Abstract
The aggregation cascade and peptide-membrane interactions of the amyloid β-peptide (Aβ) have been implicated as toxic events in the development and progression of Alzheimer’s disease. Aβ42 forms oligomers and ultimately plaques, and it has been hypothesized that these oligomeric species are the main toxic species contributing to neuronal cell death. To better understand oligomerization events and subsequent oligomer-membrane interactions of Aβ42, we performed atomistic molecular-dynamics (MD) simulations to characterize both interpeptide interactions and perturbation of model membranes by the peptides. MD simulations were utilized to first show the formation of a tetramer unit by four separate Aβ42 peptides. Aβ42 tetramers adopted an oblate ellipsoid shape and showed a significant increase in β-strand formation in the final tetramer unit relative to the monomers, indicative of on-pathway events for fibril formation. The Aβ42 tetramer unit that formed in the initial simulations was used in subsequent MD simulations in the presence of a pure POPC or cholesterol-rich raft model membrane. Tetramer-membrane simulations resulted in elongation of the tetramer in the presence of both model membranes, with tetramer-raft interactions giving rise to the rearrangement of key hydrophobic regions in the tetramer and the formation of a more rod-like structure indicative of a fibril-seeding aggregate. Membrane perturbation by the tetramer was manifested in the form of more ordered, rigid membranes, with the pure POPC being affected to a greater extent than the raft membrane. These results provide critical atomistic insight into the aggregation pathway of Aβ42 and a putative toxic mechanism in the pathogenesis of Alzheimer’s disease.
Introduction
Aggregation of proteins into amyloid deposits is a common feature of dozens of diseases (1). Alzheimer’s disease (AD) is an amyloid disease that currently affects 5.3 million people in the United States, with no current treatment to stop or slow the progression of the disease (2). AD toxicity is associated with the aggregation and accumulation of amyloid β-peptide (Aβ) in and around neural tissue (3, 4). Aβ is generated by sequential proteolysis by β- and γ-secretase within the amyloid precursor protein (APP) (4, 5, 6), and variations in the processivity of γ-secretase give rise to multiple Aβ alloforms of different lengths (7, 8). The Aβ42 alloform aggregates faster (9, 10) and is more cytotoxic than the shorter alloforms (11). Recent evidence suggests that low-molecular-weight oligomers of Aβ42 share common structures and possible mechanisms of pathogenesis, and are the most toxic agents contributing to AD (12, 13, 14, 15). In addition, neuronal cell membrane perturbation by Aβ is central to the pathology of AD (16). Understanding the mechanistic details related to the formation of low-molecular-weight oligomers and their role in membrane perturbation leading to neuronal cell death is essential for assessing the cascade of events that result in AD.
An unambiguous definition of the most cytotoxic low-molecular-weight Aβ oligomer in terms of structure and size remains elusive (17, 18). Some argue that it is not necessarily the size of the Aβ oligomer that correlates with the highest toxicity, but rather the structure and organization of the peptide. These structural states are assessed based on secondary-structure elements retained by the peptide, the density of the oligomer, and the order of events leading to fibrillization (19). The structural transition from the primarily α-helical monomer of Aβ after γ-secretase cleavage to a disordered ensemble in solution, and subsequent enrichment in β-strand structure (20) are indicative of on-pathway aggregation (21, 22) and suggest an important role for the Aβ monomer structure in oligomer and fibril formation (9, 23). Events leading to the aggregation of multiple Aβ monomers into oligomeric forms can vary (24, 25). With regard to the structural makeup and size of toxic, oligomer Aβ species, multiple pathways are possible and the structural diversity of Aβ oligomers drives the aggregation pathway (19, 26, 27). Multiple mechanisms of toxicity may exist for different Aβ oligomers and each may be related to different conformations adopted by the polymorphic Aβ (27). A structural characterization of these various oligomer species would provide necessary information about the various toxic states of Aβ; however, the details necessary for such mechanistic insight are unattainable with the resolution of current experimental techniques (28).
MD simulations are an essential complement to experimental studies and can provide key insights into the aggregation pathways of low-molecular-weight Aβ oligomers and their subsequent interactions with membranes and other proteins. Investigators have performed simulations of monomeric Aβ in various solution environments (20, 29, 30, 31, 32), as well as a few all-atom MD simulations of dimer formation (33, 34). Discrete MD, replica exchange MD, and the use of implicit solvent are techniques that have been used to characterize the structural ensembles of higher-order Aβ aggregates (34, 35, 36, 37); however, different conclusions regarding equilibrium ensembles have been drawn, most likely associated with the different force fields and sampling techniques used (16). In this study, we utilized united-atom MD simulations of full-length Aβ42 on the microsecond scale to understand and provide insight into the tetramer formation of Aβ42, and provide a foundation for studying higher-order oligomeric Aβ42 aggregate formation.
Interactions of low-molecular-weight Aβ oligomers with membranes are also of interest, given that membrane perturbation by Aβ is central to the pathology of AD (16). Aβ aggregation is accelerated in the presence of membranes, with electrostatic interactions potentially driving the peptide-membrane interactions (38, 39). However, few computational studies have examined the dynamics of low-molecular-weight oligomers, such as an Aβ42 tetramer, in the presence of a membrane. In computational studies, researchers have looked at the effect of rafts on membrane binding and dimerization of Aβ (40, 41), whereas others have utilized pre-embedded oligomeric Aβ to understand transmembrane structures (42, 43). Currently, no study has utilized united-atom MD to examine the effects of any Aβ42 oligomer of three or more peptides on membranes from a starting position that is external to the membrane. By studying the interactions of an Aβ42 tetramer binding to model membranes of a composition similar to those found in eukaryotic plasma membranes and neuronal cells (44, 45, 46), one can characterize the ability of low-molecular-weight Aβ42 species to perturb membranes.
Materials and Methods
Tetramer formation simulations
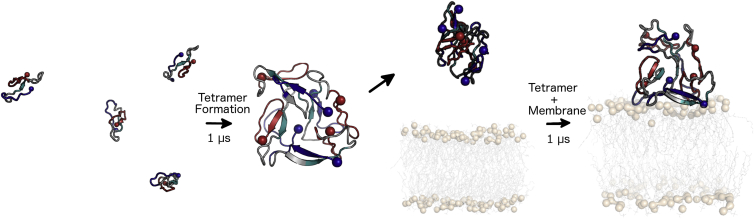
A schematic image detailing the experimental design used in this study is presented in Fig. 1. Simulations were first performed to characterize Aβ42 tetramer formation. In doing so, it was important to carefully consider the starting monomer structure that would be utilized in these simulations. The on-pathway, monomeric species sample a diverse conformational ensemble consisting of a mixture of random coil and β-strand structures, as observed in both experimental and computational studies (29, 30, 47). To obtain a monomer starting structure, the structure of Aβ42 (PDB: 1IYT) (48) was equilibrated and simulated in water and 0.150 M NaCl for 300 ns using the protocol described below. Clustering was performed based on the root mean-square deviation (RMSD) of backbone atoms to obtain a representative structure over the last 100 ns of simulation time. Four copies of the representative structure (defined as the center structure of the most populated cluster) were then placed randomly four times in a 12.7-nm cubic box, with each monomer separated by at least 1.7 nm to be beyond the nonbonded cutoff for van der Waals interactions. A minimum solute-box distance of 3.0 nm was enforced. This system design resulted in a box containing four equilibrated monomeric Aβ peptides that were separated by enough solvent and ions to negate any bias due to van der Waals interactions. Each system was then solvated with simple point charge (SPC) water (49) and 0.150 M NaCl was added, with counterions included to maintain a net neutral system.
Figure 1.
Schematic of the experimental design used to examine interpeptide interactions and their potential rearrangements in the presence of membranes. Interpreted from left to right, the far-left segment shows the initial configuration for four Aβ42 peptides placed at least 3 nm apart. Tetramer formation occurred during 1 μs MD simulations. The resulting tetramer was placed 3 nm away from either a POPC or raft model membrane and another 1 μs MD simulation was conducted. Aβ42 is shown as a cartoon, colored by region (blue, residues 1–10; gray, residues 11–16, 22–29; teal, residues 17–21; red, residues 30–42). N- and C-termini are indicated by blue and red spheres, respectively. Membranes are shown as gray sticks, with phosphorus atoms shown as tan spheres for perspective. To see this figure in color, go online.
Tetramer-membrane simulations
Representative tetramer structures were identified using RMSD clustering (as above) from each replicate in tetramer formation simulations, and placed at a center of mass (COM) distance of 3.0 nm away from pre-equilibrated palmitoyloleoylphosphatidylcholine (POPC) or raft (1:1:1 POPC/cholesterol/palmitoylsphingomyelin (PSM)) model membranes, yielding a minimum atom distance between the tetramer and membrane of at least 2.4 nm. These membranes were selected based on a previous finding that the fibrillar Aβ tetramer has a higher affinity for zwitterionic membranes than for negatively charged membranes (50), and the observed enrichment of POPC and PSM in the exofacial leaflet of the synaptic plasma membrane of neuronal cells (51). The coordinates and topologies for pure POPC were taken from Berger lipid parameters (52, 53) and agree well with experimental properties (54). Model lipid raft coordinates and topologies were obtained from a previous study by Niemela et al. (55). Cholesterol hydroxyl charges from the GROMOS96 53A6 force field (56, 57) were also utilized. Raft membrane properties, including the area per lipid (APL), bilayer thickness, and deuterium order parameters of the control system (raft membrane and solvent only), were also in agreement with properties indicating a liquid-ordered phase (55). Both membrane parameter sets have been used in recent studies of Aβ-membrane interactions (40, 56). The placement of the tetramer far from the membrane was chosen to avoid biasing the initial approach of the tetramer toward the membrane. Details regarding the tetramer-membrane distance and membrane composition are listed in Table S1 in the Supporting Material. Tetramer-membrane systems were solvated with SPC water (49) and 0.150 M NaCl. Three independent simulations, initiated from different structures generated from the three replicates of the tetramer-formation simulations described above, were carried out with each model membrane for 1 μs each, resulting in 3 μs of sampling time for the tetramer-membrane simulations of each lipid type. Two control simulations of the POPC and raft membranes were carried out in the absence of Aβ42. These membranes were solvated with SPC water and 0.150 mM NaCl, and simulated for 1 μs to assess membrane stability over long MD simulations and to serve as a control for assessing membrane perturbation effects caused by interactions with the Aβ42 tetramer. All results presented for the control membrane simulations are averages over the last 250 ns of simulation time.
General MD simulation protocol
All simulations were run using the GROMACS software package, version 4.6 (58, 59), and the GROMOS96 53A6 force field (57). GROMOS is a united-atom force field that merges nonpolar and nonaromatic hydrogens. Systems were energy-minimized using the steepest-descent method, and three replicates, defined as independent trajectories initiated with different random velocities in a canonical (NVT) ensemble, were performed separately for tetramer formation and tetramer-membrane simulations. One simulation was performed to obtain the monomer starting structure of Aβ. NVT was applied to the system for 100 ps using the Berendsen weak coupling method (60) to maintain the temperature at 310 K. An additional annealing step was added after NVT equilibration for tetramer-membrane simulations, which linearly heated the system from 100 to 310 K, at 1 bar pressure for 1 ns using the Berendsen algorithm (60). After either NVT or annealing, depending on the system, an isothermal-isobaric (NPT) ensemble was performed to maintain temperature (310 K) and pressure (1 bar) using a Nosé-Hoover thermostat (61, 62) and Parrinello-Rahman barostat (63, 64). For NVT, annealing, and NPT, position restraints were imposed on all peptide heavy atoms and all restraints were released at the outset of production MD simulations. For tetramer-membrane simulations, the peptide, membrane, and solvent (water and ions) were attached to separate thermostats. All simulations were performed using three-dimensional periodic boundary conditions. Bond lengths were constrained using Parallel Linear Constraint Solver (P-LINCS) (65), allowing an integration time step of 2 fs to be used. Cutoffs for all nonbonded interactions were set to 1.4 nm for monomer equilibration and tetramer-formation simulations, and 1.2 nm for tetramer-membrane simulations. The smooth particle mesh Ewald (PME) method (66, 67) using cubic interpolation and a Fourier grid spacing of 0.16 nm was utilized to calculate long-range electrostatic interactions. To observe interpeptide and peptide-membrane interactions, simulations of Aβ42 tetramer-formation and tetramer-membrane simulations were carried out for 1 μs, yielding a total simulation time of 9 μs. Backbone RMSD and secondary structure of Aβ42 were used to assess simulation convergence and peptide equilibration. Analysis was performed over the last 250 ns for all tetramer-formation and tetramer-membrane simulations. All averages, except for starting structure, were calculated over three replicates for that simulation set and are presented with the corresponding standard deviation. To obtain representative structures of each replicate, RMSD clustering was performed according to the method of Daura et al. (68), using the peptide backbone atoms with a cutoff of 0.3 nm for tetramer formation and 0.2 nm for selection of a monomer structure to begin the tetramer-formation simulations. The representative structures shown in all figures are the central structure of the largest cluster over the last 250 ns of simulation time. GridMAT-MD was used to calculate the APL and bilayer thickness in simulations involving membranes (69). The bilayer thickness was based on defined reference atoms of each lipid type and assigned a paired atom based on proximity on the x-y axis. The z distance between atoms was then calculated for each leaflet over time. For these calculations, the phosphorus atom in POPC and PSM was chosen, whereas a carbon atom sitting in a position similar to that of the phosphorus atom was chosen for cholesterol in the raft simulations.
Deuterium order parameters were calculated to assess the order of the sn-1 POPC lipid acyl chains along the bilayer normal using the following equation:
| (1) |
where θ is the angle between the C-D bond and bilayer normal, and the angle brackets indicate the time average over all equivalent atoms during the last 250 ns of simulation. PyMOL (70) was used to visualize snapshots and render figures. A two-tailed t-test was used for statistical analysis, with statistical significance determined as p < 0.05. Central hydrophobic core (CHC) residues were defined as residues 17–21, and C-terminal (Cterm) residues were defined as residues 29–42.
Results and Discussion
Understanding the formation of low-molecular-weight Aβ oligomers and their interactions with membranes is essential for gaining more insight into their toxicity (71, 72). Given the variability in experimental conditions and aggregation rates, and the limits of experimental resolution to fully characterize these toxic, intermediate species of Aβ, MD simulations have an important role in providing essential mechanistic details regarding the formation of low-molecular-weight oligomer structures. Simulations can also suggest possible mechanisms of the interactions of these species with model membranes and their contributions to the aggregation pathway of Aβ. In this study, we utilized united-atom MD simulations to determine the formation pathway of an Aβ42 tetramer and to subsequently place that tetramer in the presence of two model membranes: pure POPC and a cholesterol-rich raft. Aβ oligomers of this size have not been studied in the presence of model membranes and can provide essential information for understanding the type and extent of perturbations caused by peptide binding.
The approach taken in this work was driven by previous experimental work regarding low-molecular-weight oligomer structures of Aβ42 (23, 71), and discrete MD simulations of an Aβ tetramer (35). Many questions about the membrane-perturbing effects exhibited by low-molecular-weight Aβ42 oligomers remain to be resolved. By simulating a preformed tetramer in the presence of two model membranes, one can gain insight into the potential toxicity of this oligomeric species. Although it has been suggested that larger oligomers of Aβ42 exhibit the most toxicity (73), a membrane-bound tetramer was recently found to cause the highest toxicity with cultured neuronal cells as compared with monomers, dimers, and trimers of Aβ (74). With current computational power, simulation of Aβ tetramers is the most practical approach to enable studies using atomistic MD simulations of Aβ42 aggregation. The GROMOS96 53A6 force field was selected in this study because of its ability to adequately simulate monomeric Aβ relative to experimental information (75, 76). In addition, this force field selection allowed for comparison with previous studies of monomer and dimer Aβ-membrane interactions (40, 56), and the application of frequently utilized lipid parameters to simulate physiologically relevant model membranes (52, 55, 77). When random starting velocities, three replicates for each system, and clustering methods are used to generate representative structures from converged simulations, it is expected that the tetramer unit will vary slightly among the replicates, but will have a relatively similar shape when it is in solution and bound to model membranes. The representative structures of the Aβ42 tetramer in solution were chosen based on the clustering method described by Daura et al. (68), using the peptide backbone atoms with a cutoff of 0.3 nm for tetramer formation and 0.2 nm for selection of a monomer structure to begin the tetramer-formation simulations. These structures represent a relatively high percentage of the structures sampled during the last 250 ns (Fig. 2), and analysis and comparison with the second-highest cluster were also performed. No major difference in overall tetramer shape was observed visually via overlays, and the RMSD values calculated between the first and second largest clusters of formed Aβ tetramers were very low (0.33, 0.43, and 0.24 nm for replicates 1–3, respectively). Thus, utilization of the first cluster structure provided a representative Aβ42 tetramer structure in solution that could then be placed in the presence of model membranes.
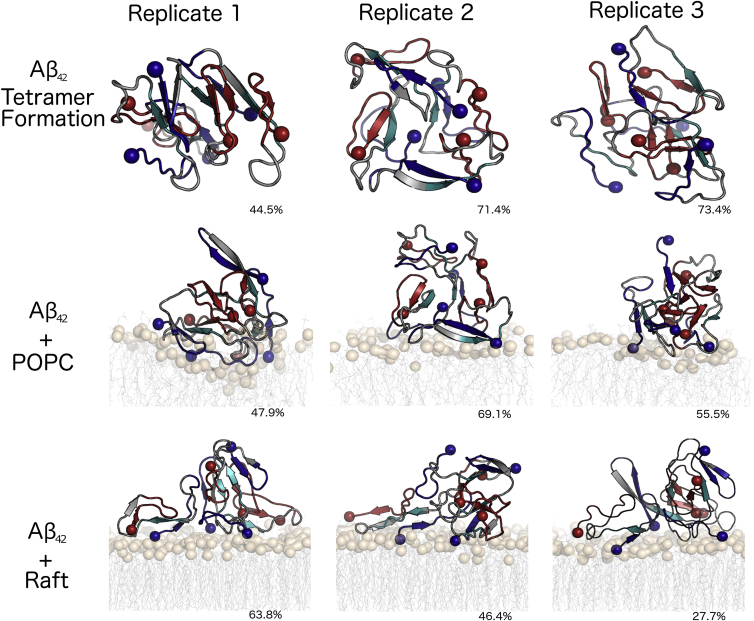
Figure 2.
Dominant morphologies of tetramer formation and tetramer-membrane interactions. The images represent the central structure of the largest cluster from the last 250 ns of each simulation, with percentages representing the cluster size (percentage of frames belonging to the cluster). The rendering of Aβ42 and the membranes is the same as in Fig. 1. To see this figure in color, go online.
Tetramer formation
An increase in the β-strand structure of Aβ42 as a function of the aggregation state is indicative of on-pathway aggregation; therefore, it is useful to study the evolution of secondary structure as it relates to the formation of an Aβ tetramer (21, 22). We first performed an analysis to quantify changes in the secondary structure of Aβ monomers as the tetramer formed. In addition, we studied tertiary and quaternary structures to better understand interpeptide interactions and any trends related to these levels of structure that might give insight into the order of events surrounding Aβ42 tetramer formation and structure. When the Aβ42 structure from PDB: 1IYT (48) was simulated in an aqueous 0.150 mM NaCl environment, it lost all α-helical structure. The resulting Aβ42 monomer that was utilized in tetramer-formation simulations consisted of 75% random coil structure and 25% β-strand structure (Table 1). An NMR study of Aβ42 monomers showed that the average β-strand structure was 20% based on Hα chemical shifts (47), indicating that the secondary-structure assignment and development observed in these simulations agreed with previous experimental and computational work (75, 76).
Table 1.
Average Secondary-Structure Content (in %), with Corresponding Standard Deviations, of Aβ42 after Tetramer Formation and after Interaction of the Tetramer with a POPC or Raft Membrane
| System | Coil | β-Strand | α-Helix |
|---|---|---|---|
| Starting structure | 75 ± 1 | 25 ± 1 | 0 ± 0 |
| Tetramer formation | 66 ± 6 | 34 ± 5 | 0 ± 0 |
| Tetramer + POPC | 68 ± 6 | 32 ± 6 | 0 ± 0 |
| Tetramer + raft | 60 ± 4 | 40 ± 4 | 0 ± 0 |
Tetramer percentages represent the structural properties of Aβ42 tetramers that formed in the indicated simulation, averaged over the final 250 ns of three replicate trajectories for a cumulative sampling time of 750 ns, with corresponding standard deviations.
Starting structure percentages represent the average secondary structure of the four Aβ42 peptides after energy minimization and before equilibration and MD simulation.
The time at which interpeptide contact initially occurred during the MD simulations of tetramer formation varied among the replicates; however, a similar trend emerged in the sequence of interpeptide contact events (Fig. S1). In all replicates, two Aβ42 peptides (denoted as peptides 1 and 2) interacted and formed a dimer, with peptides 3 and 4 binding to the dimer sequentially. Tetramer formation was complete within the first 250 ns of simulation time (Fig. S1); however, to allow observation of any potential rearrangement and increase in interpeptide β-strand structure, the simulations were continued out to 1 μs. The tetramer was determined to be stable and unchanging at this time based on RMSD time series and clustering percentage results (Fig. 2). The last 250 ns of the tetramer-formation simulations were utilized for analysis to reflect the dominant features of the tetramers that formed during these simulations. The clustering results show that a clear, dominant morphology emerged in each replicate, with the central structure of the first cluster representing 44.5%, 71.4%, and 73.4% of frames over the last 250 ns for each of the three replicates.
A statistically significant increase in average percent β-strand structure was observed for Aβ42 tetramers (34% ± 5%) compared with the starting, monomeric structures (25% ± 1%; Table 1). This structural conversion was driven by an increase in the β-strand content in the Cterm region of each peptide (41% ± 9% vs. 36% ± 5% in the starting monomer, respectively). Such an increase in β-strand content in the hydrophobic Cterm region of Aβ42 has been linked to Aβ42 fibrillation and suggests that this hydrophobic region of Aβ42 has a role in on-pathway aggregation, as shown by circular dichroism spectroscopy, electron microscopy, and kinetic experiments (9, 23). Such an increase in β-strand structure overall and in Cterm residues indicates that the tetramer unit that was formed via a united-atom MD simulation is on-pathway with known aggregation events. To assess which regions of the Aβ42 tetramer were participating in β-strand structure, we also performed an analysis of percent secondary structure per key residue region. The results are discussed further below, since the most useful comparison of these structural components is based on a comparison of the structure in solution and the membrane-bound structure (Table S2). The Aβ42 tetramer in all replicates was compact, with an average radius of gyration (Rg) of 1.6 ± 0.1 nm (Table 2) and an average self-diffusion coefficient of 2.6 ± 0.8 × 10−6 cm2 s−1 (Table S3). The initial Rg values of Aβ42 monomers were 1.0 ± 0.1 nm, in agreement with single-molecule-level fluorescence values for monomeric Aβ42 Rg (0.9 ± 0.1 nm) (78). The structural ensembles of all Aβ42 species were characterized by nonspecific interactions between the CHC and Cterm regions (Table 3; Figs. 2 and S3).
Table 2.
Average Rg Values of Aβ42 Tetramers, with Corresponding Standard Deviations
| System | Rg (nm) |
|---|---|
| Tetramer formation | 1.6 ± 0.1 |
| Tetramer + POPC | 2.1 ± 0.2 |
| Tetramer + raft | 2.4 ± 0.3 |
The average represents the Rg of Aβ42 tetramers in the indicated simulation, averaged over the final 250 ns of three replicate trajectories for a cumulative sampling time of 750 ns, with corresponding standard deviations.
The initial, monomeric peptide Rg values of Aβ42 were 1.0 ± 0.1 nm, which agrees with literature values (78).
Table 3.
Average Percentage of Intermolecular Side-Chain Contacts between Key Regions of Aβ42
| System | CHC-CHC | CHC-Cterm | Cterm-Cterm |
|---|---|---|---|
| Tetramer formation | 27 ± 25 | 26 ± 10 | 47 ± 25 |
| Tetramer + POPC | 20 ± 23 | 20 ± 24 | 59 ± 35 |
| Tetramer + raft | 43 ± 2 | 24 ± 8 | 33 ± 6 |
The average percentage represents the intermolecular contacts of Aβ42-specified region-region contacts formed in the indicated simulation, averaged over the final 250 ns of three replicate trajectories for a cumulative sampling time of 750 ns. The specified region-region contact percentage was calculated from the total number of contacts representing CHC-CHC, CHC-Cterm, and Cterm-Cterm. CHC is defined as residues 17–21, and Cterm is defined as residues 30–42.
Finally, to characterize the shapes of the formed tetramers, moments of inertia (I1, I2, I3) and eccentricity (e) were calculated from semiaxes a, b, and c as follows (79, 80):
The shape parameters from these moments of inertia can be defined by a prolate ellipsoid (rod) when I1 ≈ I2 > I3, an oblate ellipsoid (disc) when I1 ≈ I2 < I3, and a sphere when I1 ≈ I2 ≈ I3 (79, 80). Eccentricity (e) of the tetramer can also indicate its shape based on a 0 → 1 scale, with e = 0 representing a perfect sphere and e = 1 representing a rod. Eccentricity is calculated as follows:
In simulations in solution, the simulated Aβ42 tetramer structures more closely resembled an oblate ellipsoid (disc) shape with an eccentricity value of 0.79 ± 0.03 (Table 4). This finding is in contrast to the mass spectrometry (MS) results of Bernstein et al. (71), who determined a linear structure for the Aβ42 tetramer that they described as two dimer units that connected and were separated by an angle of 120°, but it is in agreement with the disc-shaped Aβ42 pentamer structure described by Ahmed et al. (81) using transmission electron microscopy and atomic force microscopy (AFM). Solution and experimental conditions could greatly influence the structure of the Aβ42 tetramer, leading to this difference in results. MS is an in vacuo technique, whereas the transmission electron microscopy and AFM experiments were performed in a hydrated, salt-containing environment. The influence of solution conditions on Aβ42 oligomerization has been confirmed by the formation of a disc-shaped Aβ42 pentamer observed in conditions similar to those utilized in these simulations (81). The formation of an oblate ellipsoid (disc-shaped) tetramer is also shown in Fig. S2 by the more compact tetramers, with all peptides being in contact with one another, as compared with the proposed linear tetramer structure (71). This compact structure has the potential for additional Aβ42 binding, given the heterogeneous (hydrophilic and hydrophobic) solvent-accessible surface area (SASA) of the Aβ42 tetramer.
Table 4.
Average Moments of Inertia (I1, I2, I3) and Eccentricity Values (e) for Aβ42 Tetramers
| System | I1 (104 amu × nm2) | I2 (104 amu × nm2) | I3 (104 amu × nm2) | e |
|---|---|---|---|---|
| Tetramer formation | 2.3 ± 0.1 | 3.2 ± 0.3 | 3.8 ± 0.3 | 0.79 ± 0.03 |
| Tetramer + POPC | 2.8 ± 0.3 | 7.0 ±1.0 | 8.0 ± 1.0 | 0.91 ± 0.01 |
| Tetramer + raft | 6.0 ± 2.0 | 55.0 ± 43.0 | 58.0 ± 44.0 | 0.97 ± 0.02 |
The average represents moment-of-inertia and eccentricity values for Aβ42 tetramers averaged over the final 250 ns of three replicate trajectories for a cumulative sampling time of 750 ns for the indicated system, with corresponding standard deviations.
Modulation of Aβ42 tetramer structure by membranes
The representative structures from the last 250 ns of each of the three replicates from the tetramer-formation simulations were placed in the presence of pure POPC or raft model membranes (Fig. 1; Table S1). Our analysis focused on the extent to which the Aβ42 tetramer disrupted the model membranes and how the membranes influenced the nature of the interpeptide interactions within the Aβ42 tetramer. In these simulations, the center of mass (COM) of the Aβ tetramer was placed equidistant from both leaflets of the bilayer. The use of periodic boundary conditions and identical lipid compositions for both leaflets avoided a bias toward interacting with one leaflet or the other. In the simulations presented here, the tetramer did not choose to interact with any specific side of the membrane. Herein, any analysis of individual leaflets refers to leaflets to which the Aβ tetramer bound and remained bound during the simulation.
The time required for the Aβ42 tetramer to bind to the POPC membrane varied across the replicates (Fig. S4), ranging from 32 to 245 ns. The Aβ42 tetramer interacted with the raft membrane quickly, with the longest replicate taking only 56 ns to bind to the membrane (Fig. S4). In all POPC and raft simulations, once the Aβ42 tetramer bound to the model membrane, it did not release from the membrane. The secondary-structure evolution of Aβ42 was of primary interest when we first studied the effects of a membrane on Aβ42 tetramer arrangements. The β-strand content of the Aβ tetramer was maintained in the presence of the POPC membrane (34% ± 5% vs. 32 ± 6% in water and bound to the membrane, respectively; Table 1). A slight increase in β-strand structure content was observed in the presence of a raft membrane (34% ± 5% vs. 40% ± 6%). To try to understand the increase in β-strand structure in the presence of the raft membrane, we considered other structural features of the tetramer. From examination of the structures, it appeared that the tetramer became more elongated in the presence of the raft membrane than in the presence of the POPC membrane. This change was borne out by the Rg values of the Aβ42 tetramer, which increased to a statistically significant degree in the presence of POPC and raft membranes as compared with the structure in the absence of membranes (Table 2). An Rg of 1.6 ± 0.1 nm was observed for the Aβ42 tetramer in solution, whereas Aβ tetramer Rg values of 2.1 ± 0.2 nm and 2.4 ± 0.3 nm were observed after binding to the POPC membrane and raft membrane, respectively (Table 2). The SASA was also calculated for the Aβ42 tetramer in solution (100 ± 2 nm2) and bound to the membrane. A slight increase in the SASA was observed for the Aβ tetramer in the presence of POPC (105 ± 5 nm2) and raft membranes (108 ± 2 nm2; Table S6). The more extended Aβ42 tetramer structure in the presence of the raft membrane could allow for an extension of inter- and intra-β-strand structure and the rearrangement observed during the simulations (Table 3). The percent β-strand structure in key regions of Aβ42 (the N-terminal (Nterm; residues 1–10), CHC, and Cterm regions) was also analyzed. There was no change in percent β-strand structure in these regions of the Aβ42 tetramer in solution or bound to POPC; however, an increase in percent β-strand structure was observed in these regions, most notably the CHC, when it was bound to raft membranes (Table S2). The increased β-strand structure in the CHC region agrees with studies showing that the CHC region is the shortest fragment of Aβ to form β-strand structure and is essential for full-length Aβ oligomer formation (82). We conclude that the raft environment propagates β-strand structure in the CHC region, whereas this propagation is not observed in the presence of POPC. These findings further highlight the role of raft membranes in influencing the structure and potential aggregation ability of an Aβ42 tetramer.
Finally, the elongation and loss of the spherical structure of the tetramer in the presence of POPC and raft membranes were observed visually (Figs. 2 and S2) and calculated using moment-of-inertia and eccentricity values. After binding to the POPC membrane, the Aβ42 tetramer became elongated and converted to a more rod-like ellipsoid structure, as demonstrated by an increase in eccentricity values (0.91 ± 0.01) as compared with the Aβ42 tetramer in solution (0.79 ± 0.03; Table 4). A considerable variation in moment-of-inertia values was observed when the Aβ42 tetramer bound to a raft membrane, showing further elongation into a prolate ellipsoid shape, with an eccentricity value of 0.97 ± 0.02 (Table 4). In the presence of both the POPC and raft membranes, the Aβ42 tetramer adopted two larger moments and one smaller moment, indicating a progression toward a more prolate ellipsoid (rod-shape) as compared with the Aβ42 tetramer in solution. In the Aβ42 tetramer and raft simulations, there was a greater degree of difference between the two larger moments and the smaller moment, influenced by the increased elongation and β-strand content of the Aβ42 tetramer when bound to the raft membrane as compared with the POPC membrane. In addition, the extent of the difference between the I2/I3 and I1 values is consistent with the degree of change in the Rg values of the Aβ42 tetramer in the presence of a raft membrane. The degree of difference between the I1/I2 and I3 values is consistent with other reported moment-of-inertia values for ellipsoid shapes (80). Irrespective of membrane type, the Aβ42 tetramer became more elongated and rod-like, which has been experimentally determined to be a property of on-pathway protofibrils of Aβ (83). This leads us to conclude that membranes serve to modulate the shape of the Aβ42 tetramer and cause a rearrangement into on-pathway, rod-shaped aggregate structures from an oblate ellipsoid Aβ42 tetramer in solution.
Given that membranes reduce the dimensionality of diffusion of molecules in solution from three dimensions to two, the self-diffusion coefficients of the Aβ42 tetramer bound to POPC and raft membranes were calculated. Significant decreases in self-diffusion coefficients were observed for the Aβ tetramer bound to POPC and raft membranes as compared with the in-solution structure in tetramer-formation simulations. The average self-diffusion coefficient calculated in the x-y plane for the Aβ42 tetramer bound to POPC was reduced by approximately half relative to its value in solution, and was further reduced by half when the Aβ42 tetramer bound to the cholesterol-rich raft (Table S3). The reduced diffusion at the membrane interface suggests strong peptide-lipid interactions and a potential for nucleation due to decreased mobility. It is hypothesized that self-diffusion is decreased in rafts, given the elongation and rearrangement of the Aβ tetramer in the presence of this model membrane. Although there are fewer specific and deep interactions of amino acid side chains with the raft membrane as compared with POPC (see Fig. 4), there are measurably more interactions and hydrogen bonds between the Aβ and the raft membrane as compared with POPC (Table S4). This finding also supports the carpeting-effect model of Aβ42-membrane interactions (50), in that the tetramer bound to both POPC and raft membranes elongated and coated the membrane surface to varying degrees as compared with structures in solution. Both membranes induced a conversion to more rod-like, on-pathway structures, and this effect was greatest in the cholesterol-rich rafts. This finding indicates that these specialized lipid microdomains that contain cholesterol are influential in the pathological aggregation pathway of Aβ, and agree with current literature linking lipid rafts to Aβ aggregation (84).
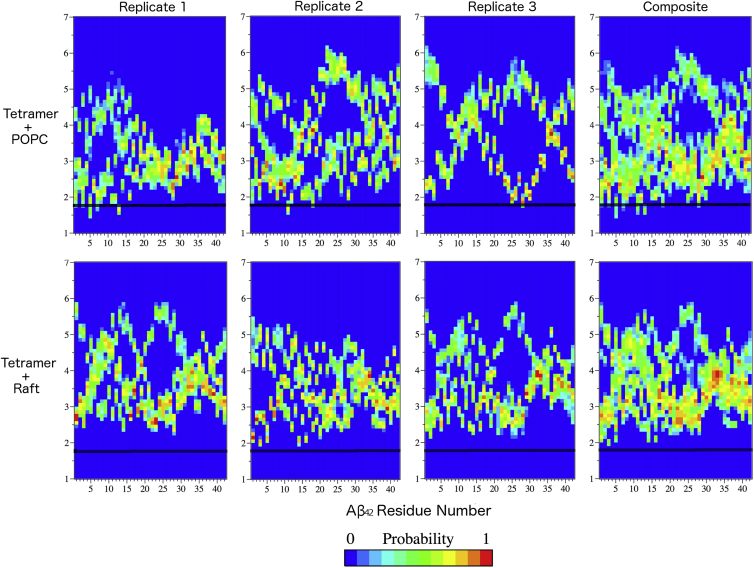
Figure 4.
Normalized frequency of the Aβ residue COM position at a distance, along the z axis (membrane normal) relative to the COM of the bilayer. The residues of Aβ are given along the x axis, and the y axis corresponds to the COM-COM distance. The black dotted line indicates the position of the phosphorus atom in the lipid headgroup. A plot for each replicate is shown, and a composite of all replicates shows the overall features of the ensembles. To see this figure in color, go online.
Lastly, a reorganization of interpeptide contacts was observed in the presence of the raft membrane. Nonspecific interactions between the CHC-CHC, CHC-Cterm, and Cterm-Cterm domains were observed in solution when the Aβ42 tetramer bound to the POPC membrane. However, after it bound to the raft membrane, CHC-CHC contacts increased measurably and consistently across the three replicates, and Cterm-Cterm interactions decreased in a corresponding manner (Table 3). This intriguing rearrangement of the Aβ42 tetramer in the presence of the raft membrane was due to the formation of two major hydrophobic nuclei (Fig. S3). In addition, the degree of elongation was more noticeable in the presence of the raft membrane, due to the rearrangement of interpeptide interactions. These simulations show that rafts cause more substantial elongation and rearrangement of the tetramer than pure POPC membranes, implicating the raft microdomains in the modulation of Aβ tetramer structure to contribute to on-pathway Aβ fibril formation.
Perturbation of POPC and raft membranes by the Aβ42 tetramer
Membrane perturbation induced by the Aβ42 tetramer was quantified in terms of deuterium order parameters (SCD), area per lipid (APL), bilayer thickness, and density profiles. An increase in SCD and bilayer thickness, coupled with a decrease in APL, indicates tighter lipid packing and subsequent elongation of lipid tails. Density profiles of the membrane were also calculated to determine whether Aβ42 tetramer binding caused an increased penetration of water into the glycerol region of the membrane or affected lipid headgroup and lipid tail properties. Control membrane systems (without any peptide) were simulated for 1 μs to serve as a control for assessing the effect of the Aβ42 tetramer on membrane perturbation. In the control simulations, the APL and bilayer thickness averaged over the last 250 ns were 62 ± 1 Å2 and 3.9 ± 0.1 nm, respectively, for POPC, and 41.2 ± 0.1 Å2 and 4.3 ± 0.2 nm for the raft membrane (Table 5). These metrics compare well with literature values (55, 85), indicating that 1 μs is adequate to achieve sufficient sampling, and that the force-field model used in this work adequately represents lipid dynamic and structural properties.
Table 5.
Membrane Properties after Tetramer Interaction
| APL (Å2) | Bilayer Thickness (nm) | |
|---|---|---|
| Tetramer + POPC | 36 ± 5 (−42%) | 4.3 ± 0.1 (+ 10%) |
| Tetramer + raft | 35 ± 3 (−15%) | 4.3 ± 0.1 (± 0%) |
Averages, with corresponding standard deviations and % difference from controls, are shown for APL and bilayer-thickness analysis metrics. The average represents the APL and thickness for the indicated simulation, averaged over the final 250 ns of three replicate trajectories for a cumulative sampling time of 750 ns. Control APL (Å2) values, averaged over the last 250 ns of simulation time of a membrane-only simulation, were 62 ± 1 (POPC) and 41.2 ± 0.1 (raft). Control bilayer-thickness values, averaged over the last 250 ns of simulation time of a membrane-only simulation, were 3.9 ± 0.1 (POPC) and 4.3 ± 0.2 (raft).
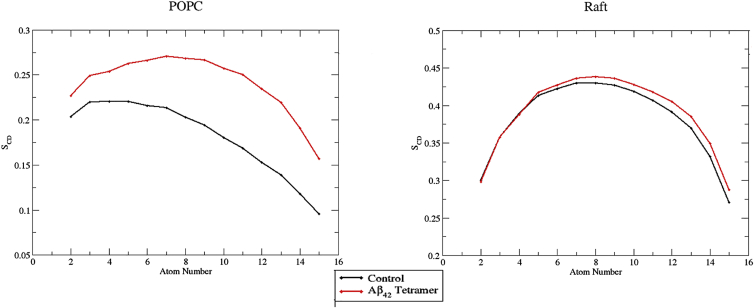
Aβ42 tetramer binding and insertion into the POPC membrane caused an average 42% decrease in APL, coupled with a 10% increase in bilayer thickness (Table 5). Deuterium order parameters were also substantially increased compared with the control POPC membrane SCD value, whereas no significant change in SCD values was observed when the control raft parameters were compared with those obtained when the Aβ42 tetramer was bound (Fig. 3). The density profiles also show that upon Aβ42 tetramer binding, the lipids packed in a way that caused the lipid tails to interdigitate at the bilayer interface (Figs. S5 and S6). In addition, the density of the interacting leaflet headgroup decreased as compared with the control and noninteracting leaflet headgroups, indicating the compactness of the lipid headgroups as a result of Aβ42 tetramer binding. Raft membranes resisted major perturbation by the Aβ42 tetramer, resulting in only a 15% decrease in APL and no change in overall bilayer thickness (Table 5). The density profiles showed no interdigitation between leaflets, and little to no change in the density of the interacting leaflet headgroup was observed as compared with the control and noninteracting leaflets (Fig. S5). These results show that the presence of cholesterol in a raft membrane attenuates major perturbations such as that observed in the pure POPC membrane upon binding of tetrameric Aβ42. The neuroprotective role of cholesterol upon Aβ42 binding has also been shown experimentally by AFM (86).
Figure 3.
Average leaflet deuterium order parameters (SCD) of the palmitoyl (sn-1) chain of POPC lipids in model membrane simulations. For clarity, only the sn-1 chain of POPC is shown; however, parameters were also analyzed for the sn-2 chain and showed similar trends. Control (no peptide present) parameters for each carbon in the lipid chain are shown in black, with the average parameter over three replicates during the last 250 ns of simulation time shown for Aβ42 tetramer (red) simulations. Standard errors of the mean values that are plotted were very small and are not included since they are not readily visible in the graphs. To see this figure in color, go online.
In addition to studying the degree of perturbation of POPC and raft membranes caused by Aβ tetramer binding, we were interested in determining whether any residues of the peptide had an increased probability of participating in peptide-membrane interactions. Distance plots, which represent the position of the center of mass (COM) of each residue in the Aβ tetramer relative to the COM of the bilayer, were generated to determine whether a certain amino acid type or position was more likely to be found at the membrane interface, and to determine the depth of insertion. Due to the large volume of data and the presence of four peptides, individual distance plots are provided for clarity, with a composite showing trends among the replicates. The positively charged residues Arg-5, Lys-16, and Lys-28, and the highly polar Nterm region of Aβ42 frequently sampled regions close to or below the phosphate region of the POPC membrane across all replicates to a greater degree than any other observed amino acid type (Figs. 4 and S7). Lipid packing and a rigid membrane-water interface have not been observed for a pure POPC membrane (87), which helps to explain the ability of these positively charged residues to insert into the lipid headgroup region of POPC and significantly disrupt the membrane integrity and interdigitation of the leaflets. There was a significant increase in lipid order, as shown by the APL of the interacting leaflet after Aβ tetramer binding, which increased the order and length of the carbon tail of the lipids (Fig. 3), pushing the ends into the lipid tails of the other leaflet. This mixing of lipid tails was not observed in the control POPC membrane. Aβ tetramer binding disrupted and caused the lipids to order to such an extent around the Aβ tetramer that the lipid tails had to occupy some of the free space between the leaflets (Figs. S5 and S6) and disrupt the other leaflet, as reflected in the overall bilayer thickness (Table 5).
Residue penetration into the raft membrane was not as deep as compared with the POPC membrane (Fig. 4), which may be a result of the raft membrane being less fluid than the POPC membrane (88). This observation is tied to the small APL of the raft membrane, indicating tight packing of phospholipid (POPC and PSM) headgroups to shield cholesterol from the water interface, thereby preventing the insertion of positively charged residues into the phosphate region of the lipid headgroups. This lack of insertion by positively charged residues into the lipid headgroup region of the raft membrane could result in a rearrangement of the Aβ42 tetramer to adjust to the presence of this more rigid membrane headgroup region as compared with POPC (Figs. 4 and S7). The influence of the headgroup rigidity of a raft on Aβ42 tetramer binding is also shown by the peripheral association of the Aβ42 tetramer with the raft membrane (Fig. 4). The lack of insertion into the phosphate region of the membrane in raft simulations of these positively charged residues could relate to the decreased perturbation of the raft membrane by the Aβ42 tetramer (Table 4; Fig. 3), and could be explained by the elongation and rearrangement of the Aβ42 tetramer as influenced by the presence of cholesterol and the rigidity of a raft membrane.
The work presented here highlights the influence of the oligomer structure and shape on peptide-membrane interactions. We hypothesize that when ordered, fibrillar Aβ interacts with a membrane, some disruption of the fibril is necessary to allow residue insertion into the bilayer. With a more amorphous Aβ tetramer, as used in this study, deeper interactions occur with POPC and raft membranes, given the ability of the less-ordered Aβ structure to rearrange. Tofoleanu et al. (89) simulated fibrillar Aβ in the presence of POPC and POPE lipids. Although POPC and POPE lipids are electrostatically neutral, they have different morphologies because of the more voluminous headgroup of POPC compared with POPE. The voluminous headgroup of POPC restricts the interactions between Aβ and the phosphate and carbonyl moieties, leading to weaker electrostatic interactions between fibrillar Aβ and POPC as compared with POPE (89). In this work, we observed strong electrostatic interactions with positively charged residues inserting into the phosphate headgroup region of POPC (Fig. 4). We hypothesize that the different types of structures used (fibrillar versus aggregate) in our study and that of Tofoleanu et al. (89) account for the differences observed.
Conclusions
Understanding the formation of low-molecular-weight Aβ42 oligomers and oligomer-membrane interactions is essential for elucidating the mechanistic details of AD toxicity caused by Aβ42. In this work, we sought to understand the aggregation mechanism of Aβ42 monomers as it relates to the formation of a tetramer and subsequent Aβ42 tetramer-membrane interactions. The Aβ42 tetramer formed in a stepwise manner, with each peptide binding individually until a relatively compact, oblate ellipsoid tetramer was formed. The Aβ42 tetramer elongated in the presence of both POPC and raft model membranes, showing the influence of membranes on tetramer structure. Rearrangement of interpeptide contacts was observed in the presence of the raft membrane, in addition to a decrease in the insertion depth of positively charged residues into the phosphate region of the membrane as compared with POPC. Aβ42 tetramer binding significantly perturbed POPC membranes, whereas the raft membrane remained relatively unperturbed by Aβ42 tetramer binding. This observation further shows the influence of cholesterol on maintaining membrane integrity in the presence of Aβ42 binding. These peptide-membrane interactions support the carpeting-effect model proposed for Aβ42 toxicity on membranes (50) and reveal the role of membrane environments in modulating Aβ fibril formation, ultimately yielding insight into the mechanism of Aβ42 toxicity and the potential for aggregation events to proceed in the presence of a membrane environment.
Author Contributions
A.M.B. and D.R.B. designed the research protocol and wrote the manuscript. A.M.B. performed the simulations and analyzed data.
Acknowledgments
A.M.B. received a Graduate Teaching Scholars Fellowship from the Virginia Tech College of Agriculture and Life Sciences. Advanced Research Computing at Virginia Tech provided computing time on the BlueRidge supercomputer.
Editor: Scott Feller.
Footnotes
Seven figures and six tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30657-9.
Supporting Material
References
- 1.Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta. 2004;1739:5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Alzheimer’s Association Alzheimer’s disease facts and figures. Alzheimers Dement. 2015;11:332–384. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J.A., Higgins G.A. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 4.Hardy J., Selkoe D.J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 5.Kamenetz F., Tomita T., Malinow R. APP processing and synaptic function. Neuron. 2003;37:925–937. doi: 10.1016/s0896-6273(03)00124-7. [DOI] [PubMed] [Google Scholar]
- 6.Lazarov O., Demars M.P. All in the family: how the APPs regulate neurogenesis. Front. Neurosci. 2012;6:81. doi: 10.3389/fnins.2012.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beel A.J., Sanders C.R. Substrate specificity of γ-secretase and other intramembrane proteases. Cell. Mol. Life Sci. 2008;65:1311–1334. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vardy E.R., Catto A.J., Hooper N.M. Proteolytic mechanisms in amyloid-beta metabolism: therapeutic implications for Alzheimer’s disease. Trends Mol. Med. 2005;11:464–472. doi: 10.1016/j.molmed.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Jarrett J.T., Berger E.P., Lansbury P.T., Jr. The carboxy terminus of the beta amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 10.Jarrett J.T., Berger E.P., Lansbury P.T., Jr. The C-terminus of the beta protein is critical in amyloidogenesis. Ann. N. Y. Acad. Sci. 1993;695:144–148. doi: 10.1111/j.1749-6632.1993.tb23043.x. [DOI] [PubMed] [Google Scholar]
- 11.Dahlgren K.N., Manelli A.M., LaDu M.J. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J. Biol. Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 12.Baglioni S., Casamenti F., Stefani M. Prefibrillar amyloid aggregates could be generic toxins in higher organisms. J. Neurosci. 2006;26:8160–8167. doi: 10.1523/JNEUROSCI.4809-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glabe C.C. Amyloid accumulation and pathogensis of Alzheimer’s disease: significance of monomeric, oligomeric and fibrillar Abeta. Subcell. Biochem. 2005;38:167–177. doi: 10.1007/0-387-23226-5_8. [DOI] [PubMed] [Google Scholar]
- 14.Kirkitadze M.D., Bitan G., Teplow D.B. Paradigm shifts in Alzheimer’s disease and other neurodegenerative disorders: the emerging role of oligomeric assemblies. J. Neurosci. Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 15.Klein W.L., Stine W.B., Jr., Teplow D.B. Small assemblies of unmodified amyloid beta-protein are the proximate neurotoxin in Alzheimer’s disease. Neurobiol. Aging. 2004;25:569–580. doi: 10.1016/j.neurobiolaging.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Nasica-Labouze J., Nguyen P.H., Derreumaux P. Amyloid β protein and Alzheimer’s disease: when computer simulations complement experimental studies. Chem. Rev. 2015;115:3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caughey B., Lansbury P.T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 18.Ross C.A., Poirier M.A. Opinion: What is the role of protein aggregation in neurodegeneration? Nat. Rev. Mol. Cell Biol. 2005;6:891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- 19.Glabe C.G. Structural classification of toxic amyloid oligomers. J. Biol. Chem. 2008;283:29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumketner A., Bernstein S.L., Shea J.-E. Amyloid β-protein monomer structure: a computational and experimental study. Protein Sci. 2006;15:420–428. doi: 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simmons L.K., May P.C., Li W.Y. Secondary structure of amyloid beta peptide correlates with neurotoxic activity in vitro. Mol. Pharmacol. 1994;45:373–379. [PubMed] [Google Scholar]
- 22.Wang Q., Wang Y., Lu H.P. Revealing the secondary structural changes of amyloid β peptide by probing the spectral fingerprint characters. J. Raman Spectrosc. 2013;44:670–674. [Google Scholar]
- 23.Bitan G., Kirkitadze M.D., Teplow D.B. Amyloid β-protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kayed R., Glabe C.G. Conformation-dependent anti-amyloid oligomer antibodies. Methods Enzymol. 2006;413:326–344. doi: 10.1016/S0076-6879(06)13017-7. [DOI] [PubMed] [Google Scholar]
- 25.Kayed R., Head E., Glabe C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 26.Deshpande A., Mina E., Busciglio J. Different conformations of amyloid beta induce neurotoxicity by distinct mechanisms in human cortical neurons. J. Neurosci. 2006;26:6011–6018. doi: 10.1523/JNEUROSCI.1189-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kayed R., Canto I., Glabe C. Conformation dependent monoclonal antibodies distinguish different replicating strains or conformers of prefibrillar Aβ oligomers. Mol. Neurodegener. 2010;5:57. doi: 10.1186/1750-1326-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teplow D.B., Lazo N.D., Stanley H.E. Elucidating amyloid beta-protein folding and assembly: A multidisciplinary approach. Acc. Chem. Res. 2006;39:635–645. doi: 10.1021/ar050063s. [DOI] [PubMed] [Google Scholar]
- 29.Brown A.M., Lemkul J.A., Bevan D.R. Simulations of monomeric amyloid β-peptide (1-40) with varying solution conditions and oxidation state of Met35: implications for aggregation. Arch. Biochem. Biophys. 2014;545:44–52. doi: 10.1016/j.abb.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 30.Sgourakis N.G., Merced-Serrano M., Garcia A.E. Atomic-level characterization of the ensemble of the Aβ(1-42) monomer in water using unbiased molecular dynamics simulations and spectral algorithms. J. Mol. Biol. 2011;405:570–583. doi: 10.1016/j.jmb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise-Scira O., Xu L., Coskuner O. Amyloid-β peptide structure in aqueous solution varies with fragment size. J. Chem. Phys. 2011;135:205101. doi: 10.1063/1.3662490. [DOI] [PubMed] [Google Scholar]
- 32.Yang M., Teplow D.B. Amyloid beta-protein monomer folding: free-energy surfaces reveal alloform-specific differences. J. Mol. Biol. 2008;384:450–464. doi: 10.1016/j.jmb.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barz B., Urbanc B. Dimer formation enhances structural differences between amyloid β-protein (1-40) and (1-42): an explicit-solvent molecular dynamics study. PLoS One. 2012;7:e34345. doi: 10.1371/journal.pone.0034345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urbanc B., Cruz L., Dokholyan N.V. Molecular dynamics simulation of amyloid β dimer formation. Biophys. J. 2004;87:2310–2321. doi: 10.1529/biophysj.104.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meral D., Urbanc B. Discrete molecular dynamics study of oligomer formation by N-terminally truncated amyloid β-protein. J. Mol. Biol. 2013;425:2260–2275. doi: 10.1016/j.jmb.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urbanc B., Cruz L., Stanley H.E. In silico study of amyloid beta-protein folding and oligomerization. Proc. Natl. Acad. Sci. USA. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T., Zhang J., Mu Y. Molecular mechanism of the inhibition of EGCG on the Alzheimer Aβ(1-42) dimer. J. Phys. Chem. B. 2013;117:3993–4002. doi: 10.1021/jp312573y. [DOI] [PubMed] [Google Scholar]
- 38.Tofoleanu F., Buchete N.V. Molecular interactions of Alzheimer’s Aβ protofilaments with lipid membranes. J. Mol. Biol. 2012;421:572–586. doi: 10.1016/j.jmb.2011.12.063. [DOI] [PubMed] [Google Scholar]
- 39.Yates E.A., Owens S.L., Legleiter J. Specific domains of Aβ facilitate aggregation on and association with lipid bilayers. J. Mol. Biol. 2013;425:1915–1933. doi: 10.1016/j.jmb.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 40.Lemkul J.A., Bevan D.R. Aggregation of Alzheimer’s amyloid β-peptide in biological membranes: a molecular dynamics study. Biochemistry. 2013;52:4971–4980. doi: 10.1021/bi400562x. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L.N., Chiu S.W., Mu Y. Amyloid β peptides aggregation in a mixed membrane bilayer: a molecular dynamics study. J. Phys. Chem. B. 2011;115:12247–12256. doi: 10.1021/jp2065985. [DOI] [PubMed] [Google Scholar]
- 42.Poojari C., Kukol A., Strodel B. How the amyloid-β peptide and membranes affect each other: an extensive simulation study. Biochim. Biophys. Acta. 2013;1828:327–339. doi: 10.1016/j.bbamem.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Strodel B., Lee J.W.L., Wales D.J. Transmembrane structures for Alzheimer’s Aβ(1-42) oligomers. J. Am. Chem. Soc. 2010;132:13300–13312. doi: 10.1021/ja103725c. [DOI] [PubMed] [Google Scholar]
- 44.Devaux P.F. Static and dynamic lipid asymmetry in cell membranes. Biochemistry. 1991;30:1163–1173. doi: 10.1021/bi00219a001. [DOI] [PubMed] [Google Scholar]
- 45.Ikeda M., Kihara A., Igarashi Y. Lipid asymmetry of the eukaryotic plasma membrane: functions and related enzymes. Biol. Pharm. Bull. 2006;29:1542–1546. doi: 10.1248/bpb.29.1542. [DOI] [PubMed] [Google Scholar]
- 46.Tsui-Pierchala B.A., Encinas M., Johnson E.M., Jr. Lipid rafts in neuronal signaling and function. Trends Neurosci. 2002;25:412–417. doi: 10.1016/s0166-2236(02)02215-4. [DOI] [PubMed] [Google Scholar]
- 47.Hou L., Shao H., Zagorski M.G. Solution NMR studies of the A β(1-40) and A β(1-42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J. Am. Chem. Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 48.Crescenzi O., Tomaselli S., Picone D. Solution structure of the Alzheimer amyloid beta-peptide (1-42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 49.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Hermans J. Interaction models for water in relation to protein hydration. Intermol. Forces. 1981;14:331–342. [Google Scholar]
- 50.Williams T.L., Serpell L.C. Membrane and surface interactions of Alzheimer’s Aβ peptide—insights into the mechanism of cytotoxicity. FEBS J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 51.Wood W.G., Igbavboa U., Eckert G.P. Cholesterol asymmetry in synaptic plasma membranes. J. Neurochem. 2011;116:684–689. doi: 10.1111/j.1471-4159.2010.07017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger O., Edholm O., Jähnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 1997;72:2002–2013. doi: 10.1016/S0006-3495(97)78845-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biocomputer Group. Using Computer Simulations to Study Biological Problems. http://wcm.ucalgary.ca/tieleman/downloads. Accessed on March 3, 2016.
- 54.Piggot T.J., Piñeiro Á., Khalid S. Molecular dynamics simulations of phosphatidylcholine membranes: a comparative force field study. J. Chem. Theory Comput. 2012;8:4593–4609. doi: 10.1021/ct3003157. [DOI] [PubMed] [Google Scholar]
- 55.Niemelä P.S., Ollila S., Vattulainen I. Assessing the nature of lipid raft membranes. PLOS Comput. Biol. 2007;3:e34. doi: 10.1371/journal.pcbi.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemkul J.A., Bevan D.R. Lipid composition influences the release of Alzheimer’s amyloid β-peptide from membranes. Protein Sci. 2011;20:1530–1545. doi: 10.1002/pro.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oostenbrink C., Villa A., van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 58.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 59.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berendsen H.J.C., Postma J.P.M., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 61.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 62.Nosé S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 2002;100:191–198. [Google Scholar]
- 63.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 64.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 65.Hess B. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008;4:116–122. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 66.Darden T., York D., Pedersen L. Particle mesh Ewald: an N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 67.Essmann U., Perera L., Pedersen L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 68.Daura X., Gademann K., Mark A.E. Peptide folding: when simulation meets experiment. Angew. Chem. Int. 1999;38:236–240. [Google Scholar]
- 69.Allen W.J., Lemkul J.A., Bevan D.R. GridMAT-MD: a grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem. 2009;30:1952–1958. doi: 10.1002/jcc.21172. [DOI] [PubMed] [Google Scholar]
- 70.Schrödinger, LLC. The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger, LLC.
- 71.Bernstein S.L., Dupuis N.F., Bowers M.T. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shankar G.M., Li S., Selkoe D.J. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat. Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sakono M., Zako T. Amyloid oligomers: formation and toxicity of Abeta oligomers. FEBS J. 2010;277:1348–1358. doi: 10.1111/j.1742-4658.2010.07568.x. [DOI] [PubMed] [Google Scholar]
- 74.Jana M.K., Cappai R., Ciccotosto G.D. Membrane-bound tetramer and trimer Aβ oligomeric species correlate with toxicity towards cultured neurons. J. Neurochem. 2016;136:594–608. doi: 10.1111/jnc.13443. [DOI] [PubMed] [Google Scholar]
- 75.Gerben S.R., Lemkul J.A., Bevan D.R. Comparing atomistic molecular mechanics force fields for a difficult target: a case study on the Alzheimer’s amyloid β-peptide. J. Biomol. Struct. Dyn. 2014;32:1817–1832. doi: 10.1080/07391102.2013.838518. [DOI] [PubMed] [Google Scholar]
- 76.Olubiyi O.O., Strodel B. Structures of the amyloid β-peptides Aβ1-40 and Aβ1-42 as influenced by pH and a D-peptide. J. Phys. Chem. B. 2012;116:3280–3291. doi: 10.1021/jp2076337. [DOI] [PubMed] [Google Scholar]
- 77.Tieleman D.P., Sansom M.S.P., Berendsen H.J.C. Alamethicin helices in a bilayer and in solution: molecular dynamics simulations. Biophys. J. 1999;76:40–49. doi: 10.1016/S0006-3495(99)77176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nag S., Sarkar B., Maiti S. Nature of the amyloid-β monomer and the monomer-oligomer equilibrium. J. Biol. Chem. 2011;286:13827–13833. doi: 10.1074/jbc.M110.199885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abel S., Sterpone F., Marchi M. Molecular modeling and simulations of AOT-water reverse micelles in isooctane: structural and dynamic properties. J. Phys. Chem. B. 2004;108:19458–19466. [Google Scholar]
- 80.Martinez A.V., Dominguez L., Straub J.E. Probing the structure and dynamics of confined water in AOT reverse micelles. J. Phys. Chem. B. 2013;117:7345–7351. doi: 10.1021/jp402270e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ahmed M., Davis J., Smith S.O. Structural conversion of neurotoxic amyloid-beta(1-42) oligomers to fibrils. Nat. Struct. Mol. Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Klimov D.K., Thirumalai D. Dissecting the assembly of Abeta16-22 amyloid peptides into antiparallel β sheets. Structure. 2003;11:295–307. doi: 10.1016/s0969-2126(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 83.Cheng I.H., Scearce-Levie K., Mucke L. Accelerating amyloid-β fibrillization reduces oligomer levels and functional deficits in Alzheimer disease mouse models. J. Biol. Chem. 2007;282:23818–23828. doi: 10.1074/jbc.M701078200. [DOI] [PubMed] [Google Scholar]
- 84.Rushworth J.V., Hooper N.M. Lipid rafts: linking Alzheimer’s amyloid-β production, aggregation, and toxicity at neuronal membranes. Int. J. Alzheimer’s Dis. 2011;2011:603052. doi: 10.4061/2011/603052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elmore D.E. Molecular dynamics simulation of a phosphatidylglycerol membrane. FEBS Lett. 2006;580:144–148. doi: 10.1016/j.febslet.2005.11.064. [DOI] [PubMed] [Google Scholar]
- 86.Seghezza S., Diaspro A., Dante S. Cholesterol drives aβ(1-42) interaction with lipid rafts in model membranes. Langmuir. 2014;30:13934–13941. doi: 10.1021/la502966m. [DOI] [PubMed] [Google Scholar]
- 87.Zhou F., Schulten K. Molecular dynamics study of a membrane-water interface. J. Phys. Chem. B. 1995;99:2194–2207. [Google Scholar]
- 88.Hancock J.F. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tofoleanu F., Brooks B.R., Buchete N.-V. Modulation of Alzheimer’s Aβ protofilament-membrane interactions by lipid headgroups. ACS Chem. Neurosci. 2015;6:446–455. doi: 10.1021/cn500277f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.