Abstract
Flavobacterium johnsoniae, a rod-shaped bacterium, glides over surfaces at speeds of ∼2 μm/s. The propulsion of a cell-surface adhesin, SprB, is known to enable gliding. We used cephalexin to generate elongated cells with irregular shapes and followed their displacement in three dimensions. These cells rolled about their long axes as they moved forward, following a right-handed trajectory. We coated gold nanoparticles with an SprB antibody and tracked them in three dimensions in an evanescent field where the nanoparticles appeared brighter when they were closer to the glass. The nanoparticles followed a right-handed spiral trajectory on the surface of the cell. Thus, if SprB were to adhere to the glass rather than to a nanoparticle, the cell would move forward along a right-handed trajectory, as observed, but in a direction opposite to that of the nanoparticle.
Introduction
Bacteria that swim are driven forward by helical filaments that rotate like propellers. The number and location of filaments vary among different bacteria, yet the core mechanism remains the same. Motile but nonswimming bacteria do not have propellers, yet they achieve self-propulsion over surfaces. Such movement is divided into two categories: 1) twitching and 2) gliding. Twitching involves the extension and retraction of type IV pili (1, 2, 3), but gliding bacteria do not use type IV pili (4, 5, 6, 7). Although the mechanism for gliding is barely understood, we know that Flavobacterium johnsoniae, which exhibits some of the fastest gliding of all known bacteria, has a powerful rotary motor (8) and a mobile cell-surface adhesin, SprB (9, 10, 11). Proteins of the type IX secretion system, which are abundant in gliding bacteria related to Flavobacterium, are required for secretion of SprB to the cell surface (12, 13, 14).
Materials and Methods
Generation of elongated cells
F. johnsoniae cells lacking RemA were grown overnight at 25°C in motility medium (MM; per liter: 1.1 g Casitone, 0.55 g yeast extract, 1.1 mM Tris (pH 7.5)) with shaking at 50 rpm. The cells were inoculated in fresh MM and grown with shaking at 50 rpm at 25°C in MM with 75 μM cephalexin, a drug that inhibits bacterial cell division. After 6 h, cells were collected, washed once with fresh MM, and then used for further analysis.
Imaging and analysis of the motion of a pole near a bend
Elongated cells were poured into a tunnel prepared by mounting a coverslip on a slide with two strips of double-stick Scotch tape, washed once with MM, and imaged. For two-dimensional tracking, a phase-contrast microscope with a Nikon Plan 40x BM NA 0.65 objective (Nikon, Melville, NY) and a Thorlabs DCC1545M-GL camera (Thorlabs, Newton, NJ) was used. Images were analyzed using a custom MATLAB (The MathWorks, Natick, MA) code. Cells with bends near one pole on either end were selected for analysis. The image of one cell was projected on the x-y plane and two lines (AB and BC) were fit on one cell (Fig. S2). The distance of the pole near the bend from the long axis of the cell was measured. The power density spectrum was calculated from the Fourier transform of the signal in Fig. 1 B. The peak of the power density spectrum was recorded as the spatial frequency, the inverse of which was the spatial period.
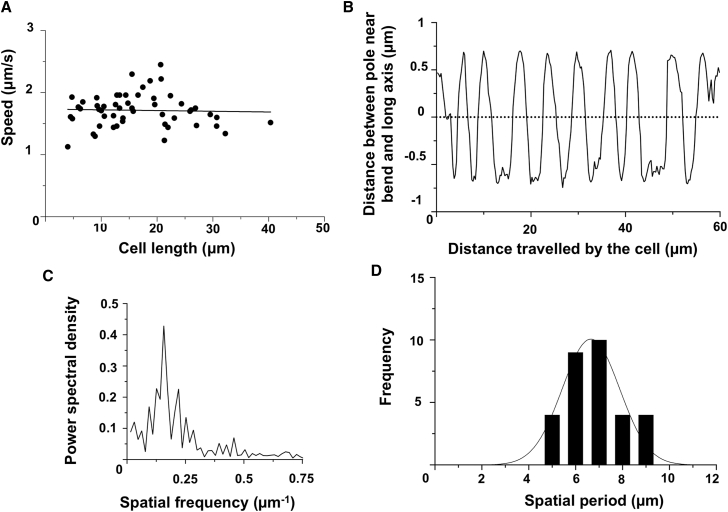
Figure 1.
Cells move sideways as they glide forward. (A) The gliding speed of 53 elongated cells was independent of cell length. Speed was plotted as dots and a linear regression was fit to the data. The coefficient of correlation was 0.03. (B) The distance of the pole near the bend of one cell, shown in Movie S1, from the long axis changed periodically with the distance traveled by the cell along its long axis. This distance was calculated using the method drawn in Fig. S2. (C) The spatial frequency of the displacement of the cell in Movie S1 moving along its long axis was 0.15 μm−1 and the spatial period was 6.6 μm. (D) Frequency distribution of the spatial periods for 31 cells. The average spatial period was 6.6 ± 1.2 μm.
To track bacteria in three dimensions, a microscope (Nikon Eclipse Ti-U) with an Apo TIRF 60x 1.49 numerical aperture (NA) objective was used in bright-field mode, with illumination by a tungsten lamp and recording by the Thorlabs camera described above. The high NA of the objective provides a shallow depth of field. Data were analyzed using a custom MATLAB code that fits a spline along the length of the cell and computes the total length of that spline. These data were used to test the model shown in Fig. 2.
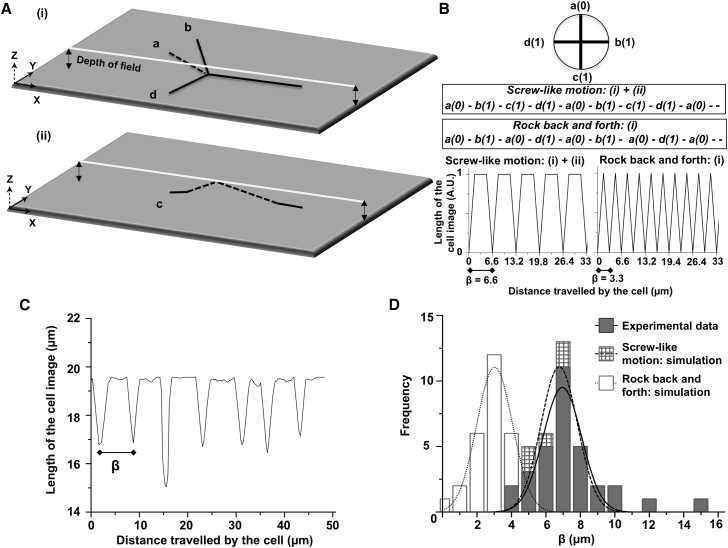
Figure 2.
Three-dimensional tracking reveals a screw-like mechanism. (A) Diagrammatic representation of a model to test whether a cell rolls like a screw or rocks back and forth. Conformations a, b, c, and d of a bent cell are depicted by black lines. Conformations b and d represent the image of the bent end of the cell when that segment lies in the microscope’s image plane (z = 0). The dashed line in (i) represents the image of the bent end of the cell when that segment is above the image plane (z > 0). The part of this segment that extends beyond the edge of the depth of field (represented by the white line) is not visible using the microscope with the high NA objective described in Materials and Methods. The dashed line in (ii) shows the part of the cell that is raised above the image plane by the cell rotation but remains within the depth of field. (B) Binary logic for the model. As a result of the decrease in the overall length of the cell image, a = 0, and b, c, and d (in which the imaged lengths are the same) = 1. Sequences for screw-like motion and rocking back and forth are boxed and the outputs for both scenarios are plotted. The periodicities for these plots are β = 6.6 and 3.3 μm, respectively. (C) Actual imaged cell length plotted as a function of distance traveled by a cell. This curve is similar to that obtained for the simulation of a cell rolling like a screw. (D) The frequency distribution of 32 β-values has a peak of 6.95 ± 1.11 μm, which is similar to the simulated frequency distribution of 32 β-values calculated from five cells that rolled like a screw while gliding forward.
Attachment of gold nanoparticles to SprB
We dissolved 2.5 μL of 20 mM freshly prepared succinimidyl 6-(3′-(2-pyridothio)propionamido)hexanoate) (#21651; Thermo Scientific, Waltham, MA) in dimethyl sulfoxide and added it to 10 μL of 1:10 diluted purified anti-SprB antibody (9). The mixture was incubated at 25°C for 30 min. After incubation, 2.5 μL of the preparation was added to 500 μL of 100 nm gold colloid (#EM.GC 100; BBI Solutions, Cardiff, UK), giving a final antibody dilution of 1/2000, and the mixture was incubated at 25°C for 2 h. After incubation, 10 μL of 1 mM O-[2-(3-mercaptopropionylamino)ethyl]-O′-methylpolyethylene glycol (#11124; Sigma-Aldrich, St. Louis, MO) was added and the preparation was incubated at 25°C for 12 h. After incubation, 100 μL of the solution was centrifuged at 1100 × g for 3 min. The supernatant fraction was discarded. Then, 40 μL of 1:10 elongated cells of an F. johnsoniae ΔremA strain was added to the pellet and mixed gently by pipetting. After incubation for 10 min, the preparation was imaged.
Imaging and analysis of gold nanoparticle motion
F. johnsoniae cells with attached nanoparticles were poured into a tunnel slide, incubated for 5 min, washed once with MM, and imaged. Some F. johnsoniae cells stuck to the glass surface by more than one adhesin and glided less vigorously than other cells. Since the cells were elongated, most of them had a slight bend along the cell body. Only cells that were fixed to the surface in a manner that prevented the shape of the bend from changing were selected for further analysis.
An evanescent wave was used to illuminate the gold nanoparticles. The experimental setup was as described by Yuan and Berg (15) except for two modifications: 1) a glass tunnel slide (described above) was used instead of a flow cell, and 2) the angle of the incident light from the 655-nm diode laser was changed so that total internal reflection occurred at the glass-water interface at the bottom of the tunnel slide, rather than at the glass-air interface at the top of the tunnel slide. The intensity of light scattered from nanoparticles 0, 1, or 2 cell diameters from the glass surface provided a calibration of the distance of particles from the glass surface (and the penetration depth of the evanescent field; Fig. S4). The nanoparticles that bound to cells stuck to the glass slide were imaged using the same setup. With this setup, the nanoparticles appeared brighter than the cell bodies (Movie S3). An image of a cell with a nanoparticle was projected in the x-y plane. The position of the nanoparticle was calculated using an ImageJ plugin, ParticleTracker. Using a custom MATLAB code, the position of the nanoparticle was measured relative to that of the cell.
To calculate movement along the z axis, the microscope was calibrated using three different nanoparticles at known positions along the z axis: sitting on the glass substrate, sitting on top of one cell, or sitting on top of two stacked cells (Fig. S4 A). A Gaussian was fit in three-dimensional space using a custom MATLAB code on the images. Essentially, the x and y coordinates corresponded to the image pixels, and the z coordinate was determined from the peak intensity. These values were used to plot the curve in Fig. S4 B. Using the curve in Fig. S4 B, the z-axis position of a nanoparticle on a cell was interpolated.
Results
To further our understanding of gliding, we used cephalexin to generate elongated cells, some of which were bent. The bend provided a marker to determine anisotropy in the movement of an otherwise cylindrically symmetric cell. Such cells were tracked in three-dimensional space, which showed that they moved forward by a screw-like mechanism. When viewed in a two-dimensional projection, SprB exhibits periodic motion (11). By attaching a gold nanoparticle to SprB, we were able to track its motion in three-dimensional space. Our results showed that SprB’s trajectory was a continuous right-handed spiral on the surface of the cell. When projected on an x-y plane, the frequency of the lateral displacement of SprB was about the same as that of the sideways movement of one pole of the cell. SprB’s motion and its adhesion to a surface resulted in rolling of a cell and gliding along its long axis. Our results show that a rod-shaped gliding bacterium works as a self-propelled screw, with SprB moving along its external threads.
Some long cells were bent, revealing periodic motion
Normally, cells of F. johnsoniae are ∼6 μm long (8). Elongated cells that continued to glide were generated by treatment with cephalexin, a technique that was demonstrated earlier by Kempf and McBride (16). Cephalexin inhibits septation, but not growth (17). Cell length, recorded after growth in cephalexin to OD600 = 0.4, ranged from 6 to 42 μm, and a large number of cells were about twice as long as normal cells (Fig. S1 in the Supporting Material). The gliding speeds of such cells were independent of cell length (Fig. 1 A). Cells with a bend near one pole were selected and the position of that pole relative to the long axis of the cell was measured. Fig. S2 shows the image of such a cell projected on the x-y plane. Two lines, AB and BC, were fit on the image. One end of line AB (labeled with a black dot in Fig. S2) was used to determine the distance between that pole and the long axis of the cell (line BC). For the cell recorded in Movie S1, the distance of the pole near the bend from the long axis of the cell changed periodically with the distance traveled by the cell along its long axis (Fig. 1 B). The spatial frequency for movement of this pole was 0.15 μm−1 and the spatial period was 6.6 μm (Fig. 1 C). A similar periodicity was observed for a population of 31 cells with a bend near one pole. The average spatial period for the population was 6.6 ± 1.2 μm (Fig. 1 D).
Cell tracking in three-dimensional space revealed a screw-like mechanism for gliding
Tracking a gliding cell when its image was projected on a two-dimensional plane did not resolve whether a cell was turning like a screw while gliding forward or it was rocking back and forth sideways. To solve this problem, a model (Fig. 2, A and B) was developed. Briefly, a cell with a bend, turning like a screw while gliding forward, would cycle sequentially through four distinct conformations (a, b, c, and d) in a pattern that would be a..b..c..d..a..b..c..d… and so on. On the contrary, in a scenario where a similar cell would rock back and forth while gliding forward, three conformations (a, b, and d) would be observed. The sequence of the three conformations in this scenario would be a..b..a..d..a..b..a..d… and so on. When imaged with a microscope with a small depth of field, cells in conformation a would appear shorter than those in conformations b, c, and d (Fig. 2 A). Binary logic was assigned to this model, with cells in conformation a assigned 0 and cells in conformations b, c, and d assigned 1. The output of this model when simulated (Fig. 2 B) showed two distinct curves. From the simulation, if the cell turned like a screw while gliding forward, the periodicity of the image-length plot, β, would be 6.6 μm. In the alternate scenario, if the cell rocked back and forth while gliding forward, β would be 3.3 μm. The observed image length (Fig. 2 C; Movie S2) varied in a manner that would be expected for the scenario in which cells turned like a screw while gliding forward. Similar patterns were observed for four more cells. The frequency distribution for β calculated from all five cells with 32 values of β had a mean of 6.95 ± 1.11 μm (Figs. 2 D and S3). This was similar to the simulated β for the scenario in which cells turned like a screw.
SprB moves along a spiral track
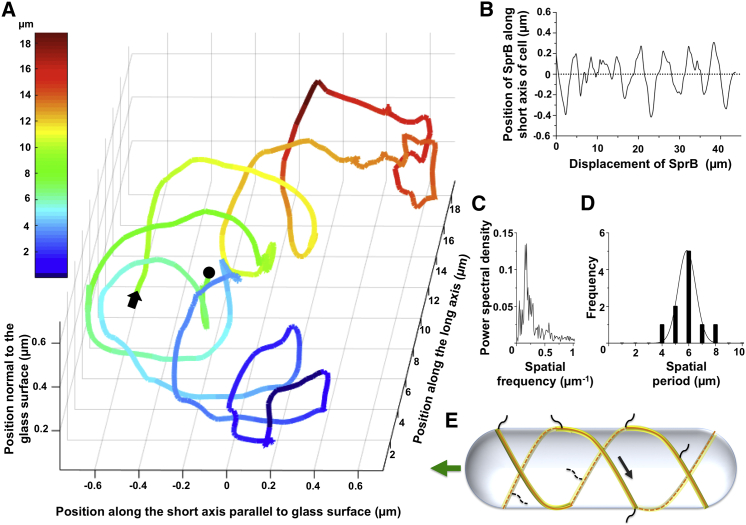
We used gold nanoparticles coated with anti-SprB antibodies as a marker for SprB. Since these antibodies can cross-react with RemA, a smaller SprB homolog (10), we used cells lacking RemA. The nanoparticles coated with anti-SprB moved spirally along the surface of such cells (Movie S3). Nanoparticles did not move on the surface of cells lacking both SprB and RemA (Movie S5). To understand the three-dimensional motion of SprB on the surface of a cylindrical cell, the cell was illuminated with an evanescent wave produced by a red laser beam internally reflected at the glass-water interface. This enabled us to use the particle intensity as a measure of distance from the glass surface. The intensities of gold nanoparticles at the surface of the glass, on top of one cell, or on top of two cells stacked together were found to decrease exponentially with distance between the nanoparticle and the glass surface (Fig. S4). This was expected from the known exponential dependence of the intensity of an evanescent wave with distance, with a decay length (penetration depth) that depends upon the angle of incidence between the laser beam and the glass-water interface (18). As is evident from the three-dimensional tracks, SprB moved along a right-handed spiral trajectory (Figs. 3 A and S6; Movies S3 and S4). The image of the cell was projected on the x-y plane and the long axis of the cell was aligned along the x axis. The distance traveled by the nanoparticle along the short axis of the cell was measured and plotted with the distance traveled along its long axis, the result of which was a periodic wave (Fig. 3 B). The spatial frequency for motion of one nanoparticle was 0.165 μm−1 and the spatial period was 6.06 μm (Fig. 3 C). A similar periodicity was observed for a population. The average spatial period of nanoparticle motion for a population of 10 cells was 5.9 ± 0.7 μm (Fig. 3 D). The spatial period for the motion of SprB is in the same range as the spatial period with which the cell rolls (Figs. 1 D and 2 D). The model shown in Fig. 3 E explains the geometry of a gliding cell. SprB moves along an external thread of a right-handed screw. Assuming that the thread is attached to the rigid framework of the cell wall (the peptidoglycan layer) and SprB can attach to the external glass surface, the thread can move relative to SprB, driving the screw-like motion of the cell.
Figure 3.
SprB moves along a spiral trajectory. (A) A three-dimensional track of SprB appears as an irregular right-handed spiral. An arrow and a dot indicate the beginning and end of the track, respectively. The data shown are from cell 1 in Movie S3. Similar tracks were visualized for cells 2 and 3 from Movie S3 and are shown in Fig. S5. The color of the track represents the position of SprB along the long axis of the cell. (B) The position of one gold nanoparticle attached to SprB (cell 1, Movie S3) changed periodically along the short axis of the cell as the nanoparticle moved along the long axis of the cell. (C) The spatial frequency of movement of one gold nanoparticle (cell 1, Movie S3) along the short axis of one cell was 0.165 μm−1 and the spatial period was 6.06 μm. (D) Frequency distribution of the spatial periods for 10 cells. The average spatial period was 5.9 ± 0.7 μm. (E) A screw-like mechanism for gliding: a track (orange) remains fixed relative to the cell. SprB (black) and a tread (yellow) are in motion relative to the cell surface. The arrow shows the right-handed motion of the SprB and tread. Interaction of SprB with an external substratum results in forward motion and right-handed rotation of the cell body.
Discussion
By working with elongated cells of irregular shape and including displacements in three-dimensional space, we have shown that the adhesin SprB moves along a right-handed spiral on the surface of a cell. When SprB binds to the substrate, the cell rolls to the right as it moves forward, as a right-handed screw would move into a wall when driven clockwise. SprB provides the adhesive, and rotary motors power the motion (8). As is evident from experiments or models presented by us and others (4, 5, 9, 10, 19, 20, 21, 22), mobile cell-surface components are essential for gliding. The biochemical features of spiral tracks on which such components are loaded and their mode of integration with the rotary motor are not known. One possible model that we previously proposed (4) suggests that similar to the way a bicycle chain is driven linearly by a rotating pedal, a rotary motor drives the linear motion of a tread that moves along a track fixed to the rigid framework of the cell. While the track is fixed onto the peptidoglycan layer, SprB and the tread are in motion relative to the cell surface. Our results show that the track from our model acts as the external thread of a screw on which a mobile tread carrying SprB is loaded. The ∼6 μm periodicity suggests that a track completes one revolution on a cell of normal size. This wavelength is maintained when cells lengthen in response to treatment with cephalexin. In order to glide for long distances over an external surface such as glass, once SprB reaches one pole, it needs to detach, and another SprB filament that is moving in the same direction needs to attach to the external surface.
Myxococcus xanthus is another rod-shaped gliding bacterium that is extensively studied. M. xanthus (phylum Proteobacteria) and F. johnsoniae (phylum Bacteroidetes) are phylogenetically far apart and they do not share similarity in the sequence of genes that are essential for gliding. A simple observation of gliding of both bacteria suggests that these bacteria might move by similar mechanisms. Yet, genetic evidence suggests that there are clear differences in their modes of gliding. Two models have been proposed to explain the gliding of M. xanthus: the focal adhesion model (23, 24) and the helical rotor model (6). In the focal adhesion model, multiprotein complexes extend from the cytoplasm to the outer membrane of a cell and attach to an external surface via focal adhesions. When these complexes are driven down the length of a cell by rotation of a cytoskeletal helix, the cell glides. In the helical rotor model, the multiprotein complexes that extend from the cytoplasm to the outer membrane do not penetrate the membrane, but they do perturb its shape. When this perturbation is driven down the length of the cell, viscous coupling to the external surface causes gliding. The mechanism in F. johnsoniae is similar to the focal adhesion model, with SprB filaments acting as focal adhesions. However, they are attached to the cell surface without having to reach from the cytoplasm to the outer membrane, and this reaching is accomplished by the rotary motors. Rotary motors similar to those in F. johnsoniae have not been observed in M. xanthus. There are obvious differences between the gliding mechanisms of F. johnsoniae and M. xanthus. F. johnsoniae is 60× faster than M. xanthus and has mobile cell-surface adhesins that are needed for attachment to an external surface. Motion of one such adhesin, SprB, enables movement of the cell. Motion of SprB is powered by a rotary motor: cells tethered to a glass surface rotate at the speed of 1–3 Hz. Presumably, linear motion of SprB is achieved by coupling the rotary motor to a tread. Interaction of SprB with an external surface results in a screw-like movement of the cell. For F. johnsoniae, a bacterium that prefers to move over and colonize external surfaces, designing a mobile and adhesive external thread and placing it such that it moves spirally on the surface of the cell appears to be the mechanism for forward motion.
Author Contributions
A.S. and H.C.B. designed the research and wrote the manuscript. A.S. performed the experiments, developed the model, and analyzed the data. A.S. and T.R. developed data analysis scripts.
Acknowledgments
We thank Pushkar P. Lele for providing a software script.
This research was supported by National Institutes of Health grant AI016478.
Editor: Kazuhiro Oiwa.
Footnotes
Six figures and five movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30654-3.
Contributor Information
Abhishek Shrivastava, Email: ashrivastava@fas.harvard.edu.
Howard C. Berg, Email: hberg@mcb.harvard.edu.
Supporting Material
In a two-dimensional plane, the cell appears to move sideways as it glides forward.
When the cell is in conformation a (Fig. 2 A), the imaged cell length is shorter. While gliding forward, the cell rolls along a right-handed spiral.
Three cells (numbered 1, 2, and 3) are shown. Data for cell 1 are shown in Fig. 3, A–C. Three-dimensional tracks for cells 2 and 3 are shown in Fig. S5.
References
- 1.Skerker J.M., Berg H.C. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedrich C., Bulyha I., Søgaard-Andersen L. Outside-in assembly pathway of the type IV pilus system in Myxococcus xanthus. J. Bacteriol. 2014;196:378–390. doi: 10.1128/JB.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biais N., Higashi D.L., Sheetz M.P. Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc. Natl. Acad. Sci. USA. 2010;107:11358–11363. doi: 10.1073/pnas.0911328107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrivastava A., Berg H.C. Towards a model for Flavobacterium gliding. Curr. Opin. Microbiol. 2015;28:93–97. doi: 10.1016/j.mib.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islam S.T., Mignot T. The mysterious nature of bacterial surface (gliding) motility: a focal adhesion-based mechanism in Myxococcus xanthus. Semin. Cell Dev. Biol. 2015;46:143–154. doi: 10.1016/j.semcdb.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 6.Nan B., Chen J., Zusman D.R. Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc. Natl. Acad. Sci. USA. 2011;108:2498–2503. doi: 10.1073/pnas.1018556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyata M., Hamaguchi T. Prospects for the gliding mechanism of Mycoplasma mobile. Curr. Opin. Microbiol. 2016;29:15–21. doi: 10.1016/j.mib.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Shrivastava A., Lele P.P., Berg H.C. A rotary motor drives Flavobacterium gliding. Curr. Biol. 2015;25:338–341. doi: 10.1016/j.cub.2014.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson S.S., Bollampalli S., McBride M.J. SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J. Bacteriol. 2008;190:2851–2857. doi: 10.1128/JB.01904-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrivastava A., Rhodes R.G., McBride M.J. Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J. Bacteriol. 2012;194:3678–3688. doi: 10.1128/JB.00588-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakane D., Sato K., Nakayama K. Helical flow of surface protein required for bacterial gliding motility. Proc. Natl. Acad. Sci. USA. 2013;110:11145–11150. doi: 10.1073/pnas.1219753110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes R.G., Samarasam M.N., McBride M.J. Flavobacterium johnsoniae gldN and gldO are partially redundant genes required for gliding motility and surface localization of SprB. J. Bacteriol. 2010;192:1201–1211. doi: 10.1128/JB.01495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrivastava A., Johnston J.J., McBride M.J. Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J. Bacteriol. 2013;195:3201–3212. doi: 10.1128/JB.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride M.J., Zhu Y. Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J. Bacteriol. 2013;195:270–278. doi: 10.1128/JB.01962-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan J., Berg H.C. Resurrection of the flagellar rotary motor near zero load. Proc. Natl. Acad. Sci. USA. 2008;105:1182–1185. doi: 10.1073/pnas.0711539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kempf M.J., McBride M.J. Transposon insertions in the Flavobacterium johnsoniae ftsX gene disrupt gliding motility and cell division. J. Bacteriol. 2000;182:1671–1679. doi: 10.1128/jb.182.6.1671-1679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishihara A., Segall J.E., Berg H.C. Coordination of flagella on filamentous cells of Escherichia coli. J. Bacteriol. 1983;155:228–237. doi: 10.1128/jb.155.1.228-237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fish K.N. Total internal reflection fluorescence (TIRF) microscopy. Curr. Protoc. Cytom. 2009;Chapter 12 doi: 10.1002/0471142956.cy1218s50. Unit12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapidus I.R., Berg H.C. Gliding motility of Cytophaga sp. strain U67. J. Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nan B., McBride M.J., Oster G. Bacteria that glide with helical tracks. Curr. Biol. 2014;24:R169–R173. doi: 10.1016/j.cub.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun M., Wartel M., Mignot T. Motor-driven intracellular transport powers bacterial gliding motility. Proc. Natl. Acad. Sci. USA. 2011;108:7559–7564. doi: 10.1073/pnas.1101101108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ridgway H.F., Lewin R.A. Characterization of gliding motility in Flexibacter polymorphus. Cell Motil. Cytoskeleton. 1988;11:46–63. doi: 10.1002/cm.970110106. [DOI] [PubMed] [Google Scholar]
- 23.Balagam R., Litwin D.B., Igoshin O.A. Myxococcus xanthus gliding motors are elastically coupled to the substrate as predicted by the focal adhesion model of gliding motility. PLOS Comput. Biol. 2014;10:e1003619. doi: 10.1371/journal.pcbi.1003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignot T., Shaevitz J.W., Zusman D.R. Evidence that focal adhesion complexes power bacterial gliding motility. Science. 2007;315:853–856. doi: 10.1126/science.1137223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In a two-dimensional plane, the cell appears to move sideways as it glides forward.
When the cell is in conformation a (Fig. 2 A), the imaged cell length is shorter. While gliding forward, the cell rolls along a right-handed spiral.
Three cells (numbered 1, 2, and 3) are shown. Data for cell 1 are shown in Fig. 3, A–C. Three-dimensional tracks for cells 2 and 3 are shown in Fig. S5.