Abstract
BACKGROUND
Level 1 evidence demonstrates increased overall survival with cisplatin-based neoadjuvant chemotherapy for patients with muscle-invasive urothelial cancer. Utilization remains low, however, in part because neoadjuvant chemotherapy is not effective for every patient. To identify patients most likely to benefit, we evaluated germline pharmacogenomic markers for association with neoadjuvant chemotherapy sensitivity in two large cohorts of urothelial cancer patients.
PATIENTS AND METHODS
Patients receiving neoadjuvant cisplatin-based chemotherapy for muscle-invasive urothelial cancer were eligible. Nine germline single nucleotide polymorphisms (SNPs) potentially conferring platinum sensitivity were tested for association with complete pathologic response to neoadjuvant chemotherapy (pT0) or elimination of muscle-invasive cancer (<pT2).
RESULTS
205 patients were analyzed—59 in the discovery set and 146 in an independent replication cohort—from three institutions. pT0 (26%) and <pT2 (50%) rates were consistent across discovery and replication populations. Using a multivariate recessive genetic model, rs244898 in RARS (odds ratio [OR]=6.8 [95% CI 1.8–28.9], P=0.006) and rs7937567 in GALNTL4 (OR=4.8 [95% CI 1.1–22.6], P=0.04) were associated with pT0 in the discovery set. Despite these large effects, neither associated with achievement of pT0 in the replication. A third SNP, rs10964552, was associated with <pT2 in the discovery set, but also failed to replicate.
CONCLUSION
Germline SNPs previously associated with platinum sensitivity were not associated with neoadjuvant chemotherapy response in a large replication cohort of urothelial cancer patients. These results emphasize the need for replication when evaluating pharmacogenomic markers, and demonstrate that multi-institutional efforts are feasible and will be necessary to achieve advances in urothelial cancer pharmacogenomics.
Keywords: bladder cancer, urothelial cancer, cisplatin sensitivity, polymorphisms, pharmacogenomics
MicroAbstract
To identify urothelial cancer patients most likely to benefit from neoadjuvant chemotherapy, we evaluated germline pharmacogenomic markers for association with response in 205 patients across three institutions. pT0 (26%) and <pT2 (50%) rates were consistent across respective discovery and replication cohorts. Despite large effects for three polymorphisms in the discovery set, none associated with achievement of pT0 or <pT2 on replication. Multi-institutional efforts are feasible and will be necessary to achieve advances in urothelial cancer precision medicine.
INTRODUCTION
Despite level 1 evidence demonstrating survival benefit for cisplatin-based neoadjuvant chemotherapy in urothelial cancer1–3, its utilization has historically been low4–6. Indeed, cisplatin-based neoadjuvant chemotherapy is not effective for every patient—approximately half will demonstrate disease down-staging to non-muscle-invasive disease, and approximately one-third will achieve a complete pathologic response1,7–9. However, in those that do achieve a complete pathologic response (pT0), overall survival is dramatically improved independent of initial clinical stage or other clinical factors: 85% of those attaining pT0 are alive at 5 years, compared to 45% of those not achieving a complete response1. The likelihood of achieving pT0 is about 2.5 times higher with receipt of neoadjuvant chemotherapy1.
These data invite the proposition that the neoadjuvant setting is an ideal clinical niche in which to investigate predictive chemotherapy-response biomarkers, with the goals being better patient selection leading to an improved therapeutic index10. Patients unlikely to respond to cisplatin-based therapy could proceed directly to cystectomy or be considered for novel neoadjuvant treatments.
Our project sought to apply rapidly-evolving genomic knowledge to this question, with the hypothesis that germline genetic polymorphisms are potentially important predictors of cisplatin response in urothelial cancer. Most prior studies in bladder cancer have focused on tumor genomics (i.e., somatic mutations such as p53 and ERCC1/2) rather than germline genetic variation (inherited DNA polymorphisms) as determinants of chemotherapy response. Yet the importance of germline polymorphisms in governing drug levels/disposition, toxicity, and response has long been recognized in oncology (TPMT polymorphisms with 6-mercaptopurine and UGT1A1 polymorphisms with irinotecan are salient examples)11. In bladder cancer, we previously examined a large list of germline polymorphisms from candidate genes hypothesized to have effects on cisplatin sensitivity and tested these in a heterogeneous population of platinum-treated patients12. While several SNPs were correlated with response, the findings were not replicated12, and the model did not focus on the uniquely relevant neoadjuvant setting.
Given the key role of cisplatin in the treatment of urothelial cancer, the question of genetic predisposition to cisplatin-based chemotherapy deserves attention as one of high clinical importance. In this study, we sought to identify and replicate novel germline polymorphisms of interest of cisplatin response in two large populations of urothelial cancer patients receiving cisplatin-based neoadjuvant chemotherapy, with pathologic disease response in the surgical specimen as the primary endpoint.
PATIENTS AND METHODS
Patients
Members of the institutions participating in this project (Fox Chase Cancer Center [FCCC], Memorial Sloan Kettering Cancer Center [MSKCC], and The University of Chicago [Chicago]) collected germline DNA samples and clinical follow-up data from urothelial cancer patients treated with neoadjuvant chemotherapy, under respective institutional review board-approved protocols, including a study funded and designed specifically for this purpose (clinicaltrials.gov#NCT01206426). To be included, patients must have had muscle-invasive urothelial carcinoma (≥cT2) and must have received ≥3 cycles of chemotherapy in the neoadjuvant setting consisting of a regimen with either gemcitabine/cisplatin (GC) or methotrexate/vinblastine/doxorubicin/cisplatin (MVAC), and must have received definitive surgery (bladder, upper tract, and urethra primarys permitted). Patients with pure variant histologies were excluded (mixed histologies were included as long as the predominant component was urothelial carcinoma). Patients with clinically apparent positive nodes prior to neoadjuvant chemotherapy were excluded. Germline DNA was isolated from peripheral blood (Chicago, FCCC) or saliva (MSKCC). In assembling discovery and replication cohorts, enrolled patients with germline DNA that was already extracted and ready for analysis were included in the first (discovery) cohort (all from MSKCC). Remaining patients were by definition included in the replication cohort, including patients from Chicago, FCCC, and MSKCC patients not included in the discovery cohort.
SNP Selection
Prior germline investigation of platinum sensitivity has centered primarily on candidate genes—genes hypothesized to modulate cisplatin sensitivity because of their putative role in the drug’s mechanism of action. These efforts have largely focused on genes involved in DNA repair13,14. Such studies, including those in urothelial cancer, have been unable to consistently replicate any germline polymorphisms. We therefore intended to apply a different approach to the question by using genome-wide methods to select SNPs for testing – thus not confining analysis to the supposition that important platinum sensitivity SNPs are located in ‘traditional’ candidate genes.
We previously utilized and refined a novel cell-based genome-wide method to identify germline genetic variants governing chemotherapy susceptibility15, specifically for platinum drugs16,17. This in vitro model employs well-genotyped lymphoblastoid cell lines (LCLs) from healthy individuals in the International HapMap Project18, which were then treated with platinum to produce individual “sensitivity phenotypes”. Then, genome-wide association studies (GWAS) were performed to associate platinum susceptibility with specific SNPs. Associating SNPs represent potentially novel genetic determinants of platinum sensitivity, identified from across the genome (unbiased approach) and often in genomic regions not previously implicated.
We selected 10 SNPs having the highest quality associations from these prior studies for testing in the current study. Five of these (rs2191934, rs9527419, rs244903, rs7210837, rs3893319) were strongly associated in a large cell-based genome-wide meta-analysis of n=608 human germline DNA samples treated with platinum compounds to determine sensitivities17. All five were among the top statistical signals, with rs2191934 (meta P= 8.3 × 10−5) and rs9527419 (meta P=5.8 × 10−6) specifically found to (distantly) regulate the expression of GSTT1, ERCC6, and ERCC2, respectively, although the SNPs themselves were not located in any of these genes17 which is potentially why they were missed by traditional candidate gene analyses. Separately, rs7937567 was identified and replicated in a prior cell-based genome-wide study16, and is located in an intron of GALNTL4 which was implicated twice in separate cell-based platinum sensitivity studies16,19, the latter of which also implicated rs2136241 (CDCA1 promoter SNP) and rs1649942 (intron of NRG3) both of which we selected. rs1649942 was also shown to be significantly associated with survival in a clinical cohort of carboplatin-treated ovarian cancer patients20. rs10964552, located in MLLT3, was selected as it was found as a top signal in a prior cell-based genome-wide study and was shown to regulate expression of HIST1H3A, a histone component, with higher HIST1H3A levels associated with platinum resistance16. Finally, rs6870861 was associated with platinum sensitivity in both a large cell-based genome-wide study and a cohort of head and neck cancer patients21, and SNPs in linkage disequilibrium were shown to trans-regulate SLC22A5 which is a member of the organic cation transporter family intensively studied in platinum handling21.
Genotyping
Genotyping was performed using MassARRAY iPLEX system (Sequenom, Inc.). For rs244903, a SNP in complete linkage disequilibrium in Caucasians (rs244898) was genotyped as the proxy due to design limitations with rs244903. One SNP (rs7210837) was unable to be successfully genotyped because of primer failure, leaving 9 SNPs tested in all patients. As an assessment of DNA quality, call rates across the tested SNPs in the discovery and replication sets exceeded 97.7% and 98.8%, respectively.
Phenotype Definition and Association Analysis
We evaluated the association of the pre-selected germline pharmacogenomic markers with neoadjuvant therapy outcome. For each platinum susceptibility SNP of interest, pathologic disease response at surgery was compared between individuals based on genotype at that SNP, allowing identification of genotypes associated with cisplatin susceptibility/resistance. Association with complete pathologic response (pT0 rate) at surgery was the primary endpoint. Down-staging after neoadjuvant chemotherapy (rate of <pT2) was the secondary endpoint. Surgical staging was assigned by dedicated pathologists for clinical purposes with no knowledge of patients’ genomic information. Personnel performing genotyping were blinded to surgical outcomes until after assigning genotypes.
Statistical Analysis
In the discovery cohort, univariate and multivariate logistic regression analyses were conducted to investigate association between each SNP and pT0/<pT2 rates. Recessive, dominant, and additive genetic models were tested. Since replication in a second, independent population was conducted, the P value for nominal significance in the discovery set was not adjusted for multiple testing correction and was chosen as P<0.05, the threshold for a SNP to be considered promising and thus carried forward for testing in the replication set. Before replication testing, formal sample size analysis was undertaken to determine replication cohort size needed to have adequate power to replicate SNPs. Using 10,000 simulated datasets, it was calculated that ≥134 patients in the replication cohort would provide 80% power to detect effects of SNPs selected independently from the discovery cohort at P=0.05, assuming odds ratios and minor allele frequencies equivalent to those in the discovery.
It was pre-specified that in the validation, even if the genetic relationship did not necessarily follow the same (e.g. recessive) model, we would still examine for genetic effects by using log-additive and dominant models; the power of these analyses was expected to be even greater.
In both the discovery and replication sets, secondary analysis was also pre-specified to test the association between response rate and combination of SNPs (number of favorable genotypes carried in each patient), using a trend test. In this, the number of favorable genotypes carried by each patient was considered. In the simplest 2-SNP model (which was ultimately employed), each patient was coded as carrying either both, one and only one, or neither favorable genotype, and incorporated as t=(2, 1, 0) in an additive model22.
To eliminate confounding via population stratification23, only samples from self-identified Caucasians were included in the primary analyses (discovery and replication). All SNPs were in Hardy-Weinberg equilibrium.
RESULTS
Discovery Population
For first analysis of the nine pre-identified SNPs, platinum sensitivity (as defined by pT0 and <pT2 rates, respectively) was tested in a single-institution discovery cohort of 59 patients. The clinical features of this cohort are shown in Table 1 (left side). The pathologic complete response (pT0) rate in this cohort was 25.4%. Fifty-two percent (52%) of patients were down-staged (<pT2) at surgery after receiving chemotherapy.
Table 1.
Clinical Characteristics of the Discovery and Validation Populations
| Discovery Cohort (N=59) | Validation Cohort (N=146) | |||
|---|---|---|---|---|
| Variable | Categories | N (%) | Categories | N (%) |
| Age | Median Range |
64 years 31 – 86 |
Median Range |
65 years 32 – 83 |
| Gender | Male Female |
40 (68%) 19 (32%) |
Male Female |
104 (71%) 42 (29%) |
| Institution | MSKCC | 59 (100%) | MSKCC Fox Chase Univ Chicago |
92 (63%) 33 (23%) 21 (14%) |
| Primary Site | Bladder | 59 (100%) | Bladder Upper Tract Synchronous |
141 (97%) 4 (3%) 1 (<1%) |
| Clinical Stage |
≥cT2, N0 | 59 (100%) | ≥cT2, N0 | 146 (100%) |
| Treatment | GC GC+sunitinib MVAC |
55 (93%) 3 (5%) 1 (2%) |
GC GC+sunitinib DD GC GC→gem/carbo DD MVAC |
89 (61%) 7 (5%) 7 (5%) 1 (<1%) 42 (29%) |
| Pathologic Response |
pT0 <pT2 ≥pT2 |
15 (25%) 16 (27%) 28 (48%) |
pT0 <pT2 ≥pT2 |
38 (26%) 35 (24%) 73 (50%) |
Note: To eliminate confounding via population stratification, only samples from self-identified Caucasians were included in the primary analyses (discovery and replication).
Abbreviations: GC = gemcitabine/cisplatin; MVAC = methotrexate/vinblastine/adriamycin/cisplatin; gem = gemcitabine; carbo = carboplatin; DD = dose dense; MSKCC = Memorial Sloan Kettering Cancer Center
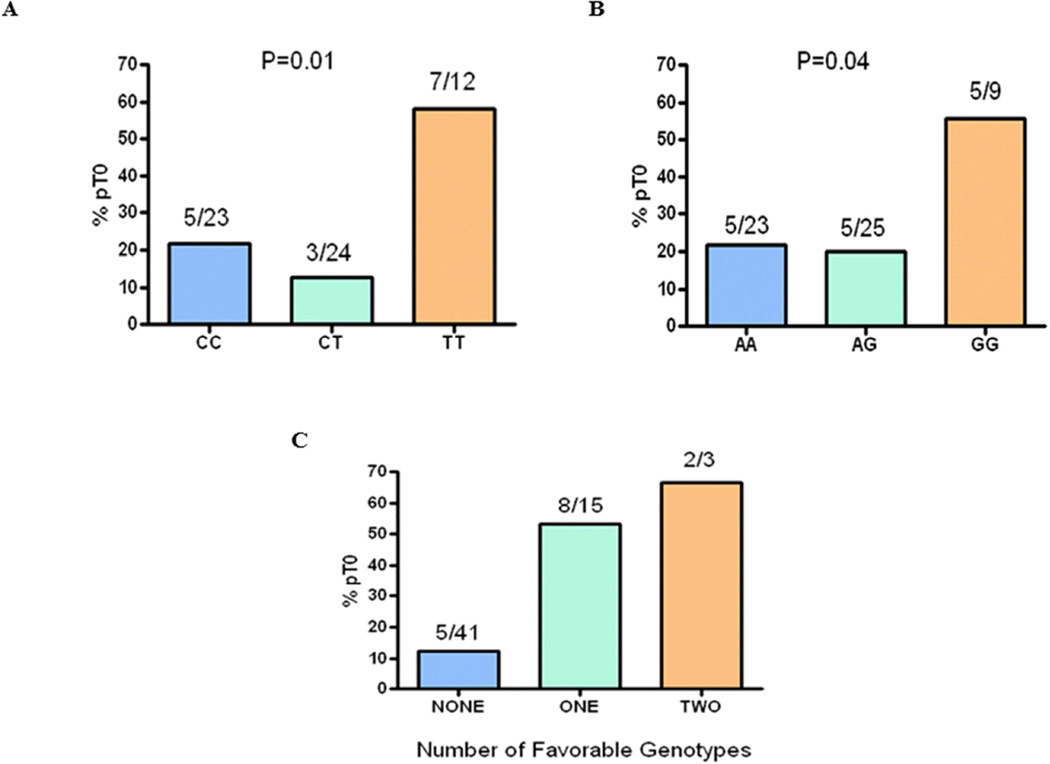
Using a recessive genetic model, rs244898 in RARS (odds ratio [OR] 6.8 [95% CI 1.8–28.9], univariate P=0.006) and rs7937567 in GALNTL4 (OR 4.8 [95% CI 1.1–22.6], univariate P=0.04) were associated with likelihood of achieving pT0. For each SNP, patients carrying the favorable genotype achieved pT0 in >56% of cases (Figure 1A and 1B). Demonstrating the apparent independent nature of the two SNPs, patients carrying either favorable genotype had pT0 OR=8.5 (95% CI 2.5–31.8, P=0.0008). The combined effect of testing for both SNPs was also highly informative, as 2 of the 3 patients with both favorable genotypes achieved pT0 (67%), compared to 8 of 15 patients with one favorable genotype (53%), and only 5 of 41 achieving pT0 among those who lacked both favorable genotypes (12%) (Figure 1C). The negative predictive value considering both SNPs in the discovery cohort was 88%.
Figure 1. Positive Associations of SNPs in the Discovery Population (n=59).
(A) Association of rs244898 with pT0 rate, using a recessive genetic model. Patients carrying both favorable alleles (TT) had a pT0 rate of 58%. rs244898 is an intron SNP in RARS with a frequency in Caucasians of 0.44. rs244898 is in linkage disequilibrium with an exon missense SNP for this gene.
(B) Association of rs7937567 with pT0 rate, using a recessive genetic model. Patients carrying both favorable alleles (GG) had a pT0 rate of 56%. rs7937567 is an intron SNP in GALNTL4 with a frequency in Caucasians of 0.42.
(C) Two-SNP model incorporating both rs244898 and rs7937567 examining the effect of zero, one, or two favorable genotypes on pathologic response rate (pT0). Patients with either the rs244898 TT genotype or the rs7937567 GG genotype had a >50% rate of pT0. Patients lacking both favorable genotypes were highly unlikely to achieve pT0 (12%).
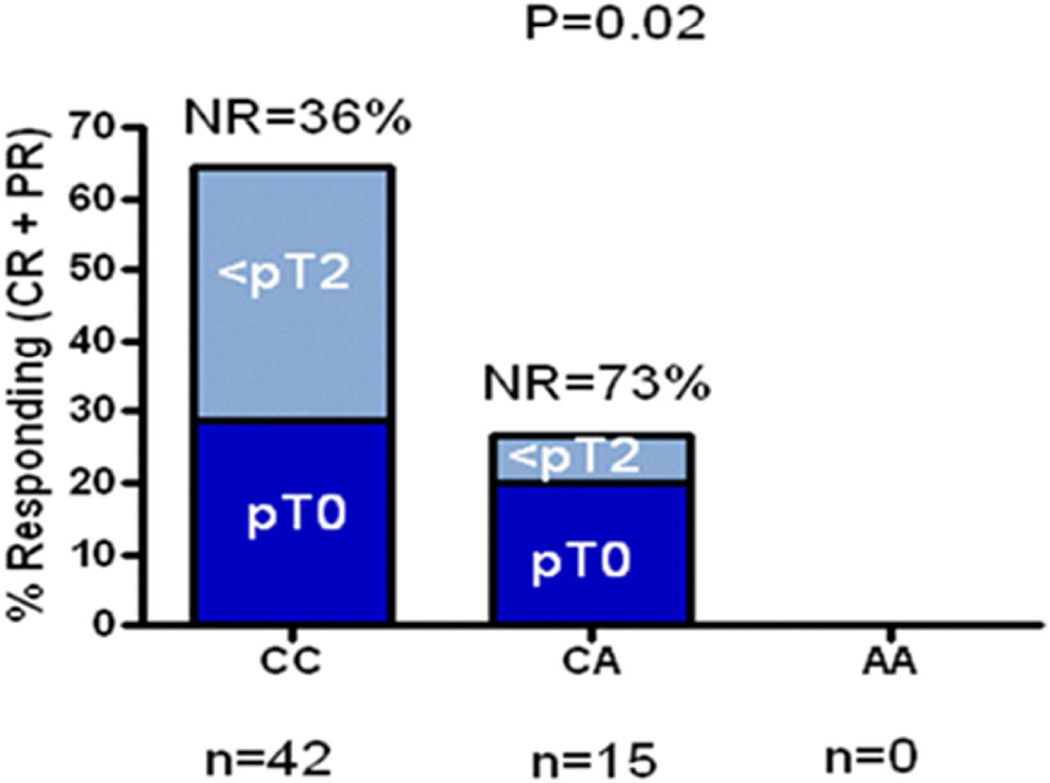
For analysis of the secondary endpoint (<pT2), one SNP was significantly associated with down-staging at time of cystectomy (using an additive genetic model): rs10964552 (in MLLT3) with OR 5.0 (95% CI 1.4–20.5), univariate P=0.02. Expressed another way, the likelihood of non-response to cisplatin-based neoadjuvant therapy was significantly higher in patients carrying the A allele for rs10964552 (Figure 2).
Figure 2. Positive SNP Association with Down-staging at Cystectomy After Neoadjuvant Chemotherapy in the Discovery Population (n=59).
The likelihood of non-response to cisplatin-based neoadjuvant therapy was significantly higher in patients carrying the A allele of rs10964552. rs10964552 is an intron SNP in MLLT3 with a frequency in Caucasians of 0.17.
The full SNP association results including the direction of the clinical effect by allele for both the primary (pT0) and secondary (<pT2) clinical endpoints are shown in Table 2.
Table 2.
Summary of Genotype Association Results in the Discovery Cohort
| SNP | Recessive allele (frequency)* |
Association with pT0† | Association with <pT2 (downstaging)‡ |
||
|---|---|---|---|---|---|
| OR | P value | OR | P value | ||
| rs10964552 | A (0.13) | NA | NA | 0.20 | 0.017 |
| rs1649942 | C (0.20) | 0.00 | 0.994 | 1.02 | 0.963 |
| rs2136241 | C (0.46) | 0.82 | 0.795 | 1.08 | 0.832 |
| rs2191934 | T (0.47) | 0.42 | 0.300 | 1.26 | 0.535 |
| rs244898 | T (0.41) | 6.82 | 0.006 | 1.60 | 0.192 |
| rs3893319 | G (0.05) | NA | NA | 1.93 | 0.471 |
| rs6870861 | C (0.11) | 0.00 | 0.992 | 0.38 | 0.131 |
| rs7937567 | G (0.38) | 4.75 | 0.040 | 0.95 | 0.885 |
| rs9527419 | T (0.14) | NA | NA | 1.09 | 0.871 |
allele frequency in the discovery cohort of this study
OR= odds ratio
using recessive genetic model. OR are calculated using the major allele as the reference (comparator). NA: for these results, OR and P values are not provided since these were test instances where there were either zero or only one homozygous recessive patient present in the cohort. OR and P values for these situations in a recessive model were therefore undependable.
using additive genetic model; the OR is expressed for the impact of carriage of increasing copies of the minor allele
Replication Population
The two SNPs associated with pT0 and the third SNP associated with down-staging to <pT2 were then tested in the multi-institutional independent validation cohort of 146 patients (we were able to recruit even more patients than the 134 required by the minimum power calculation threshold for replication). The clinical features of the replication population are shown in Table 1 (right side). Rates of pT0 and <pT2 were 26% and 50%, respectively, comparable to the discovery population.
For analysis of the primary endpoint in the replication population, regression was performed on pT0 based on whether patients carried the favorable genotypes—TT for rs244898 and GG for rs7937567. Though each SNP had an odds ratio of effect of approximately 5 on pT0 in the discovery set, neither was associated with achievement of pT0 in the replication set (rs244898 replication cohort OR=1.1, P=0.79; rs7937567 replication cohort OR=0.6, P=0.42). The two SNPs combined (rs244898 and rs7937567 in one model) were also not significant.
The third SNP (rs10964552) which was associated with pathologic down-staging to <pT2 in the discovery set, also failed to replicate (replication cohort OR=0.9, P=0.69).
Given the possibility that differences in treatment regimens may have confounded replication (a much higher percentage of patients received dose dense MVAC in the replication cohort compared to the nearly exclusively GC-treated discovery cohort), we performed subanalysis of only the GC-treated validation cohort patients and nonetheless did not find replication of any SNPs. Genotype frequencies were not significantly different between the discovery and replication cohorts: rs244898 discovery/replication CC=0.39/0.26, CT=0.41/0.54, TT=0.20/0.20; rs7937567 AA=0.40/0.36, AG=0.44/0.50, GG=0.16/0.14; and rs10964552 CC=0.74/0.72, CA=0.26/0.26, AA=0.00/0.02.
DISCUSSION
Given that the chemotherapy survival benefit occurs in a minority of patients but that all patients are exposed to very substantial toxicities, clinical benefit in this population of patients would be markedly enhanced if we could restrict chemotherapy to patients most likely to benefit. The era of genomics offers a ripe avenue for this type of pursuit. Indeed, recent advances have begun to address this problem, identifying tumor-based genomic markers predictive of cisplatin-based chemotherapy response in bladder cancer24–28, including an elegant investigation performed in the neoadjuvant setting29. It is likely that both somatic and germline factors govern chemotherapy responses.
We investigated germline SNPs identified via unbiased GWAS approaches. These SNPs all had published evidence of association with platinum responsiveness, some with strong mechanistic plausibility16,17,19–21. Despite intriguing associations for three of these SNPs in the discovery population of the present study, none were replicated in our well-powered validation cohort.
Several reasons for this were considered. First, while the discovery and replication cohorts were similar in nearly all measured demographics, the discovery population was derived from a single institution, while the replication cohort was assembled from three centers. We viewed the latter as a strength, and indeed replication across institutions would have increased generalizability. However, lack of replication may reflect unmeasured clinical differences between cohorts, including patient or practice differences. There was a higher proportion of (dose dense) MVAC use in the validation set and this could have hindered replication through a mechanism by which platinum-specific genetic effects may be obscured in a four-drug regimen, although post-hoc analysis of only the GC-treated validation cohort patients did not find replication. Failure of replication could simply demonstrate that findings in the discovery cohort were spurious associations. This is certainly possible given the small size of the discovery cohort. (Perhaps consistent with this idea was the finding that the discovery SNPs associated with pT0 were not also found associated with pathologic downstaging (<pT2) in the discovery set). Although the SNPs tested were previously associated in other models, the relevance of those prior cell-based models to clinical treatment-response remains unproven.
Strengths of our study include the carefully defined and highly relevant clinical endpoint of complete pathologic response, prospective identification of samples for inclusion, and execution of a formal power analysis pre-determining the required sample size for replication prior to replication testing (to decrease the likelihood that negative findings were a result of an under-powered analysis).
Our goal is to improve overall survival while also sparing potential toxicity in individuals unlikely to benefit from therapies. One means of accomplishing this will be the development of newer, perhaps better-tolerated drugs. Simultaneously, we should continue to strive for a better understanding of genetic factors governing platinum-based chemotherapy in urothelial cancer, since even the advent of new therapies is only likely to add to this traditional backbone of therapy for this challenging disease.
CONCLUSIONS
We investigated germline SNPs implicated as potentially governing platinum responsiveness, but were unable to replicate association of these SNPs with achievement of pathologic response after neoadjuvant cisplatin-based chemotherapy for urothelial cancer in >200 treated patients from three institutions. Our results emphasize the importance of replication when evaluating pharmacogenomic markers. We demonstrated that multi-institutional collaborations are feasible and necessary to achieve advances in urothelial cancer pharmacogenomics. Through this existing collaboration, we are indeed now pursuing a follow-on genome-wide study to identify new germline polymorphisms of platinum chemotherapy response in urothelial carcinoma.
Clinical Practice Points.
Level 1 evidence demonstrates increased overall survival with cisplatin-based neoadjuvant chemotherapy for patients with muscle-invasive urothelial cancer. Utilization remains low, however, in part because neoadjuvant chemotherapy is not effective for every patient.
To improve selection of patients most likely to benefit, we evaluated germline pharmacogenomic markers for association with neoadjuvant chemotherapy sensitivity in two large cohorts of urothelial cancer patients.
Nine germline single nucleotide polymorphisms (SNPs) potentially conferring platinum sensitivity were tested for association with complete pathologic response to neoadjuvant chemotherapy (pT0) or elimination of muscle-invasive cancer (<pT2).
205 patients were analyzed—59 in the discovery set and 146 in an independent replication cohort—from three institutions.
pT0 (26%) and <pT2 (50%) rates were consistent across discovery and replication populations.
Using a multivariate recessive genetic model, rs244898 in RARS (odds ratio [OR]=6.8 [95% CI 1.8–28.9], P=0.006) and rs7937567 in GALNTL4 (OR=4.8 [95% CI 1.1–22.6], P=0.04) were associated with pT0 in the discovery set.
Despite these large effects, neither associated with achievement of pT0 in the replication. A third SNP, rs10964552, was associated with <pT2 in the discovery set, but also failed to replicate.
These results emphasize the need for replication when evaluating pharmacogenomic markers, and demonstrate that multi-institutional efforts are feasible and will be necessary to achieve advances in urothelial cancer pharmacogenomics.
Acknowledgments
Support for this study was provided by a Cancer Research Foundation Young Investigator Award (PHO) and by the Ken Carmel Family Cancer Research Fund (KO). PHO was also supported by a Paul Calabresi Career Development Award in Clinical Oncology (NIH K12 CA139160-01A1). Additional MSKCC support was provided by the Wiener Research and Therapeutics Research Program in Bladder Cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no relevant conflicts of interest to disclose.
REFERENCES
- 1.Grossman HB, Natale RB, Tangen CM, et al. Neoadjuvant chemotherapy plus cystectomy compared with cystectomy alone for locally advanced bladder cancer. N Engl J Med. 2003;349(9):859–866. doi: 10.1056/NEJMoa022148. [DOI] [PubMed] [Google Scholar]
- 2.Levano S, Ginz H, Siegemund M, et al. Genotyping the butyrylcholinesterase in patients with prolonged neuromuscular block after succinylcholine. Anesthesiology. 2005;102(3):531–535. doi: 10.1097/00000542-200503000-00009. [DOI] [PubMed] [Google Scholar]
- 3.International Collaboration of T, Medical Research Council Advanced Bladder Cancer Working P, European Organisation for R et al. International phase III trial assessing neoadjuvant cisplatin, methotrexate, and vinblastine chemotherapy for muscle-invasive bladder cancer: long-term results of the BA06 30894 trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29(16):2171–2177. doi: 10.1200/JCO.2010.32.3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowan NG, Chen Y, Downs TM, et al. Neoadjuvant chemotherapy use in bladder cancer: a survey of current practice and opinions. Advances in urology. 2014;2014:746298. doi: 10.1155/2014/746298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raj GV, Karavadia S, Schlomer B, et al. Contemporary use of perioperative cisplatin-based chemotherapy in patients with muscle-invasive bladder cancer. Cancer. 2011;117(2):276–282. doi: 10.1002/cncr.25429. [DOI] [PubMed] [Google Scholar]
- 6.David KA, Milowsky MI, Ritchey J, Carroll PR, Nanus DM. Low incidence of perioperative chemotherapy for stage III bladder cancer 1998 to 2003: a report from the National Cancer Data Base. The Journal of urology. 2007;178(2):451–454. doi: 10.1016/j.juro.2007.03.101. [DOI] [PubMed] [Google Scholar]
- 7.Dash A, Pettus JAt, Herr HW, et al. A role for neoadjuvant gemcitabine plus cisplatin in muscle-invasive urothelial carcinoma of the bladder: a retrospective experience. Cancer. 2008;113(9):2471–2477. doi: 10.1002/cncr.23848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choueiri TK, Jacobus S, Bellmunt J, et al. Neoadjuvant dose-dense methotrexate, vinblastine, doxorubicin, and cisplatin with pegfilgrastim support in muscle-invasive urothelial cancer: pathologic, radiologic, and biomarker correlates. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(18):1889–1894. doi: 10.1200/JCO.2013.52.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plimack ER, Hoffman-Censits JH, Viterbo R, et al. Accelerated methotrexate, vinblastine, doxorubicin, and cisplatin is safe, effective, and efficient neoadjuvant treatment for muscle-invasive bladder cancer: results of a multicenter phase II study with molecular correlates of response and toxicity. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(18):1895–1901. doi: 10.1200/JCO.2013.53.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Donnell PH. Bladder cancer pharmacogenomics: recent insights and future perspectives. Pharmacogenomics. 2012;13(14):1553–1556. doi: 10.2217/pgs.12.145. [DOI] [PubMed] [Google Scholar]
- 11.O'Donnell PH, Ratain MJ. Germline pharmacogenomics in oncology: decoding the patient for targeting therapy. Molecular oncology. 2012;6(2):251–259. doi: 10.1016/j.molonc.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher DJ, Vijai J, Hamilton RJ, et al. Germline single nucleotide polymorphisms associated with response of urothelial carcinoma to platinum-based therapy: the role of the host. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2013;24(9):2414–2421. doi: 10.1093/annonc/mdt225. [DOI] [PubMed] [Google Scholar]
- 13.Marsh S. Pharmacogenomics of taxane/platinum therapy in ovarian cancer. International journal of gynecological cancer : official journal of the International Gynecological Cancer Society. 2009;19(Suppl 2):S30–S34. doi: 10.1111/IGC.0b013e3181c10513. [DOI] [PubMed] [Google Scholar]
- 14.Wei SZ, Zhan P, Shi MQ, et al. Predictive value of ERCC1 and XPD polymorphism in patients with advanced non-small cell lung cancer receiving platinum-based chemotherapy: a systematic review and meta-analysis. Medical oncology. 2011;28(1):315–321. doi: 10.1007/s12032-010-9443-1. [DOI] [PubMed] [Google Scholar]
- 15.Huang RS, Duan S, Bleibel WK, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(23):9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Donnell PH, Gamazon E, Zhang W, et al. Population differences in platinum toxicity as a means to identify novel genetic susceptibility variants. Pharmacogenetics and genomics. 2010;20(5):327–337. doi: 10.1097/FPC.0b013e3283396c4e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler HE, Gamazon ER, Stark AL, et al. Genome-wide meta-analysis identifies variants associated with platinating agent susceptibility across populations. The pharmacogenomics journal. 2013;13(1):35–43. doi: 10.1038/tpj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International HapMap C. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 19.Huang RS, Duan S, Shukla SJ, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. American journal of human genetics. 2007;81(3):427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang RS, Johnatty SE, Gamazon ER, et al. Platinum sensitivity-related germline polymorphism discovered via a cell-based approach and analysis of its association with outcome in ovarian cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(16):5490–5500. doi: 10.1158/1078-0432.CCR-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ziliak D, O'Donnell PH, Im HK, et al. Germline polymorphisms discovered via a cell-based, genome-wide approach predict platinum response in head and neck cancers. Translational research : the journal of laboratory and clinical medicine. 2011;157(5):265–272. doi: 10.1016/j.trsl.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Human heredity. 2002;53(3):146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 23.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Human molecular genetics. 2008;17(R2):R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellmunt J, Paz-Ares L, Cuello M, et al. Gene expression of ERCC1 as a novel prognostic marker in advanced bladder cancer patients receiving cisplatin-based chemotherapy. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2007;18(3):522–528. doi: 10.1093/annonc/mdl435. [DOI] [PubMed] [Google Scholar]
- 25.Takata R, Katagiri T, Kanehira M, et al. Predicting response to methotrexate, vinblastine, doxorubicin, and cisplatin neoadjuvant chemotherapy for bladder cancers through genome-wide gene expression profiling. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(7):2625–2636. doi: 10.1158/1078-0432.CCR-04-1988. [DOI] [PubMed] [Google Scholar]
- 26.Takata R, Katagiri T, Kanehira M, et al. Validation study of the prediction system for clinical response of M-VAC neoadjuvant chemotherapy. Cancer science. 2007;98(1):113–117. doi: 10.1111/j.1349-7006.2006.00366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JK, Havaleshko DM, Cho H, et al. A strategy for predicting the chemosensitivity of human cancers and its application to drug discovery. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):13086–13091. doi: 10.1073/pnas.0610292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams PD, Cheon S, Havaleshko DM, et al. Concordant gene expression signatures predict clinical outcomes of cancer patients undergoing systemic therapy. Cancer research. 2009;69(21):8302–8309. doi: 10.1158/0008-5472.CAN-09-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Allen EM, Mouw KW, Kim P, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer discovery. 2014;4(10):1140–1153. doi: 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]