Abstract
Charged multivesicular body protein 2B (CHMP2B) – a component of the endosomal complex required for transport-III (ESCRT-III) – is responsible for the vital membrane deformation functions in autophagy and endolysosomal trafficking. A dominant mutation in CHMP2B (CHMP2BIntron5) is associated with a subset of heritable frontotemporal dementia – frontotemporal dementia linked to chromosome 3 (FTD-3). ESCRT-III recruits Vps4, an AAA-ATPase that abscises the membrane during various cellular processes including autophagy and intraluminal vesicle formation. CHMP2BIntron5 results in a C-terminus truncation removing an important Vps4 binding site as well as eliminating the normal autoinhibitory resting state of CHMP2B. CHMP2B is expressed in most cell types but seems to be especially vital for proper neuronal function. CHMP2BIntron5-mediated phenotypes include misregulation of transmembrane receptors, accumulation of multilamellar structures, abnormal lysosomal morphology, down regulation of a brain-specific micro RNA (miRNA-124), abnormal dendritic spine morphology, decrease in dendritic arborization, and cell death. Currently, transgenic-fly, -mouse, and -human cell lines are being used to better understand the diverse phenotypes and develop therapeutic approaches for the CHMP2BIntron5-induced FTD-3.
Keywords: Autophagy, CHMP2BIntron5, endolysosome, ESCRT-III, frontotemporal dementia, FTD-ALS
1. Introduction
Frontotemporal dementia (FTD) is the second most common neurodegenerative disease in patients under 65 after Alzheimer’s disease. It is a heterogeneous disease caused by many different genetic mutations, exhibiting different molecular pathologies, and presenting different clinical symptoms (Pan and Chen, 2013). However, all subtypes are characterized by a general degeneration of the frontal and anterior temporal lobes. Different subtypes of FTD may be described based on their clinical presentation or their genetic basis. Moreover, one genetic cause of FTD may have several distinct clinical presentations. At the clinical level, the most prevalent subtype is behavioral variant FTD (bvFTD), which accounts for about 70% of cases and is characterized by impaired executive function but well-maintained memory abilities at early stages of the disease (Pan and Chen, 2013; Piguet et al., 2011). Another clinical subtype is primary progressive aphasia (PPA), which has several subtypes of its own, distinguished based on what aspect of language is most affected: nonfluent variant PPA (nfvPPA), semantic dementia, (SD), and logopenic varient PPA (lvPPA; (Gorno-Tempini et al., 2011). FTD also overlaps with a number of similar disorders including: progressive supranuclear palsy (PSP) syndromes, corticobasal degeneration (CBD), and argyrophilic disease (AGD; (Pan and Chen, 2013). Furthermore, FTD can also be described as existing on a continuum with amyotrophic lateral sclerosis (ALS) termed the FTD-ALS spectrum (De Silva et al., 2015; Gascon and Gao, 2014; Lattante et al., 2015; Ling et al., 2013; Talbot and Ansorge, 2006).
One FTD subtype, frontotemporal dementia linked to chromosome 3 (FTD-3), is a heritable form of FTD associated with a dominant mutation - a single nucleotide mutation in the splicing site - that results in the production of different mutant proteins in charged multivesicular body protein 2B (CHMP2B, Vps2 in yeast). The first in-depth analysis of one of the mutant isoforms (CHMP2BIntron5) produced by this mutation occurred just 10 years ago (Skibinski et al., 2005). CHMP2B is an important subunit of the endosomal complex required for transport III (ESCRT-III), which has membrane deformation activity that is relevant in many cellular functions. FTD-3 has an average age of onset of 57, ranging from 46 to 67. However, accurately determining an age of onset is often difficult due to subtle initial symptoms that grow progressively more pronounced. These symptoms initially consist of slight personality changes including disinhibition, lack of empathy, and inappropriate emotional responses. Dyscalculia and deficits in visuospatial function are also sometimes experienced. The more pronounced later-stage symptoms can include apathy, aggressive behavior, and motor deficits such as Parkinsonian symptoms (Lunau et al., 2012; Snowden et al., 2002). Thus, FTD-3 usually falls within a bvFTD diagnosis but may also overlap with motor neuron-related pathology on the FTD-ALS spectrum (Ling et al., 2013).
Although mutations in several different genes can cause FTD (see (Benussi et al., 2015), this review will focus on the recent findings on the CHMP2BIntron5 mutant isoform. Though it is a relatively uncommon mutation in FTD patients (Benussi et al., 2015; Roberson, 2012), it is still widely studied due to its robust pathology. Because of this, the CHMP2BIntron5 mutant isoform and its resulting pathology are well characterized. Although FTD has recently been labeled as a part of the FTD-ALS spectrum (Gascon and Gao, 2014; Lattante et al., 2015; Ling et al., 2013), CHMP2B mutations are considered to result in a disorder most close to FTD on this spectrum (Ling et al., 2013); therefore FTD caused by the CHMP2BIntron5 mutation and its role in autophagy and other processes will be the focus of this review.
2. Mutations causing FTD
2.1 Non-CHMP2B mutations in FTD
Generally patients suffering from all types of FTD, of which FTD-3 is a subtype, are diagnosed with degenerative atrophy in the frontal and anterior temporal lobes (Sieben et al., 2012). However, FTD has numerous monogenic causes, most of which are caused by mutations in genes other than CHMP2B (Benussi et al., 2015; Roberson, 2012). The most common mutations associated with FTD are in C9ORF72 (chromosome 9 open reading frame 72; up to 50% of familial case; (Majounie et al., 2012), PGRN (progranulin; 5–20% of cases; (Rademakers et al., 2012), and MAPT (microtubulin associated protein Tau; 5–20% of cases; (Rademakers et al., 2004; Sieben et al., 2012). The mutation in C9ORF72 is an expansion of a hexanucleotide repeat. Normally there are 20 or fewer GGGGCC repeats, but affected patients have from 30 up to thousands of repeats; this increase in repeat number causes protein aggregates and neuronal inclusions (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Stewart et al., 2012). There are several documented PGRN mutations that either decrease the expression of or truncate the protein product, progranulin, which is an important growth factor (Baker et al., 2006; Cruts et al., 2006). The MAPT mutants create protein aggregates causing a change in the balance between two isoforms of the TAU protein, which is required for microtubule stabilization. There have been over 40 mutations in MAPT noted which cause a variety of neurodegenerative diseases, including FTD (Hutton et al., 1998; Sieben et al., 2012).
Mutations in CHMP2B, FUS, and VCP are much more rare, each accounting for less than 1% of FTD cases. Another relatively uncommon cause of FTD is a mutation in TARDBP, which produces the transactive response DNA-binding protein 43 (TDP-43). Mutations in TARDBP cause FTD-Ubiquitin (FTD-U) by forming TDP-43 aggregates in the cytoplasm, though normal TDP-43 is mostly localized in the nucleus (Benussi et al., 2015; Neumann et al., 2006). Also displaying TDP pathology, are FTD and ALS patients with rare variants of TANK-binding kinase 1 (TBK1) and optineurin (OPTN; (Pottier et al., 2015). TBK1 is a kinase with many substrates, including an ESCRT-I subunit and OPTN, which is involved in antibacterial autophagy (Gijselinck et al., 2015). It has recently been seen that several different mutations to TREM2 encoding triggering receptor expressed on myeloid cells 2 protein, are associated with bvFTD (Borroni et al., 2014). For more information on the heterogeneous genetic causes of FTD and ALS see (Sieben et al., 2012) and (Benussi et al., 2015).
2.2 CHMP2B mutations in FTD
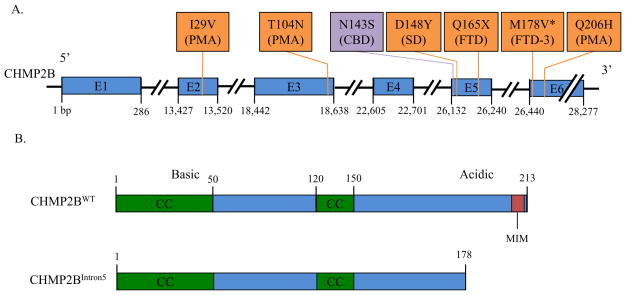
Several different CHMP2B mutations have been identified in FTD-3 patients. The first, producing the CHMP2BIntron5 mutant isoform, was discovered in a large Danish family with a history of FTD-3. The mutation is a G to C substitution in the acceptor splice site of exon 6 resulting in the inclusion of the 201 base pair intron 5, which contains a stop codon. Thus, upon translation, it yields a 36 amino acid truncation of the C-terminus (Figure 1). The last 36 amino acids are replaced with a single valine residue from the fifth intron. CHMP2BDelta10 is an alternative mRNA transcript generated from the same mutation. It results from the use of a cryptic splice site located 10 base pairs near the start of exon 6. The resulting protein is also missing the final 36 amino acids of the normal protein, however 29 random amino acids are added to the C-terminus rather than a single valine (Skibinski et al., 2005). CHMP2BQ165X is an unrelated mutation resulting in a 49 amino acid C-terminus truncation; it was found in a Belgian family and appears to result in similar pathology to the CHMP2BIntron5 mutant isoform (van der Zee et al., 2008).
Figure 1.
Mutations in the CHMP2B gene causing FTD and other neurodegenerative diseases. A. There are several CHMP2B mutations that have been found to cause diseases on the FTD-ALS spectrum (I29V, T104N, D148Y, Q165X, M178V, and Q206H; orange boxes) as well as other neurodegenerative disorders; corticobasal degeneration caused by N143S (purple box). B. Functional wild type CHMP2B domains and altered CHMP2BIntron5 protein. Wild type CHMP2B is a 213 amino acid protein with two coiled-coil (CC) domains as well as a microtubule interacting and transport interacting motif (MIM). The N-terminus contains basic alpha helices and the C-terminus contains acidic alpha helices, leading to an auto-inhibitory self-binding. CHMP2BIntron5 loses much of the acidic C-terminus, decreasing its autoinhibition and removing the MIM, which is a Vps4 binding site. E, Exon; PMA, primary muscular atrophy; CBD, corticobasal degeneration; SD, semantic dementia; FTD, frontotemporal dementia; * denotes mutation causing CHMP2BIntron5 and CHMP2BDelta10.
2.3 CHMP2B mutations in other diseases
Numerous other mutations in CHMP2B are associated with neurodegenerative diseases both within and distinct from the FTD-ALS spectrum. Within the FTD-ALS spectrum, a C to A substitution resulting in a T104N substitution, an A to G substitution resulting in a I29V substitution, and an A to G substitution resulting in a Q206H substitution are all associated with primary muscular atrophy (PMA; Figure 1A; (Cox et al., 2010; Isaacs et al., 2011). Also, a G to T substitution resulting in a D148Y mutation that is associated with SD (Skibinski et al., 2005). Another neurodegenerative disease related to but not part of the FTD-ALS spectrum caused by a CHMP2B mutation is corticobasal degeneration caused by an A to G substitution resulting in a N143S mutation (Figure 1A; (van der Zee et al., 2008). Furthermore, a rare copy number variation of CHMP2B has been found in early-onset familial Alzheimer’s disease (Hooli et al., 2014).
Additionally, CHMP2B and other ESCRT proteins play a role in cell proliferation due to their role in regulating growth signaling pathways through membrane receptor regulation, thus they also appear to play a role in cell proliferation and possibly cancer. One ESCRT-II component, Vps25, plays an important anti-tumorigenesis role. Mutants of VPS25 cause a lack of downregulation of Notch receptors after signaling, leading to excessive cell proliferation and tumor growth (Herz et al., 2006; Thompson et al., 2005). Moreover, ectopic expression of CHMP2BIntron5 in Drosophila eyes causes misregulation of Notch receptors resulting in overgrowth of eye tissue (Cheruiyot et al., 2014).
3. CHMP2B: Structure and Function
3.1 Structure of CHMP2B
CHMP2B contains two coiled coil domains near the N-terminus (amino acid residues 1–50 and 120–150) and a microtubule-interacting and transport (MIT)- interacting motif (MIM) domain at the C-terminus (Figure 1B; (Ghanim et al., 2010). Coiled coil domains usually are important for providing structural support for the protein as well as interacting with other proteins, for example, they are found in kinesin motor proteins to connect the heads to the tail and are able to bear a very large load (Surkont et al., 2015). MIM domains are present in all ESCRT-III components and are composed of a single helix that binds between two of the three helices in an MIT domain, in this case in the MIT domain of Vps4 (Ghazi-Tabatabai et al., 2009). The MIM domain of CHMP2B is important for the recruitment of Vps4, which leads to membrane abcission and ESCRT-III depolymerization. It also facilitates the binding of many different effector proteins to activated ESCRT-III subunits (Hurley, 2010; Schmidt and Teis, 2012). Vps4 binding to the MIM of CHMP2B is also able to release the standard autoinhibition of the protein, allowing it to polymerize (Bodon et al., 2011). CHMP2B normally exists in an autoinhibiting state in the cytosol with the acidic C-terminus bound to its basic N-terminus. Because of this ability to self bind end to end, CHMP2B can also form polymers (Bajorek et al., 2009). The formation of helical CHMP2B polymers, which only occurs at the membrane, creates protrusions in the cell membrane when CHMP2B is overexpressed (Bodon et al., 2011). However, under physiological conditions it appears that expansion and compression of spiral-like polymers of CHMP4 (Snf7 – ESCRT-III component) play a critical role in membrane deformation and fission to generate MVBs (Chiaruttini et al., 2015; Henne et al., 2013).
3.2 Function of wild type CHMP2B
CHMP2B is an important subunit of ESCRT-III (Ghazi-Tabatabai et al., 2009). ESCRT-III is a membrane deformation complex responsible for many cellular processes including the formation of intraluminal vesicles (ILVs) within multivesicular bodies (MVBs) in the endolysosomal pathway, autophagy, membrane repair, the formation of extracellular vesicles, nuclear membrane modulation, cytokinesis, and retrovirus budding (Odorizzi, 2015; Olmos et al., 2015; Scheffer et al., 2014). ESCRT-III is instrumental in all of these processes because of its membrane scission function. Specifically, CHMP2B’s primary function in ESCRT-III is to recruit Vps4, an AAA-ATPase, to induce the disassembly of the complex; Vps4 is also the membrane scission machinery (Hanson and Cashikar, 2012). For this recruitment to occur, ESCRT-III must first assemble, because it is an ephemeral complex unlike the stable ESCRT-0, ESCRT-I and ESCRT-II.
The first step in the assembly of ESCRT-III is the activation of CHMP6 (Vps20) by ESCRT-II, which serves to nucleate the complex. This is followed by the oligomerization of CHMP4 (A-C; Snf7), which is then capped by CHMP3 (Vps24). CHMP3 recruits CHMP2 (A and B), completing the assembly of the core ESCRT-III proteins. CHMP2 provides a binding site for the three accessory components of ESCRT-III, CHMP1 (A and B; Did2), CHMP5 (Vps60), and Ist1. CHMP2 also recruits the Vps4 complex, which both pinches off the membrane and induces disassembly of ESCRT-III by binding the MIM of CHMP2B (Hurley, 2010; Schmidt and Teis, 2012; Schuh and Audhya, 2014). This order of assembly has been precisely characterized, but the mechanics of the membrane scission have not yet been defined (Henne et al., 2013).
The function of ESCRT-III in the formation of ILVs in MVBs is responsible for the endosomal sorting of transmembrane proteins. ESCRTs 0-II function in the initial formation of ILVs: ESCRT-0 sorts membrane proteins by binding to the membrane and the ubiquitinated proteins on it, ESCRTs I and II also binds to ubiquitinated proteins as well as causing the inward budding of the vesicle. ESCRT-III further deforms the membrane and recruits Vps24, which abscises the bud (Hurley, 2010; Schmidt and Teis, 2012). The most noteworthy cargo for these MVBs are transmembrane receptors, many of which have been shown to be dependent on ESCRT functioning for proper sorting and degeradation (Ahmad et al., 2009; Rusten et al., 2012). Some ESCRT genes are even labeled as tumor suppressor genes because of their sequestering of growth factors and other receptors such as Notch and EGFR (Herz et al., 2006; Rodahl et al., 2009; Sun et al., 2015).
Another role of ESCRT-III is in macroautophagy (referred to as autophagy). Autophagy is a cellular process that includes the degradation of cytosolic protein aggregates and organelles such as mitochondria. Autophagy was originally thought to occur only during starvation and stress. Currently, a large body of literature has established that a basal level of autophagy occurs in normally functioning cells (Ahmad and Lee, 2014). A vital step in the autophagy pathway is the formation of the autophagosome – a large double membrane vesicle containing the cargo destined for autophagic degradation. ESCRT-III may be important for proper fusion of the autophagosome with the lysosome, making it an essential protein for maintaining homeostasis through basal autophagy (Lee and Gao, 2008). Loss of Snf7-2, an ESCRT-III component, causes the accumulation of autophagosomes, indicating the importance of ESCRT functioning in autophagy (Lee et al., 2007). The loss of ESCRT genes in several different animals including C. elegans, Drosophila, and mammalian models causes autophagosome accumulation. This may be due to both the lack of fusion of autophagosomes to lysosomes as well as the increased formation of autophagosomes through activation of the kinase JNK which activates autophagy (Rusten et al., 2012).
Because autophagy is responsible for many cellular processes, such as the maintenance of membrane receptors in signaling pathways, the trafficking of organelles for degradation, and clearing protein aggregates, it has numerous effects on cells. However, these effects are more pronounced in neurons, likely due to their post-mitotic status (Ahmad and Lee, 2014). For example, the loss of basal autophagy causes neurodegeneration, even when any pathological protein aggregates are lacking (Hara et al., 2006).
More recently, new research has outlined more neuron-specific roles for CHMP2B, most likely through its participation as an ESCRT-III component. CHMP2B has been found to colocalize with PSD-95, a postsynaptic marker, and is consistently present at dendritic branch points. An shRNA-mediated knockdown of CHMP2B causes substantial reductions in long dendritic arborization and a decrease in number of dendritic spines with a particular reduction in the number of mushroom spines. These structural deficits were coupled with a decrease in long term potentiation, indicating that CHMP2B plays a role in the formation and maintenance of synapses (Chassefeyre et al., 2015). However, in cultured hippocampal neurons, it has been reported that a CHMP2B knockdown resulted in an increase in neuronal spine density, but importantly, this was also coupled with a decrease in mushroom spines (Belly et al., 2010). This would still likely result in decreased neural plasticity because mushroom spines are responsible for the formation of long-term synapses (Bourne and Harris, 2007). These neuron-specific functions have provided additional insights into the role of mutant CHMP2B in neurodegenerative disorders.
4. CHMP2BIntron5 causes diverse defects
The histopathological analysis of FTD-3 patients with the CHMP2BIntron5 mutant isoform showed frontal cortices with accumulation of enlarged vacuoles marked with late-endosome receptors. This pathology was seen throughout the neocortex including the hippocampus and insula, and was absent in GRN and MAPT mutants, AD patients, and prion disease patients (Urwin et al., 2010). Since the first study in 2005 identifying CHMP2BIntron5 as the genetic basis of FTD-3, numerous studies in model systems: Drosophila, human and mouse neuronal cell lines, mouse primary neurons, cultured human fibroblasts, and mouse, have largely recreated the cellular and behavioral FTD symptoms. Further, these studies have provided insights into CHMP2BIntron5-mediated deficits at molecular, cellular, and behavioral levels.
Interestingly, ectopic expression of CHMP2BIntron5 and CHMP2B knockdown can show similar deficits. For instance, both knockdown of CHMP2B and ectopic expression of CHMP2BIntron5 cause a decrease in mushroom spines in cultured neurons (Belly et al., 2010; Chassefeyre et al., 2015). Even though CHMP2BIntron5 causes an increase in number of spines, there is also a reduced size of dendritic spines in cultured hippocampal neurons due to a targeted loss of mushroom and stubby spines (Belly et al., 2010). The CHMP2B knockdown and CHMP2BIntron5 have similar phenotypes because CHMP2BIntron5 can possibly create an effective knockdown by titrating endogenous CHMP2B and other ESCRT-III components from their normal interactions on a nascent MVB. Indeed, in mouse cortical neurons, CHMP2BIntron5 was shown to have stronger interaction with ESCRT-III components when compared to CHMP2BWT (Lee et al., 2007). Furthermore, CHMP2BIntron5 and endogenous CHMP2B have been shown to form co-labeled aggregates in transfected hippocampal neurons (Belly et al., 2010). The reduction in mushroom spines could result in mental deficits such as those seen in FTD because spines with mushroom morphology are the most likely to form strong and lasting synapses (Bourne and Harris, 2007).
Due to the C-terminus truncation, CHMP2BIntron5 lacks its MIM domain (a Vps4 binding site) and C-terminus acidic alpha helices (Figure 1B). Lack of the MIM domain prevents Vps4-ESCRT-III binding-induced dissociation of the ESCRT-III complex (Bodon et al., 2011). This lack of dissociation of the ESCRT-III complex leads to accumulation of endosomes and autophagosomes. This has been demonstrated in Drosophila (Lee et al., 2007) and mouse models of CHMP2BIntron5 (Clayton et al., 2015; Ghazi-Noori et al., 2012), CHMP2BIntron5 transfected neuroblastomas (van der Zee et al., 2008) and FTD-3 patient fibroblasts and neurons (Urwin et al., 2010). Furthermore, similar pathology has been seen in a distinct C-terminus truncation in an FTD patient (Q165X;(van der Zee et al., 2008) showing that the lack of Vps4 binding to the MIM domain is likely the cause of pathology. It has also been shown that CHMP2BIntron5, despite its C-terminus truncation, can polymerize at the membrane to form helical structures that create membrane protrusions. This polymerization also occurs with CHMP2BWT but is potentially more pathologically relevant for CHMP2BIntron5 because it lacks the normal autoinhibition that is present in the wild type protein. This pathological mechanism is often neutralized in studies due to fluorescent tagging at the N-terminus, which eliminates the ability of CHMP2B to polymerize (Bodon et al., 2011). The majority of CHMP2BIntron5 phenotypes are considered a downstream consequence of the disruption of endolysosomal pathway.
One of the most significant consequences of the defective endolysosomal pathway is misregulation of receptor turnover, which usually results in upregulation of receptor signaling. Fibroblasts from CHMP2BIntron5 patients were shown to have decreased receptor sequestering and showed improper formation of some ILVs in MVBs. CHMP2BIntron5 transfected neuroblastomas showed reduced endosome-lysosome fusion as well as a decrease in degradation of EGFR (Urwin et al., 2010). Studies using Drosophila models of CHMP2BIntron5-mediated FTD showed upregulation of Toll (Ahmad et al., 2009), Notch (Cheruiyot et al., 2014), and TGF-β and JNK signaling (West et al., 2015). FTD-GRN and bvFTD patients and a CHMP2BIntron5 mouse model had an increase in the AMPA receptor subunits Gria2, 3, and 4 (Gascon et al., 2014). CHMP2BIntron5 mice showed social deficits and an age-dependent down regulation of miR-124, which caused up-regulation of Gria2, 3, and 4. Similar molecular changes were seen in induced pluripotent stem cell (iPSC) derived neurons and bvFTD patient brain samples. Furthermore, overexpression of miR-124 and Gria2 knockdowns partially recued the social deficits in mice (Gascon et al., 2014).
CHMP2B also plays a role in the autophagosome-lysosome pathway. Consequently, CHMP2BIntron5-mediated deficits include abnormal aggregation of autophagosome-related membranes and proteins. This causes accumulation of autophagosomes and amphisomes that contain ubiquitinated proteins that can cause neurodegeneration (Filimonenko et al., 2007; Lee et al., 2007; Rusten et al., 2012). Expression of CHMP2BIntron5 may also inhibit sealing of the phagophore in the formation of autophagosomes because it produces aberrant multilamellar structures. This was seen in CHMP2BIntron5 expressing HeLa cells and a necrosis phenotype resulting from this phenotype was seen in CHMP2BIntron5 expressing Drosophila eyes (Lu et al., 2013).
Mouse models of FTD-3 have also been useful in exploring this autophagy phenotype. Two mouse models expressing human CHMP2BIntron5 have been created using different methods but both display many of the same phenotypes (Gascon et al., 2014; Ghazi-Noori et al., 2012). Both mouse models displayed increased p62 (an autophagy marker) levels in much of the brain as well as ubiquitin deposits. Because of these autophagy-related aggregates, both models also showed recruitment of astroglia to the site resulting in astrogliosis (Clayton et al., 2015; Gascon et al., 2014; Ghazi-Noori et al., 2012). An additional endolysosomal and autophagy related phenotype observed in the mouse neurons was the swelling of axons due to accumulation of membrane bound vessels of both of these origins (Ghazi-Noori et al., 2012).
Recently, these CHMP2BIntron5 expressing mice from (Ghazi-Noori et al., 2012) were found to contain large autofluorescent aggregates distinct from normal age-related accumulation of lipofuscin. They were found throughout the neurons, astrocytes, and microglia in the cortex and thalamus and worsened with age. They did not colocalize with p62, but were able to be marked by late endosomal markers and had similar morphology to MVBs. This seems to indicate a lysosomal storage pathology in CHMP2BIntron5 expressing HeLa cells as well as FTD-3 patient frontal neurons, but not AD patients (Clayton et al., 2015).
5. Future Perspectives
Since the pathological function of CHMP2BIntron5 in FTD-3 was first characterized just ten years ago (Skibinski et al., 2005), the field has made substantial progress in investigating the mechanism of this neurodegenerative disorder. However, many questions remain unanswered. For example, research using the Drosophila model to conduct a genetic screen identified several dominant enhancers (i.e. loss of one copy of a modifier gene causes a worsened phenotype) of CHMP2BIntron5 phenotype (Ahmad et al., 2009). Characterization of the most robust modifiers from this screen has shown that CHMP2BIntron5 pathology can result from faulty MVB formation leading to misregulation of Notch (Cheruiyot et al., 2014), Toll (Ahmad et al., 2009), TGF-β and JNK signaling (West et al., 2015), and Syntaxin13 (Lu et al., 2013). This work creates the impetus to characterize other modifiers through genetic screens, especially recessive modifiers. Because many of the known modifiers function through signaling pathways, an area of further investigation is the identification of how many signaling pathways are affected by ESCRT-III pathology and the relative contribution of these pathways to the FTD-3 symptoms.
The CHMP2BIntron5-mediated misregulation of diverse signaling pathways raises a broader question of why this mutant isoform only leads to FTD-3 in late, pre-senile adulthood when ESCRTs clearly regulate processes involved in early development (Rusten et al., 2012; Xu et al., 2012). It is also curious why only the brain, and only a subset of neurons, is affected by loss of ESCRT function, which controls essential processes in nearly all cell types. What makes the neurons in the frontal and anterior temporal lobes more sensitive to this ESCRT pathology?
Another future direction is to further elucidate and consolidate the role of ESCRTs and CHMP2B in autophagy and endolysosomal pathways. The ability to control and target CHMP2BIntron5 expression in animal models allows identification of interacting proteins in these processes. Recent work has used a transgenic eye expression model to show that syntaxin13 functions in the autophagy pathway (Lu et al., 2013) and Rab8 mediates signaling pathways through the endolysosomal pathway (West et al., 2015). Further studies could be done in this system to identify targets for ESCRT-III mediated pathology and to further clarify the interactions that are required for the function of these pathways. A further use of such a transgenic model is to investigate the effect of CHMP2BIntron5 on other physiological processes relevant to neurodegeneration, such as circadian rhythms. This is relevant after recent reports that circadian deficits are a common symptom for patients with many neurodegenerative disorders including FTD (Anderson et al., 2009)for recent reviews see (Musiek et al., 2015) and (Videnovic et al., 2014).
There are now at least two mouse models of CHMP2BIntron5-mediated FTD-3 (Gascon et al., 2014; Ghazi-Noori et al., 2012). So far, both models have mainly been used to describe the pathology created by the mutant isoform in more detail (Clayton et al., 2015; Gascon et al., 2014; Ghazi-Noori et al., 2012). However, recent results from Gascon et al. have suggested an interesting connection between CHMP2BIntron5 and the miRNA pathways as a potential cause of the phenotypes seen in FTD-3 (Gascon et al., 2014). These models can now be used in more translational studies to develop treatments for FTD.
6. Conclusions
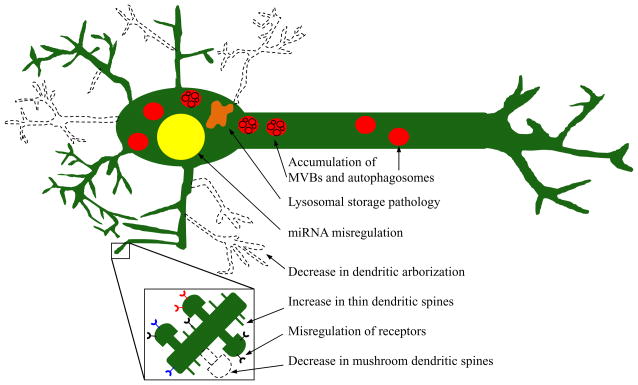
CHMP2BIntron5 causes a variety of molecular and cellular defects largely by perturbing intracellular vesicle formation due to the lack of the MIM domain for Vps4 binding. This includes the pathology it was originally characterized for, the improper trafficking of MVBs and autophagosomes to the lysosome. However, there are also many other deficits related to CHMP2BIntron5 expression, especially seen in neurons. These include alterations in miRNA expression, changes in neural spine number and type causing weakened synaptic transmission, and a decrease in dendritic arborization (Figure 2). Drosophila and mouse models have arisen to use this mutation to model specific aspects of neurodegenerative diseases and to further identify the molecular mechanisms of CHMP2BIntron5 pathology.
Figure 2.
CHMP2BIntron5 causes a diverse array of neuronal pathologies. The known molecular and structural bases of this neurodegeneration include: misregulation of miRNA, accumulation of autophagosomes and MVBs, abnormally sized lysosomes, a decrease in dendritic arborization, an overall increase in dendritic spines but with a decrease in mushroom spines, and misregulation of receptors such as Notch, and EGFR. These pathologies combined not only weaken the cell, but also leads to death of many frontal and temporal neurons.
Highlights.
We review the structure and function of CHMP2B.
We review the pathology caused by the CHMP2BIntron5 mutation.
We discuss the future of current CHMP2BIntron5 model systems.
Acknowledgments
The authors would like to thank Peter Joshua Kavaler for comments on the manuscript. STA is supported by an investigator grant of Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P20GM0103423).
Abbreviations
- CHMP2B
charged multivesicular body protein 2B
- ESCRT-III
endosomal complex required for transport-III
- FTD
frontotemporal dementia
- iPSC
induced pluripotent stem cell
- bvFTD
behavioral variant FTD
- PPA
primary progressive aphasia
- nfvPPA
nonfluent variant PPA
- SD
semantic dementia
- lvPPA
logopenic variant PPA
- PSP
progressive supranuclear palsy
- CBD
corticobasal degeneration
- AGD
argyrophilic disease
- ALS
amyotrophic lateral sclerosis
- FTD-3
FTD linked to chromosome 3
- C9ORF72
chromosome 9 open reading frame 72
- PGRN
progranulin
- MAPT
microtubulin associated protein Tau
- TDP-43
transactive response DNA-binding protein 43
- FTD-U
FTD-ubiquitin
- PMA
primary muscular atrophy
- MIT
microtubule-interacting and transport
- MIM
MIT-interacting motif
- MVB
multi-vesicular body
- ILV
intraluminal vesicle
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad ST, et al. Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc Natl Acad Sci U S A. 2009;106:12168–73. doi: 10.1073/pnas.0903134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad ST, Lee J-A. Molecular Mechanisms Underlying the Role of Autophagy in Neurodegenerative Diseases. 2014:45–59. [Google Scholar]
- Anderson KN, et al. Disrupted sleep and circadian patterns in frontotemporal dementia. Eur J Neurol. 2009;16:317–23. doi: 10.1111/j.1468-1331.2008.02414.x. [DOI] [PubMed] [Google Scholar]
- Bajorek M, et al. Structural basis for ESCRT-III protein autoinhibition. Nature Structural & Molecular Biology. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Belly A, et al. CHMP2B mutants linked to frontotemporal dementia impair maturation of dendritic spines. J Cell Sci. 2010;123:2943–54. doi: 10.1242/jcs.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benussi A, Padovani A, Borroni B. Phenotypic Heterogeneity of Monogenic Frontotemporal Dementia. Front Aging Neurosci. 2015;7:171. doi: 10.3389/fnagi.2015.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodon G, et al. Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem. 2011;286:40276–86. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroni B, et al. Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol Aging. 2014;35:934 e7–10. doi: 10.1016/j.neurobiolaging.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr Opin Neurobiol. 2007;17:381–6. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Chassefeyre R, et al. Regulation of postsynaptic function by the dementia-related ESCRT-III subunit CHMP2B. J Neurosci. 2015;35:3155–73. doi: 10.1523/JNEUROSCI.0586-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheruiyot A, et al. Expression of mutant CHMP2B, an ESCRT-III component involved in frontotemporal dementia, causes eye deformities due to Notch misregulation in Drosophila. FASEB J. 2014;28:667–75. doi: 10.1096/fj.13-234138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N, et al. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell. 2015;163:866–79. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton EL, et al. Frontotemporal dementia caused by CHMP2B mutation is characterised by neuronal lysosomal storage pathology. Acta Neuropathol. 2015 doi: 10.1007/s00401-015-1475-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LE, et al. Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS) PLoS One. 2010;5:e9872. doi: 10.1371/journal.pone.0009872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- De Silva D, et al. Motor function and behaviour across the ALS-FTD spectrum. Acta Neurol Scand. 2015 doi: 10.1111/ane.12471. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon E, Gao FB. The emerging roles of microRNAs in the pathogenesis of frontotemporal dementia-amyotrophic lateral sclerosis (FTD-ALS) spectrum disorders. J Neurogenet. 2014;28:30–40. doi: 10.3109/01677063.2013.876021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon E, et al. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat Med. 2014;20:1444–51. doi: 10.1038/nm.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim M, et al. CHMP2B mutations are rare in French families with frontotemporal lobar degeneration. J Neurol. 2010;257:2032–6. doi: 10.1007/s00415-010-5655-8. [DOI] [PubMed] [Google Scholar]
- Ghazi-Noori S, et al. Progressive neuronal inclusion formation and axonal degeneration in CHMP2B mutant transgenic mice. Brain. 2012;135:819–32. doi: 10.1093/brain/aws006. [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, et al. Evolution and assembly of ESCRTs. Biochem Soc Trans. 2009;37:151–5. doi: 10.1042/BST0370151. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, et al. Loss of TBK1 is a frequent cause of frontotemporal dementia in a Belgian cohort. Neurology. 2015;85 doi: 10.1212/WNL.0000000000002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorno-Tempini ML, et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:337–62. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz H, et al. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooli BV, et al. Rare autosomal copy number variations in early-onset familial Alzheimer’s disease. Molecular Psychiatry. 2014;19:676–681. doi: 10.1038/mp.2013.77. [DOI] [PubMed] [Google Scholar]
- Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–87. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton M, et al. Association of missense and 5’-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- Isaacs AM, et al. Frontotemporal dementia caused by CHMP2B mutations. Current Alzheimer Research. 2011;8:246–251. doi: 10.2174/156720511795563764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattante S, et al. Defining the genetic connection linking amyotrophic lateral sclerosis (ALS) with frontotemporal dementia (FTD) Trends Genet. 2015;31:263–73. doi: 10.1016/j.tig.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Lee JA, et al. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–7. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Lee JA, Gao FB. ESCRT, autophagy, and frontotemporal dementia. BMB Reports. 2008;41:827–832. doi: 10.5483/bmbrep.2008.41.12.827. [DOI] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, et al. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol Cell. 2013;52:264–71. doi: 10.1016/j.molcel.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunau L, et al. Presymptomatic cerebral blood flow changes in CHMP2B mutation carriers of familial frontotemporal dementia (FTD-3), measured with MRI. BMJ Open. 2012;2:e000368. doi: 10.1136/bmjopen-2011-000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, et al. Frequency of the C9orf72 hecanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurology. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musiek ES, Xiong DD, Holtzman DM. Sleep, circadian rhythms, and the pathogenesis of Alzheimer disease. Exp Mol Med. 2015;47:e148. doi: 10.1038/emm.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Odorizzi G. Membrane manipulations by the ESCRT machinery. F1000Res. 2015;4:516. doi: 10.12688/f1000research.6319.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos Y, et al. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–9. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Chen X. Clinical, neuropathology and molecular genetics of frontotemporal dementia: a mini-review. Translational Neurodegeneration. 2013;2 doi: 10.1186/2047-9158-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piguet O, et al. Behavioural-variant frontotemporal dementia: diagnosis, clinical staging, and management. The Lancet Neurology. 2011;10:162–172. doi: 10.1016/S1474-4422(10)70299-4. [DOI] [PubMed] [Google Scholar]
- Pottier C, et al. Whole-genome sequencing reveals important role for TBK1 and OPTN mutations in frontotemporal lobar degeneration without motor neuron disease. Acta Neuropathol. 2015;130:77–92. doi: 10.1007/s00401-015-1436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers R, Cruts M, van Broeckhoven C. The role of tau (MAPT) in frontotemporal dementia and related tauopathies. Hum Mutat. 2004;24:277–95. doi: 10.1002/humu.20086. [DOI] [PubMed] [Google Scholar]
- Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol. 2012;8:423–34. doi: 10.1038/nrneurol.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED. Mouse models of frontotemporal dementia. Ann Neurol. 2012;72:837–49. doi: 10.1002/ana.23722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodahl LM, et al. The role of ESCRT proteins in attenuation of cell signalling. Biochem Soc Trans. 2009;37:137–42. doi: 10.1042/BST0370137. [DOI] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Stenmark H. Shaping development with ESCRTs. Nat Cell Biol. 2012;14:38–45. doi: 10.1038/ncb2381. [DOI] [PubMed] [Google Scholar]
- Scheffer LL, et al. Mechanism of Ca(2)(+)-triggered ESCRT assembly and regulation of cell membrane repair. Nat Commun. 2014;5:5646. doi: 10.1038/ncomms6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Teis D. The ESCRT machinery. Current Biology. 2012;22:R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh AL, Audhya A. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol. 2014;49:242–61. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieben A, et al. The genetics and neuropathology of frontotemporal lobar degeneration. Acta Neuropathol. 2012;124:353–72. doi: 10.1007/s00401-012-1029-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37:806–8. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- Snowden JS, Neary D, Mann DMA. Frontotemporal dementia. British Journal of Psychiatry. 2002;180:140–143. doi: 10.1192/bjp.180.2.140. [DOI] [PubMed] [Google Scholar]
- Stewart H, et al. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol. 2012;123:409–17. doi: 10.1007/s00401-011-0937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, et al. Unravelling the pivotal role of Alix in MVB sorting and silencing of the activated EGFR. Biochemical Journal. 2015;466:475–487. doi: 10.1042/BJ20141156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkont J, et al. Coiled-coil length: Size does matter. Proteins. 2015 doi: 10.1002/prot.24932. [DOI] [PubMed] [Google Scholar]
- Talbot K, Ansorge O. Recent advances in the genetics of amyotrophic lateral sclerosis and frontotemporal dementia: common pathways in neurodegenerative disease. Hum Mol Genet. 2006;15(2):R182–7. doi: 10.1093/hmg/ddl202. [DOI] [PubMed] [Google Scholar]
- Thompson BJ, et al. Tumor suppressor properties of the ESCRT-II complex component Vps25 in Drosophila. Dev Cell. 2005;9:711–20. doi: 10.1016/j.devcel.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Urwin H, et al. Disruption of endocytic trafficking in frontotemporal dementia with CHMP2B mutations. Hum Mol Genet. 2010;19:2228–38. doi: 10.1093/hmg/ddq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee J, et al. CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum Mol Genet. 2008;17:313–22. doi: 10.1093/hmg/ddm309. [DOI] [PubMed] [Google Scholar]
- Videnovic A, et al. ‘The clocks that time us’–circadian rhythms in neurodegenerative disorders. Nature Reviews Neurology. 2014;10:689–693. doi: 10.1038/nrneurol.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RJ, et al. Rab8, POSH, and TAK1 regulate synaptic growth in a Drosophila model of frontotemporal dementia. J Cell Biol. 2015;208:931–47. doi: 10.1083/jcb.201404066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. Population-specific regulation of Chmp2b by Lbx1 during onset of synaptogenesis in lateral association interneurons. PLoS One. 2012;7:e48573. doi: 10.1371/journal.pone.0048573. [DOI] [PMC free article] [PubMed] [Google Scholar]