22 abolish MreB polymers (28, 31), the effect of sublethal concentrations on MreB structure is not known. We measured MreB polymer length and the amount of MreB that appears to be on the surface, and we found that these quantities appeared to be inversely correlated with the concentration of A22 in wild-type cells when moderate amounts of the drug are used (Fig. S6). Lengths ranged from 500 ± 10 nm for untreated cells to 440 ± 10 nm for cells at our maximum treatment level of 1 μg/ml. As the concentration of A22 is increased, MreB becomes localized to the membrane and more fluorescence is found in the cytoplasm. These results are consistent with previous work showing that MreB bound to A22 weakens interprotofilament contacts, thus decreasing the stability of MreB polymers (27, 28).
We next examined A22 resistance and polymer length in each of the 14 mreB point mutants. A22 resistance positively correlated with polymer length, likely due to specific amino acid substitutions leading to polymer stabilization and a reduction in the turnover of MreB monomers (Fig. 4 B). It has been hypothesized that the ADP to A22 exchange happens in the post–ATP-hydrolysis monomeric state (32). This would stabilize MreB mutations that promote an ATP-bound conformation and would increase A22 resistance. The most A22-resistant mutant we isolated was E143A, which was previously proposed to be deficient for ATP hydrolysis (31). The cell diameter is 930 ± 40 nm, identical to that of the unmutated strain. This strain shows robust growth for A22 concentrations less than 100 μg/ml. Above this level, A22 begins to bind nonspecifically to other proteins, which increases lethality (29). Future work on the MreBE143A mutant could shed light on MreB function as it is currently thought that the turnover of MreB monomers is physiologically necessary (17).
Correlation analysis between MreB polymeric properties and cell morphology
Our fluorescence analysis of labeled cells yields a number of quantitative metrics of cell shape and MreB polymer conformation. These include cell width, cell length, cell volume, polymer number, polymer size, polymer helical pitch angle relative to the cell centerline, and the fraction of MreB that appears to reside on the inner membrane. We used a correlation analysis of these quantitative metrics to investigate which properties of MreB were most predictive of changes in cellular morphology (Fig. 5).
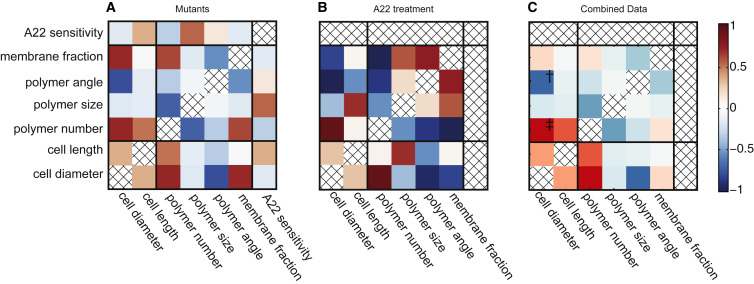
Figure 5.
Cell shape was modified using either point mutants in mreBmsfGFP or treatment with sublethal concentrations of A22. Maps of Pearson correlation coefficients between cell shape metrics and measured MreB properties were created for each set of conditions, for the (A) mutants (15 conditions), (B) A22 treatment (6 conditions), and (C) combined data sets (19 conditions). In (C), † indicates p = .023 and ‡ indicates p < .001, after accounting for multiple comparison. To see this figure in color, go online.
The two strongest correlations with cell diameter in both data sets are MreB helical pitch angle and MreB polymer number. To analyze the significance of correlations that are consistent between the two data sets, we combined the data from both treatment cases and recalculated the correlation (Fig. 4 C). The largest correlations were between cell diameter and MreB polymer angle (−0.69, p < .05) and between cell diameter and polymer number (0.76, p < 0.001). Both show significance after accounting for the effect of multiple comparisons.
MreB pitch angle is highly predictive of cell diameter for both A22 and mutant experiments
The correlation between MreB polymer number and cell diameter is likely due to the coupling between cell volume and number of MreB proteins. Cell volume is strongly correlated with cell diameter, and cells with more MreB monomers create more detectable polymers. This results in the significant correlation between polymer number and diameter. The correlation coefficients between MreB polymer number per cell and cell diameter are 0.70 and 0.92 for the mutant and A22 treatment data sets, respectively. We also observed a reduction in the total number of MreB polymers as polymer length increased, with an inverse correlation coefficient value of −0.71 and −0.64 for the mutant and A22 treatment data sets, respectively. This is expected as a larger percentage of MreB is present in the longer polymers, decreasing the total number of short polymers. Thus, polymer length and number correlations confirm previous findings that MreB assembly is important but these properties are not specifically informative with respect to cell diameter control.
In contrast, the unexpected inverse correlation between MreB pitch angle and cell width across all conditions tested yields new insight, to our knowledge, into shape control. We observed an inverse correlation between MreB helical pitch angle and cell diameter, with correlation coefficients of −0.78 and −0.95 for the mutant and A22 treatment data sets, respectively. The sign of this correlation indicates that a reduction in pitch angle leads to an increase in the cell width. Interestingly the average handedness of the polymers changed from the right-handed (93 ± 1°) to left-handed (84 ± 3°) crossing 90° as cells get wider. The increase in cell diameter with larger helical pitch angles is in agreement with theoretical work by Jian et al. (9), and the observed change in handedness is in agreement with previous work showing that the growth twist of E. coli switches handedness with increasing concentration of A22 (33).
A22-specific correlations
We also observed correlations specific to the A22 treatment. We measured an inverse correlation coefficient of −0.57 between MreB polymer length and cell diameter in the A22 data treatment, but the same correlation was insignificant in the mutants (see Materials and Methods for significance determination). The inconsistencies between the correlations from A22 treatment and the mutants are likely due to the mechanism by which A22 effects MreB polymers, namely reducing the concentration of MreB monomers competent for polymerization. In the mutant data set, we see that mutants with longer polymers are more resistant to A22 with a correlation coefficient of 0.62. Mutations that lead to longer polymers stabilize MreB in its polymeric form and counteract the effects of A22.
The correlation between the fraction of MreB that is membrane-bound and cell diameter shows opposite correlations in the two data sets. For the A22 treatment, there is an increase in the cytoplasmic portion of MreB as cell diameter increases, with a correlation coefficient of −0.80. This is consistent with previous findings that A22 increases the diffuse MreB monomer pool by preventing polymerization (27). In the MreB mutant data set, the reverse trend is seen and the membrane bound MreB fraction positively correlates with cell diameter with a value of 0.69. This contradiction suggests that the proportion of MreB localized on the membrane is not a direct contributor to the determination of cell diameter.
Discussion
MreB’s role in localizing the cell wall insertion machinery has been previously shown to be necessary for establishment and maintenance of rod shape. However, the mechanism via which rod-shaped bacteria establish specific diameters has remained unclear. Bacteria such as E. coli and B. subtilis exhibit a chiral growth twist that is determined by the organization of the PG cell wall. Before this study, we hypothesized that the helical pitch angle of MreB could help organize cell wall synthesis in this manner (13). Our simulations suggested that the chiral order of the peptidoglycan would primarily alter cell diameter. At that time, we had no way of making measurements of functional polymers, nor a way to alter the helical pitch angle of MreB to test this hypothesis. In this study, we addressed both of those limitations by generating a minimally perturbative MreB fluorescent fusion, using both A22 treatment and mutagenesis to alter MreB, and using automated 3D image analysis to quantify both polymer and cell shape characteristics. Our analysis of correlations between all biophysical parameters shows that cell diameter has a significant correlation with polymer angle. This correlation is robust to different types of treatments and has a correlation coefficient of −0.95 in the A22 data set and −0.78 in the mutants. Both treatments lead to similar cell diameters for a measured angle (Fig. 4 C), and a significant correlation is observed across data sets. The similarity across two independently derived sets of MreB perturbations supports the conclusion that the helical pitch angle of MreB is a key determinant of cell diameter and that alterations to MreB effect cell shape through the MreB helical conformation. Importantly, these results indicate that rather than only acting as a scaffold to cluster various aspects of the cell growth machinery, MreB forms extended polymers and the biophysical properties of MreB polymers dictate specific aspects of morphology.
We hypothesize that the mutations and A22 treatment alter the mechanics of the interaction between neighboring MreB monomers and/or between the MreB polymers and the cell membrane, resulting in a change in the angle of the helical fragments relative to the curvature of the cylindrical cell (15). In cells treated with A22, the drug interacts with nucleotide-bound MreB to prevent nucleotide hydrolysis and destabilize filaments (28, 32). A22 treatment and mutations perturbing the nucleotide-binding pocket of MreB have the potential to alter the geometry of the polymer as it has been shown that the angle between adjacent MreB monomers depends on the state of the bound nucleotide (34). In addition, mutations in the MreB-MreB binding surface or near the membrane-binding domain can directly affect the structure of the polymer and higher-order filaments. These types of structural changes have been shown to affect important parameters in the determination of the helical conformation of MreB, such as the stiffness, bending angles, and twisting angles (15, 35, 36, 37), which could result in polymers with different helical pitch angles inside the cell.
Though we have shown the role of MreB in determining steady-state diameter, bacteria also dynamically adjust their morphology in response to environmental and internal conditions, e.g., cells grown in minimal media grow into thinner rods. It will be interesting to see if there are similar principles connecting MreB and cell shape in these situations. In addition to MreB, there are a number of MreB-associated proteins involved in cell growth. How these proteins interact with MreB and influence its polymeric structure remains unclear. The division machinery also likely plays a role in the determination of cell diameter (38), and it may be possible to investigate this by performing experiments similar to those described here with the tubulin homolog FtsZ.
Acknowledgments
The authors thank Kerwyn Huang, Tom Silhavy, Ned Wingreen, and members of the Shaevitz and Gitai labs for helpful discussions. Funding for this work was provided by the NIH (T32GM00738840, RO1GM107384) and the NSF (PHY0844466, PHY1521553).
Editor: Julie Biteen
Footnotes
Nine figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30580-X.
Supporting Material
References
- 1.Viswanathan V.K. Sizing up microbes. Gut Microbes. 2012;3:483–484. doi: 10.4161/gmic.22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang F., Huang K.C. How and why cells grow as rods. BMC Biol. 2014;12:54. doi: 10.1186/s12915-014-0054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Höltje J.V. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scheffers D.J., Pinho M.G. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaevitz J.W., Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harb. Perspect. Biol. 2010;2:a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White C.L., Gober J.W. MreB: pilot or passenger of cell wall synthesis? Trends Microbiol. 2012;20:74–79. doi: 10.1016/j.tim.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 7.van Teeffelen S., Wang S., Gitai Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. USA. 2011;108:15822–15827. doi: 10.1073/pnas.1108999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendezú F.O., de Boer P.A. Conditional lethality, division defects, membrane involution, and endocytosis in mre and mrd shape mutants of Escherichia coli. J. Bacteriol. 2008;190:1792–1811. doi: 10.1128/JB.01322-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang H., Si F., Sun S.X. Mechanical control of bacterial cell shape. Biophys. J. 2011;101:327–335. doi: 10.1016/j.bpj.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ursell T.S., Nguyen J., Huang K.C. Rod-like bacterial shape is maintained by feedback between cell curvature and cytoskeletal localization. Proc. Natl. Acad. Sci. USA. 2014;111:E1025–E1034. doi: 10.1073/pnas.1317174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billings G., Ouzounov N., Huang K.C. De novo morphogenesis in L-forms via geometric control of cell growth. Mol. Microbiol. 2014;93:883–896. doi: 10.1111/mmi.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Y., Sun M., Shaevitz J.W. Direct measurement of cell wall stress stiffening and turgor pressure in live bacterial cells. Phys. Rev. Lett. 2011;107:158101. doi: 10.1103/PhysRevLett.107.158101. [DOI] [PubMed] [Google Scholar]
- 13.Wang S., Furchtgott L., Shaevitz J.W. Helical insertion of peptidoglycan produces chiral ordering of the bacterial cell wall. Proc. Natl. Acad. Sci. USA. 2012;109:E595–E604. doi: 10.1073/pnas.1117132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen L.T., Gumbart J.C., Jensen G.J. Coarse-grained simulations of bacterial cell wall growth reveal that local coordination alone can be sufficient to maintain rod shape. Proc. Natl. Acad. Sci. USA. 2015;112:E3689–E3698. doi: 10.1073/pnas.1504281112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang S., Wingreen N.S. Cell shape can mediate the spatial organization of the bacterial cytoskeleton. Biophys. J. 2013;104:541–552. doi: 10.1016/j.bpj.2012.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swulius M.T., Jensen G.J. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J. Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reimold C., Defeu Soufo H.J., Graumann P.L. Motion of variable-length MreB filaments at the bacterial cell membrane influences cell morphology. Mol. Biol. Cell. 2013;24:2340–2349. doi: 10.1091/mbc.E12-10-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joseleau-Petit D., Liébart J.C., D’Ari R. Unstable Escherichia coli L forms revisited: growth requires peptidoglycan synthesis. J. Bacteriol. 2007;189:6512–6520. doi: 10.1128/JB.00273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbing K., Brent R. Current Protocols in Molecular Biology. Wiley; Hoboken, NJ: 2002. Media preparation and bacteriological tools; p. 1.1. [DOI] [PubMed] [Google Scholar]
- 21.Kass M., Witkin A., Terzopoulos D. Snakes: active contour models. Int. J. Comput. Vis. 1987;1:321–331. [Google Scholar]
- 22.Helmuth, J. A., and I. F. Sbalzarini. 2009. Deconvolving active contours for fluorescence microscopy images. Proc. Advances in Visual Computing, Part. 1, 5th, Las Vegas, NV. 5875:544–553.
- 23.Bendezú F.O., Hale C.A., de Boer P.A.J. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landgraf D., Okumus B., Paulsson J. Segregation of molecules at cell division reveals native protein localization. Nat. Methods. 2012;9:480–482. doi: 10.1038/nmeth.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domínguez-Escobar J., Chastanet A., Carballido-López R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 26.Garner E.C., Bernard R., Mitchison T. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gitai Z., Dye N.A., Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 28.van den Ent F., Izoré T., Löwe J. Bacterial actin MreB forms antiparallel double filaments. eLife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takacs C.N., Poggio S., Jacobs-Wagner C. MreB drives de novo rod morphogenesis in Caulobacter crescentus via remodeling of the cell wall. J. Bacteriol. 2010;192:1671–1684. doi: 10.1128/JB.01311-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monds R.D., Lee T.K., Huang K.C. Systematic perturbation of cytoskeletal function reveals a linear scaling relationship between cell geometry and fitness. Cell Reports. 2014;9:1528–1537. doi: 10.1016/j.celrep.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dye N.A., Pincus Z., Theriot J.A. Mutations in the nucleotide binding pocket of MreB can alter cell curvature and polar morphology in Caulobacter. Mol. Microbiol. 2011;81:368–394. doi: 10.1111/j.1365-2958.2011.07698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bean G.J., Flickinger S.T., Amann K.J. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry. 2009;48:4852–4857. doi: 10.1021/bi900014d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tropini C., Lee T.K., Huang K.C. Principles of bacterial cell-size determination revealed by cell-wall synthesis perturbations. Cell Reports. 2014;9:1520–1527. doi: 10.1016/j.celrep.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Colavin A., Hsin J., Huang K.C. Effects of polymerization and nucleotide identity on the conformational dynamics of the bacterial actin homolog MreB. Proc. Natl. Acad. Sci. USA. 2014;111:3585–3590. doi: 10.1073/pnas.1317061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews S.S., Arkin A.P. A mechanical explanation for cytoskeletal rings and helices in bacteria. Biophys. J. 2007;93:1872–1884. doi: 10.1529/biophysj.106.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soufo H.J., Graumann P.L. Bacillus subtilis MreB paralogues have different filament architectures and lead to shape remodelling of a heterologous cell system. Mol. Microbiol. 2010;78:1145–1158. doi: 10.1111/j.1365-2958.2010.07395.x. [DOI] [PubMed] [Google Scholar]
- 37.Kruse T., Møller-Jensen J., Gerdes K. Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J. 2003;22:5283–5292. doi: 10.1093/emboj/cdg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aaron M., Charbon G., Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 2007;64:938–952. doi: 10.1111/j.1365-2958.2007.05720.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.