Abstract
Dysfunction of prefrontal cortex (PFC) inhibitory neurons that express the calcium-binding protein parvalbumin or the neuropeptide somatostatin in schizophrenia may be related to disturbances in the migration, phenotypic specification, and/or maturation of these neurons. These pre- and postnatal developmental stages are regulated in a cell type-specific manner by various transcription factors and co-activators, fibroblast growth factor receptors (FgfR), and other molecular markers. Consequently, we used quantitative PCR to quantify mRNA levels for these developmental regulators in the PFC of 62 schizophrenia subjects in whom parvalbumin and somatostatin neuron disturbances were previously reported, and in antipsychotic-exposed monkeys. Relative to unaffected comparison subjects, subjects with schizophrenia exhibited elevated mRNA levels for 1) the transcription factor MafB, which is expressed by parvalbumin and somatostatin neurons as they migrate from the medial ganglionic eminence to the cortex, 2) the transcriptional coactivator PGC-1α, which is expressed postnatally by parvalbumin neurons to maintain parvalbumin levels and inhibitory function, and 3) FgfR1, which is required for the migration and phenotypic specification of parvalbumin and somatostatin neurons. Elevations in these markers were most prominent in younger schizophrenia subjects and were not present in antipsychotic-exposed monkeys. Finally, expression levels of other important developmental regulators (i.e. Dlx1, Dlx5, Dlx6, SATB1, Sip1/Zeb2, ST8SIA4, cMaf, Nkx6.2, and Arx) were not altered in schizophrenia. The over-expression of a subset of molecular markers with distinct roles in the pre- and postnatal development of parvalbumin and somatostatin neurons might reflect compensatory mechanisms to sustain the development of these neurons in the face of other insults.
Keywords: GABA, prenatal, ontogeny, inhibitory, postmortem
1. Introduction
Disturbances in the subpopulations of inhibitory (GABA) neurons that express the calcium-binding protein parvalbumin or the neuropeptide somatostatin have been commonly reported in the prefrontal cortex (PFC) from subjects with schizophrenia (Curley et al., 2011; Fung et al., 2010; Hashimoto et al., 2003; Mellios et al., 2009; Morris et al., 2008; Volk et al., 2012). Cortical parvalbumin and somatostatin neurons share a common site of prenatal origin in the medial ganglionic eminence in humans (Fertuzinhos et al., 2009; Hansen et al., 2013; Ma et al., 2013; Zecevic et al., 2011), and residual evidence of disruptions in early developmental processes such as phenotypic specification and migration has been reported in these neurons in postmortem brain tissue in schizophrenia (Hashimoto et al., 2003; Joshi et al., 2012). These data suggest that dysfunction of cortical parvalbumin and somatostatin neurons in schizophrenia may result, at least in part, from insults during development; such insults could occur as early as the prenatal period, initiating processes that interfere with the birth, migration, cell-type specification, and/or maturation of these neurons (Volk and Lewis, 2013, 2014). Consistent with this hypothesis, deficits in the transcription factor Lhx6, which regulates the migration and differentiation of parvalbumin and somatostatin neurons during prenatal development (Fertuzinhos et al., 2009; Georgiev et al., 2012; Jakovcevski et al., 2011; Liodis et al., 2007; Neves et al., 2013; Zhao et al., 2008), have been reported in the prefrontal cortex of two cohorts of schizophrenia subjects (Volk et al., 2014; Volk et al., 2012).
The various stages of pre- and postnatal development of cortical parvalbumin and somatostatin neurons are also regulated by the expression of a diverse array of transcription factors and co-activators (e.g., Dlx1, Dlx5, Dlx6, MafB, cMaf, Sox6, Nkx6.2, Zeb2/Sip1/Zfhx1b, Arx, PGC-1α), chemokine receptors (e.g., CXCR4, CXCR7), and molecular markers of other functions (e.g, Fgfr1, SATB1, and polysialyltransferases such as ST8SIA4) (Anderson et al., 1997; Azim et al., 2009; Batista-Brito et al., 2009; Cobos et al., 2005; Cobos et al., 2006; Colasante et al., 2008; Cowell et al., 2007; Denaxa et al., 2012; Fogarty et al., 2007; Krocher et al., 2014; Lucas et al., 2010; McKinsey et al., 2013; Meechan et al., 2012; Muller et al., 2008; Sanchez-Alcaniz et al., 2011; Sousa et al., 2009; van et al., 2013; Wang et al., 2010; Wang et al., 2011). Consequently, knowledge of the status of these developmental regulators in schizophrenia may provide insight into disturbances in the molecular processes that are disrupted in early life and the resulting impact on cortical parvalbumin and somatostatin neuron development. Therefore, we quantified transcript levels of these developmental regulators in the PFC of a large cohort of schizophrenia subjects with known disturbances in parvalbumin and somatostatin neurons (Hashimoto et al., 2003; Morris et al., 2008; Volk et al., 2014; Volk et al., 2012).
2. Materials and methods
2.1 Human subjects
Brain specimens were obtained during routine autopsies conducted at the Allegheny County Medical Examiner's Office after consent was obtained from next-of-kin. An independent committee of experienced research clinicians made consensus DSMIV (American Psychiatric, 1994) diagnoses for each subject using structured interviews with family members and review of medical records (Volk et al., 2010); the absence of a psychiatric diagnosis was confirmed in unaffected comparison subjects using the same approach. To control for experimental variance, subjects with schizophrenia or schizoaffective disorder (n=62) were matched individually to one unaffected comparison subject for sex and as closely as possible for age and RNA integrity number (RIN; Agilent Bioanalyzer) (Supplemental Table S1). Samples from subjects in a pair were processed together throughout all stages of the study. The mean age, postmortem interval, RNA integrity number (RIN), and tissue freezer storage time did not differ between subject groups (t(122) ≤0.45, p ≥0.65) (Table 1). Mean (± standard deviation) brain pH was different between the schizophrenia (6.6 ± 0.3) and unaffected subject groups (6.7 ± 0.2; t(122) =2.6, p=0.01), but the difference was quite small and of uncertain significance. All procedures were approved by the University of Pittsburgh's Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research.
Table 1.
Summary of demographic and postmortem characteristics of human subjects
| Parameter | Healthy Comparison | Schizophrenia |
|---|---|---|
| N | 62 | 62 |
| Sex | 47M / 15F | 47M / 15F |
| Race | 52W / 10B | 46W / 16B |
| Age (years) | 48.7 ± 13.8 | 47.7 ± 12.7 |
| Postmortem Interval (hours) | 18.8 ± 5.5 | 19.2 ± 8.5 |
| Freezer Storage Time (months) | 132.8 ± 56.8 | 129.0 ± 61.3 |
| Brain pH | 6.7 ± 0.2 | 6.6 ± 0.3 |
| RNA Integrity Number | 8.2 ± 0.6 | 8.1 ± 0.6 |
For brain pH, t(122) =2.6, p=0.01. For all others, t(122) ≤0.45, p ≥0.65. Values are group means ± standard deviation.
2.2 Quantitative PCR
Standardized amounts of gray matter from PFC area 9 were collected in TRIzol reagent in a manner that ensured minimal white matter contamination and excellent RNA preservation (Volk et al., 2013). Standardized dilutions of total RNA for each subject were used to synthesize cDNA. All primer pairs (Supplemental Table S2) demonstrated high amplification efficiency (>94%) across a wide range of cDNA dilutions and specific single products in dissociation curve analysis. Quantitative PCR was performed using the comparative cycle threshold (CT) method with Power SYBR Green dye and the ViiA-7 Real-Time PCR System (Applied Biosystems) as previously described (Volk et al., 2011). Three reference genes (beta actin, cyclophilin A, and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) that we previously reported to be stably expressed in the present cohort of schizophrenia and comparison subjects (Volk et al., 2015). were used to normalize target mRNA levels. The difference in CT (dCT) for each target transcript was calculated by subtracting the geometric mean CT for the three reference genes from the CT of the target transcript (mean of four replicate measures). Because dCT represents the log2-transformed expression ratio of each target transcript to the reference genes, the relative level of the target transcript for each subject is reported as 2−dCT (Vandesompele et al., 2002; Volk et al., 2010).
2.3 Antipsychotic-exposed monkeys
Young adult, male, long-tailed monkeys (Macaca fascicularis) received oral doses of haloperidol, olanzapine or placebo (n=6 monkeys per group) twice daily for 17–27 months, as previously described (Dorph-Petersen et al., 2005). RNA was isolated from PFC area 9, and quantitative PCR was conducted for the same three reference genes and target genes described above (Supplemental Table S2), with all monkeys from a triad processed together on the same plate. All animal studies followed the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee.
2.4 Statistical analysis
The ANCOVA model we report includes mRNA level as the dependent variable, diagnostic group as the main effect, and age, postmortem interval, brain pH, RIN, and freezer storage time as covariates. Because each schizophrenia subject was individually matched to an unaffected subject to account for the parallel processing of tissue samples from a pair and to balance diagnostic groups for sex and age, a second ANCOVA model with subject pair as a blocking factor and including postmortem interval, brain pH, RIN, and freezer storage time was also used. Because both models produced similar results, only the results from unpaired ANCOVA model are reported. Subsequent analyses of differences in mRNA levels between schizophrenia subjects grouped by presence of a comorbid substance use disorder, psychotropic medications at time of death, and tobacco use at time of death and suicide as manner of death were conducted using the unpaired ANCOVA models. For the antipsychotic-exposed monkey study, an ANOVA model with mRNA level as the dependent variable, treatment group as the main effect, and triad as a blocking factor was employed.
3. Results
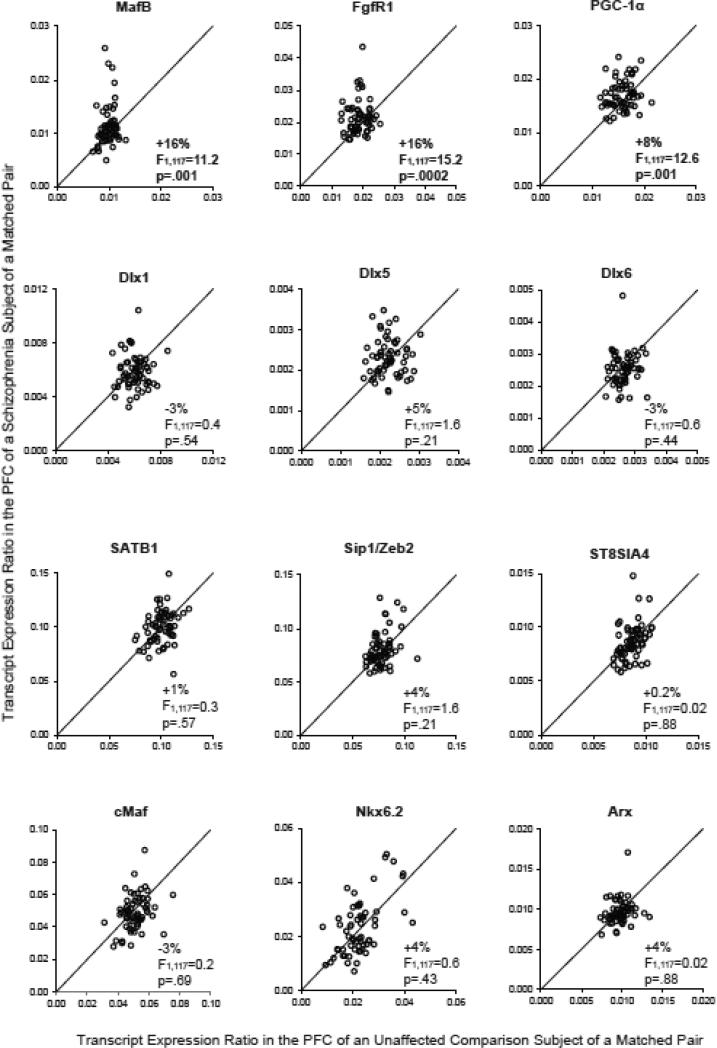
Among the developmental regulators of cortical parvalbumin and somatostatin neurons examined in this study (Figure 1), transcript levels for MafB (+16%; F(1,117)=11.2, p=.001), FgfR1 (+16%; F(1,117)=15.2, p=.0002), and PGC-1α (+8%; F(1,117)=12.6, p=.001) were significantly higher in the PFC of schizophrenia subjects relative to unaffected comparison subjects. In contrast, none of the other transcripts examined (i.e. Dlx1, Dlx5, Dlx6, SATB1, Sip1/Zeb2, ST8SIA4, cMaf, Nkx6.2, and Arx) differed between the subject groups (for all, F(1,117)<1.6, p>.20).
Figure 1. Quantitative PCR analysis of transcript levels of regulators of cortical parvalbumin and somatostatin neuron development in the PFC in schizophrenia.
Transcript levels for each schizophrenia subject relative to the matched unaffected comparison subject in a subject pair are indicated by open circles. Data points to the left of the unity line indicate higher mRNA levels in the schizophrenia subject relative to the unaffected comparison subject in a matched pair.
Transcript levels for MafB, FgfR1, and PGC-1α did not differ between schizophrenia subjects as a function of use of antipsychotics (all F(1,55)≤3.6, p≥0.06), antidepressants (all F(1,55)≤2.2, p≥0.14), benzodiazepines and/or valproate (all F(1,55)≤2.6, p≥0.11), or tobacco use (all F(1,47)≤1.0, p≥0.32) at time of death, or in those with a diagnosis of a substance use disorder (all F(1,55)≤1.2, p≥0.28). Although mRNA levels for MafB (F(1,55)=0.003, p=.95) and FgfR1 (F(1,55)=0.1, p=.72) did not differ between schizophrenia subjects with suicide or natural/accidental manner of death, PGC-1α mRNA levels were significantly lower (−12%; F(1,55)=4.1, p=0.048) in schizophrenia subjects with suicide as manner of death relative to schizophrenia subjects with natural/accidental deaths.
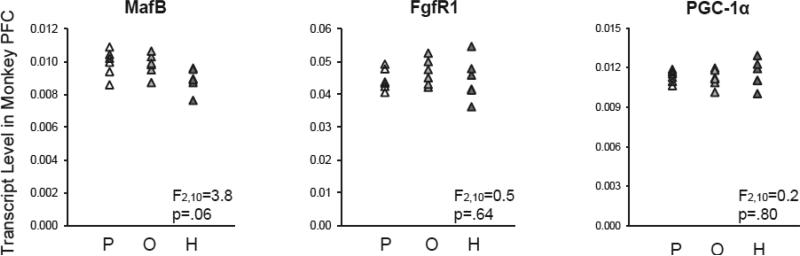
Transcript levels for MafB (F(2,10)=3.8, p=.06), FgfR1 (F(2,10)=0.5, p=.64), and PGC-1α (F(2,10)=0.2, p=.80) in the PFC also did not differ among monkeys exposed chronically to haloperidol, olanzapine, or placebo (Figure 2). However, posthoc analyses using Fisher's least significant difference found that MafB mRNA levels were lower in the PFC of haloperidol-exposed monkeys relative to placebo-exposed monkeys (−10%; p=.03) and to olanzapine-exposed monkeys (−9%; p=.047), suggesting that exposure to haloperidol may partially mask the disease effect on MafB mRNA levels in schizophrenia subjects.
Figure 2. Transcript levels for developmental regulators of cortical parvalbumin and somatostatin neurons in PFC area 9 of antipsychotic-exposed monkeys.
Quantitative PCR analysis revealed no statistically significant differences in mRNA levels for MafB, PGC-1α, or FgfR1 in monkeys exposed chronically to placebo (P), olanzapine (O), or haloperidol (H). However, posthoc analyses using Fisher's least significant difference revealed that MafB mRNA levels were lower in the PFC of haloperidol-exposed monkeys relative to placebo-exposed monkeys (−10%; p=.03) and olanzapine-exposed monkeys (−9%; p=.047). Mean values are indicated by horizontal black bars.
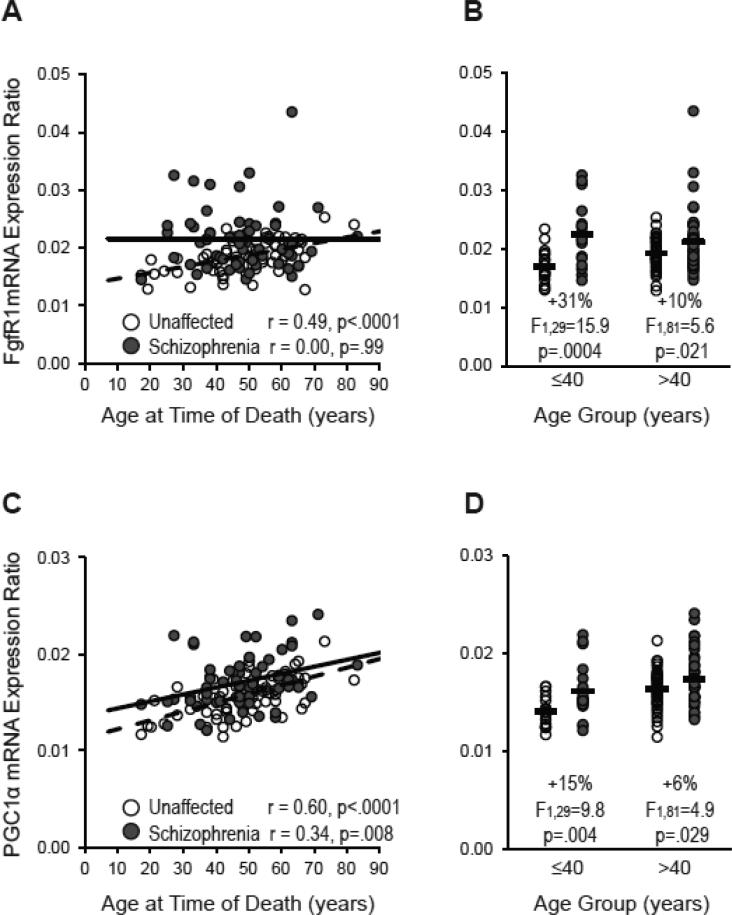
Interestingly, FgfR1 mRNA levels were positively correlated with age in unaffected comparison subjects (r=.49, p<.0001) but not in schizophrenia subjects (r=0.00, p=.99; Figure 3). Furthermore, the elevation in FgfR1 mRNA levels relative to unaffected comparison subjects was three-fold higher in schizophrenia subjects age 40 and under (+31%; F(1,29)=15.9, p=.0004; n=18 subject pairs) than in schizophrenia subjects over age 40 (+10%; F(1,81)=5.6, p=.021; n=44 subject pairs) (Figure 3). In contrast, mRNA levels for PGC-1α increased with age in schizophrenia subjects (r=.34, p=.008), unaffected comparison subjects (r=.60, p<.0001), and across all subjects (r=.42, p<.0001) (Figure 3). The elevation in PGC-1α mRNA levels relative to unaffected comparison subjects was also higher in schizophrenia subjects age 40 and under (+15%; F(1,29)=9.8, p=.004) than in schizophrenia subjects over age 40 (+6%; F(1,81)=4.9, p=.029; Figure 3). Finally, MafB mRNA levels did not correlate with age in schizophrenia subjects (r=−.13, p=.32), unaffected comparison subjects (r=−.02, p=.87), or across all subjects (r=−.10, p=.29).
Figure 3. Relationship between FgfR1 and PGC-1α mRNA levels and age in schizophrenia and unaffected comparison subjects.
Relative FgfR1 (A) and PGC-1α (C) mRNA levels are plotted against age in schizophrenia subjects (dark gray circles and solid black linear regression line; n=62) and unaffected comparison subjects (open circles and dashed black linear regression line; n=62). B and D. Schizophrenia subjects were divided into two groups age 40 and under and over 40 [range (with mean ± standard deviation): 17-40 years (32.4 ± 6.0 years) and 41-83 years (53.9 ± 8.8 years)]. Relative FgfR1 (B) and PGC-1α (D) mRNA levels for schizophrenia subjects within these subgroups were then compared to their matched unaffected comparison subjects. Black bars note the average mRNA expression level in the schizophrenia and corresponding comparison subjects.
Consistent with their expression specifically in cortical parvalbumin and somatostatin neurons (Cobos et al., 2006), MafB mRNA levels were correlated with mRNA levels for GAD67 (r=.40, p=.001), parvalbumin (r=.29, p=.024), and somatostatin (r=.26, p=.042) in the PFC of unaffected comparison subjects (Supplementary Figure S1). Furthermore, consistent with a role in the postnatal development of cortical parvalbumin neurons (Cowell et al., 2007; Dougherty et al., 2014; Lucas et al., 2010), PGC-1α mRNA levels in unaffected comparison subjects were positively correlated with mRNA levels for parvalbumin (r=.32, p=.012) and GAD67 (r=.24, p=.058) (Supplementary Figure S2), although the correlation with GAD67 did not quite achieve statistical significance. However, neither MafB nor PGC-1α mRNA levels were correlated with these GABA neuron-related markers in schizophrenia subjects (r<|.20|, p>.12). Finally, FgfR1 mRNA levels did not correlate with any GABA neuron-related markers in either unaffected comparison or schizophrenia subjects (r<|.22|, p>.085).
4. Discussion
In this study, we determined whether molecular markers that are critical for the pre- and postnatal development of cortical parvalbumin and somatostatin neurons were altered in the PFC of subjects with schizophrenia. Several developmental regulators with diverse roles in the ontogeny and maturation of cortical parvalbumin and somatostatin neurons, such as the transcription factor Mafb, the transcriptional co-activator PGC-1α, and fibroblast growth factor receptor 1 (FgfR1), showed higher transcript levels in schizophrenia subjects. In contrast, mRNA levels for the other nine developmental regulators examined did not differ between subject groups. Furthermore, higher transcript levels for MafB, PGC-1α, and FgfR1 did not appear to be attributable to exposure to antipsychotic medications or other comorbid factors, and evidence from the antipsychotic-exposed monkeys suggests that haloperidol may partially mask the disease effect of higher MafB mRNA levels. Transcript levels for MafB and PGC-1α were correlated with GABA neuron markers in unaffected comparison subjects, consistent with their cell type-specific localization to inhibitory neurons. However, the correlations between the mRNA levels for MafB and PGC-1α and the GABA neuron markers were lost in the schizophrenia subjects. Taken together, these findings suggest that higher mRNA levels for important developmental regulators of cortical parvalbumin and somatostatin neurons may be related to the disrupted development of these neurons in schizophrenia.
Studies to date of transcript levels of developmental markers that regulate the earliest prenatal stages of cortical parvalbumin and somatostatin neuron ontogeny have produced a complex pattern of results in schizophrenia. For example, the transcription factor Lhx6 and the chemokine receptors CXCR4 and CXCR7 are involved in the regulation of parvalbumin and somatostatin neuron migration from the medial ganglionic eminence to the cerebral cortex during prenatal development (Fertuzinhos et al., 2009; Georgiev et al., 2012; Jakovcevski et al., 2011; Li et al., 2008; Liodis et al., 2007; Neves et al., 2013; Sanchez-Alcaniz et al., 2011; Tanaka et al., 2010; Vogt et al., 2014; Wang et al., 2011; Zhao et al., 2008). We recently reported lower Lhx6 mRNA levels and higher CXCR4 and CXCR7 mRNA levels in the PFC of the same cohort of schizophrenia subjects (Volk et al., 2015; Volk et al., 2014; Volk et al., 2012). These findings raise the questions of whether and how altered expression of Lhx6 and CXCR4 and CXCR7 may contribute to the previously reported evidence of disturbances in the migration and phenotypic specification of cortical parvalbumin and somatostatin neurons in schizophrenia (Hashimoto et al., 2003; Joshi et al., 2012). Interestingly, transduction of CXCR7 into Lhx6 mutant cells in the murine medial ganglionic eminence partially rescues the cortical lamination pattern of parvalbumin and somatostatin neurons that is disrupted in Lhx6 mutant mice (Liodis et al., 2007; Neves et al., 2013; Vogt et al., 2014; Zhao et al., 2008). Taken together, these findings suggest the existence of compensatory mechanisms to support the migration and final cortical positioning of parvalbumin and somatostatin neurons in schizophrenia. Consistent with this hypothesis, in the same cohort of schizophrenia subjects, we now report higher mRNA levels for the transcription factor MafB, which is similarly expressed by the vast majority of parvalbumin and somatostatin neurons during prenatal migration to the cortex and continues to be expressed in this cell-type specific manner postnatally (Cobos et al., 2006). Thus, higher MafB mRNA levels may also represent a compensatory response to preserve the migration of parvalbumin and somatostatin neurons during prenatal development; however, mouse models examining the effects of higher MafB levels on cortical parvalbumin and somatostatin neuron development are needed to further test this hypothesis. Furthermore, other transcription factors that are expressed in the medial ganglionic eminence and govern prenatal ontogeny of cortical parvalbumin and somatostatin neurons (e.g., i.e. Dlx5, Dlx6, Nkx6.2, Zeb2/Sip1, Arx (Cobos et al., 2006; Fogarty et al., 2007; McKinsey et al., 2013; Sousa et al., 2009; van et al., 2013; Wang et al., 2010); see also our prior study on Sox6 (Volk et al., 2012), were unchanged in the PFC in schizophrenia, indicating that the disease process affecting these developmental regulators is molecularly-specific.
Several developmental regulators that play a prominent role in the postnatal maturation, trophic support, and adult survival of cortical parvalbumin and somatostatin neurons are also abnormally expressed in schizophrenia. For example, the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) is expressed postnatally by cortical parvalbumin neurons where it is required to achieve a fully mature state including maintenance of parvalbumin levels and inhibitory function (Cowell et al., 2007; Lucas et al., 2010). PGC-1α also regulates the intrinsic excitability of parvalbumin neurons (Dougherty et al., 2014), and over-expression of PGC-1α induces higher levels of a broad array of synaptic and metabolic markers (Lucas et al., 2014) and higher parvalbumin levels (Lucas et al., 2010). Thus, PGC-1α expression would be expected to be correlated with that of activity-dependent gene products in PV neurons. Indeed, PGC-1α mRNA levels were correlated with mRNA levels for GAD67 and parvalbumin in unaffected comparison subjects. However, we found higher PGC-1α mRNA levels in the PFC of the same cohort of schizophrenia subjects in which we previously reported lower parvalbumin and GAD67 mRNA levels (Volk et al., 2014; Volk et al., 2012). Furthermore, PGC-1α mRNA levels were not correlated with mRNA levels for GAD67 or parvalbumin in schizophrenia subjects, and another study reported a highly similar dissociation in schizophrenia (McMeekin et al., 2015). Thus, elevated PGC-1α levels in schizophrenia appear to be an inadequate compensatory response to support cortical parvalbumin neuron excitability in the illness.
Fibroblast growth factor receptor 1 (Fgfr1) expression prenatally by telencephalic radial glial progenitor cells, which play an essential role in neuronal migration, and postnatally by astrocytes, which provide trophic support to interneurons, is required for prenatal migration and postnatal phenotypic specification of cortical parvalbumin and somatostatin neurons (Muller et al., 2008; Smith et al., 2014). In the present study, we report higher FgfR1 mRNA levels in the PFC in schizophrenia. In contrast to PGC-1α, FgfR1 mRNA levels were not correlated with other GABA neuron-related markers, which may reflect that FgfR1, while important for trophic support and development of cortical parvalbumin and somatostatin neurons, is expressed postnatally by astrocytes (Muller et al., 2008; Smith et al., 2014). Interestingly, the magnitude of the increases in mRNA levels for FgfR1 (and PGC-1α) was higher in younger than in older subjects with schizophrenia. However, it is unclear if these differences reflect an accelerated age-related increase in expression of these markers in young adulthood or a disruption of the aging process later in life in the disorder. It is also important to note that additional studies directly examining the effects of overexpression of FgfR1 on the development of cortical parvalbumin and somatostatin neurons are needed.
Finally, the transcription factor Dlx1, which is expressed postnatally by somatostatin neurons, is required to maintain postnatal GAD67 expression, and is required for cortical somatostatin neurons to survive the preadolescent period in mice (Cobos et al., 2005). Although lower Dlx1 mRNA levels were previously reported in conjunction with deficits in GAD67 mRNA levels in the orbitofrontal cortex in schizophrenia (Joshi et al., 2012), we did not detect a difference in Dlx1 mRNA levels in the PFC of schizophrenia subjects in whom we had previously reported deficits in GAD67 mRNA levels (Curley et al., 2011; Volk et al., 2012).
Taken together, these findings shed some new light on a complex picture of developmental pathology of cortical parvalbumin and somatostatin neurons in schizophrenia. A diversity of molecular markers, including transcription factors, transcriptional co-activators, fibroblast growth factors receptors, and chemokine receptors, with distinct roles in the prenatal development, postnatal maturation, trophic support and survival of cortical parvalbumin and somatostatin neurons are overexpressed in the PFC of the same schizophrenia subjects in whom disturbances in PFC parvalbumin and somatostatin neurons have been previously reported. In contrast, the transcription factor Lhx6 is under-expressed while other developmental regulators are unchanged in the disorder. Further understanding of the potential role of these critical regulators of cortical parvalbumin and somatostatin neuron development in the disease process of schizophrenia requires additional investigations. First, do alterations in these developmental regulators occur very early in life when the pre- and/or postnatal development of parvalbumin and somatostatin neurons could be affected in individuals with schizophrenia? Or do these disturbances emerge later in life when the maintenance, function, and phenotype of parvalbumin and somatostatin neurons could be impacted in the disorder? Our present and recent findings (Hoftman and Lewis, 2011; Hoftman et al., 2015), which suggest that these alterations are most prominent in younger individuals with schizophrenia, could not however determine whether the abnormalities are also present prenatally. Second, animal models that combine 1) induced disturbances in cortical parvalbumin and somatostatin neuron development (e.g., Lhx6 homozygous null mutation mice) and 2) overexpression of these developmental regulators are needed to further understand any potential compensatory effects on cortical parvalbumin and somatostatin neuron development. Finally, the upstream pathogenetic mechanisms that may disrupt the prenatal ontogeny of cortical parvalbumin and somatostatin neurons and lead to a cascade of molecular changes, including the overexpression of developmentally-relevant molecules, in schizophrenia continues to remain fertile ground for further study.
Supplementary Material
Acknowledgements
The authors wish to acknowledge Elizabeth Sengupta, MA for her work in processing the tissue sections and designing primers.
Role of funding source
This study was supported by grants from the National Institutes of Health (MH100066 to Dr. Volk; MH043784 and MH051234 to Dr. Lewis).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
David A. Lewis currently receives investigator-initiated research support from Pfizer. In 2013-2015, he served as a consultant in the areas of target identification and validation and new compound development to Autifony, Bristol-Myers Squibb, Concert Pharmaceuticals and Sunovion. All other authors have nothing to disclose.
Contributors
Dr. Volk oversaw all aspects of the design and implementation of the study and was the primary author of the manuscript. Ms. Edelson contributed to the design of the study, collected the data, and assisted with the interpretation of the data. Dr. Lewis contributed to the design of the study and the preparation of the manuscript. All authors contributed to and have approved the final manuscript.
References
- American Psychiatric A. DSM-IV. Diagnostic and Statistical Manual of Mental Disorders. Fourth Edition American Psychiatric Association; Washington, D.C.: 1994. [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JLR. Interneuron migration from basal forebrain to neocortex: Dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Azim E, Jabaudon D, Fame RM, Macklis JD. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat Neurosci. 2009;12(10):1238–1247. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Rossignol E, Hjerling-Leffler J, Denaxa M, Wegner M, Lefebvre V, Pachnis V, Fishell G. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63(4):466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8(8):1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex 16 Suppl. 2006;1:i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Colasante G, Collombat P, Raimondi V, Bonanomi D, Ferrai C, Maira M, Yoshikawa K, Mansouri A, Valtorta F, Rubenstein JL, Broccoli V. Arx is a direct target of Dlx2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci. 2008;28(42):10674–10686. doi: 10.1523/JNEUROSCI.1283-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell RM, Blake KR, Russell JW. Localization of the transcriptional coactivator PGC-1alpha to GABAergic neurons during maturation of the rat brain. J Comp Neurol. 2007;502(1):1–18. doi: 10.1002/cne.21211. [DOI] [PubMed] [Google Scholar]
- Curley AA, Arion D, Volk DW, Asafu-Adjei JK, Sampson AR, Fish KN, Lewis DA. Cortical deficits of glutamic acid decarboxylase 67 expression in schizophrenia: Clinical, protein, and cell type-specific features. Am J Psychiatry. 2011;168(9):921–929. doi: 10.1176/appi.ajp.2011.11010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denaxa M, Kalaitzidou M, Garefalaki A, Achimastou A, Lasrado R, Maes T, Pachnis V. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Rep. 2012;2(5):1351–1362. doi: 10.1016/j.celrep.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorph-Petersen KA, Pierri JN, Perel JM, Sun Z, Sampson AR, Lewis DA. The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharmacology. 2005;30(9):1649–1661. doi: 10.1038/sj.npp.1300710. [DOI] [PubMed] [Google Scholar]
- Dougherty SE, Bartley AF, Lucas EK, Hablitz JJ, Dobrunz LE, Cowell RM. Mice lacking the transcriptional coactivator PGC-1alpha exhibit alterations in inhibitory synaptic transmission in the motor cortex. Neuroscience. 2014;271:137–148. doi: 10.1016/j.neuroscience.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fertuzinhos S, Krsnik Z, Kawasawa YI, Rasin MR, Kwan KY, Chen JG, Judas M, Hayashi M, Sestan N. Selective depletion of molecularly defined cortical interneurons in human holoprosencephaly with severe striatal hypoplasia. Cereb Cortex. 2009;19(9):2196–2207. doi: 10.1093/cercor/bhp009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marin O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27(41):10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2010;167(12):1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- Georgiev D, Gonzalez-Burgos G, Kikuchi M, Minabe Y, Lewis DA, Hashimoto T. Selective expression of KCNS3 potassium channel alpha-subunit in parvalbumin-ontaining GABA neurons in the human prefrontal cortex. PLoS One. 2012;7(8):e43904. doi: 10.1371/journal.pone.0043904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, Kriegstein AR. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16(11):1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, Sampson AR, Lewis DA. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoftman GD, Lewis DA. Postnatal developmental trajectories of neural circuits in the primate prefrontal cortex: identifying sensitive periods for vulnerability to schizophrenia. Schizophr Bull. 2011;37(3):493–503. doi: 10.1093/schbul/sbr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoftman GD, Volk DW, Bazmi HH, Li S, Sampson AR, Lewis DA. Altered cortical expression of GABA-related genes in schizophrenia: illness progression vs developmental disturbance. Schizophr Bull. 2015;41(1):180–191. doi: 10.1093/schbul/sbt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcevski I, Mayer N, Zecevic N. Multiple origins of human neocortical interneurons are supported by distinct expression of transcription factors. Cereb Cortex. 2011;21(8):1771–1782. doi: 10.1093/cercor/bhq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi D, Fung SJ, Rothwell A, Weickert CS. Higher gamma-aminobutyric acid neuron density in the white matter of orbital frontal cortex in schizophrenia. Biol Psychiatry. 2012;72(9):725–733. doi: 10.1016/j.biopsych.2012.06.021. [DOI] [PubMed] [Google Scholar]
- Krocher T, Rockle I, Diederichs U, Weinhold B, Burkhardt H, Yanagawa Y, Gerardy-Schahn R, Hildebrandt H. A crucial role for polysialic acid in developmental interneuron migration and the establishment of interneuron densities in the mouse prefrontal cortex. Development. 2014;141(15):3022–3032. doi: 10.1242/dev.111773. [DOI] [PubMed] [Google Scholar]
- Li G, Adesnik H, Li J, Long J, Nicoll RA, Rubenstein JL, Pleasure SJ. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J Neurosci. 2008;28(5):1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liodis P, Denaxa M, Grigoriou M, Akufo-Addo C, Yanagawa Y, Pachnis V. Lhx6 activity is required for the normal migration and specification of cortical interneuron subtypes. J Neurosci. 2007;27(12):3078–3089. doi: 10.1523/JNEUROSCI.3055-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas EK, Dougherty SE, McMeekin LJ, Reid CS, Dobrunz LE, West AB, Hablitz JJ, Cowell RM. PGC-1alpha provides a transcriptional framework for synchronous neurotransmitter release from parvalbumin-positive interneurons. J Neurosci. 2014;34(43):14375–14387. doi: 10.1523/JNEUROSCI.1222-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas EK, Markwardt SJ, Gupta S, Meador-Woodruff JH, Lin JD, Overstreet-Wadiche L, Cowell RM. Parvalbumin deficiency and GABAergic dysfunction in mice lacking PGC-1alpha. J Neurosci. 2010;30(21):7227–7235. doi: 10.1523/JNEUROSCI.0698-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Wang C, Wang L, Zhou X, Tian M, Zhang Q, Zhang Y, Li J, Liu Z, Cai Y, Liu F, You Y, Chen C, Campbell K, Song H, Ma L, Rubenstein JL, Yang Z. Subcortical origins of human and monkey neocortical interneurons. Nat Neurosci. 2013;16(11):1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio LA, Huylebroeck D, Higashi Y, Rubenstein JL. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron. 2013;77(1):83–98. doi: 10.1016/j.neuron.2012.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeekin LJ, Lucas EK, Meador-Woodruff JH, McCullumsmith RE, Hendrickson RC, Gamble KL, Cowell RM. Cortical PGC-1alpha-Dependent Transcripts are Reduced in Postmortem Tissue From Patients With Schizophrenia. Schizophr Bull. 2015 doi: 10.1093/schbul/sbv184. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meechan DW, Tucker ES, Maynard TM, LaMantia AS. Cxcr4 regulation of interneuron migration is disrupted in 22q11.2 deletion syndrome. Proc Natl Acad Sci U.S.A. 2012;109(45):18601–18606. doi: 10.1073/pnas.1211507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Huang HS, Baker SP, Galdzicka M, Ginns E, Akbarian S. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol Psychiatry. 2009;65(12):1006–1014. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Morris HM, Hashimoto T, Lewis DA. Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cereb Cortex. 2008;18(7):1575–1587. doi: 10.1093/cercor/bhm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller SK, Fagel DM, Stevens HE, Rabenstein RL, Maragnoli ME, Ohkubo Y, Picciotto MR, Schwartz ML, Vaccarino FM. Deficiency in inhibitory cortical interneurons associates with hyperactivity in fibroblast growth factor receptor 1 mutant mice. Biol Psychiatry. 2008;63(10):953–962. doi: 10.1016/j.biopsych.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Neves G, Shah MM, Liodis P, Achimastou A, Denaxa M, Roalfe G, Sesay A, Walker MC, Pachnis V. The LIM homeodomain protein Lhx6 regulates maturation of interneurons and network excitability in the mammalian cortex. Cereb Cortex. 2013;23(8):1811–1823. doi: 10.1093/cercor/bhs159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Alcaniz JA, Haege S, Mueller W, Pla R, Mackay F, Schulz S, Lopez-Bendito G, Stumm R, Marin O. Cxcr7 controls neuronal migration by regulating chemokine responsiveness. Neuron. 2011;69(1):77–90. doi: 10.1016/j.neuron.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Smith KM, Maragnoli ME, Phull PM, Tran KM, Choubey L, Vaccarino FM. Fgfr1 inactivation in the mouse telencephalon results in impaired maturation of interneurons expressing parvalbumin. PLoS One. 2014;9(8):e103696. doi: 10.1371/journal.pone.0103696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cereb Cortex 19 Suppl. 2009;1:i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka DH, Mikami S, Nagasawa T, Miyazaki J, Nakajima K, Murakami F. CXCR4 is required for proper regional and laminar distribution of cortical somatostatin-, calretinin-, and neuropeptide Y-expressing GABAergic interneurons. Cereb Cortex. 2010;20(12):2810–2817. doi: 10.1093/cercor/bhq027. [DOI] [PubMed] [Google Scholar]
- van d.B.V., Stappers E, Vandesande B, Dimidschstein J, Kroes R, Francis A, Conidi A, Lesage F, Dries R, Cazzola S, Berx G, Kessaris N, Vanderhaeghen P, van Ijcken W, Grosveld FG, Goossens S, Haigh JJ, Fishell G, Goffinet A, Aerts S, Huylebroeck D, Seuntjens E. Directed migration of cortical interneurons depends on the cell-autonomous action of Sip1. Neuron. 2013;77(1):70–82. doi: 10.1016/j.neuron.2012.11.009. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):34.31–34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt D, Hunt RF, Mandal S, Sandberg M, Silberberg SN, Nagasawa T, Yang Z, Baraban SC, Rubenstein JL. Lhx6 directly regulates Arx and CXCR7 to determine cortical interneuron fate and laminar position. Neuron. 2014;82(2):350–364. doi: 10.1016/j.neuron.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Chitrapu A, Edelson JR, Lewis DA. Chemokine receptors and cortical interneuron dysfunction in schizophrenia. Schizophr Res. 2015;167(1-3):12–17. doi: 10.1016/j.schres.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Edelson JR, Lewis DA. Cortical inhibitory neuron disturbances in schizophrenia: role of the ontogenetic transcription factor Lhx6. Schizophr Bull. 2014;40(5):1053–1061. doi: 10.1093/schbul/sbu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Eggan SM, Lewis DA. Alterations in metabotropic glutamate receptor 1alpha and regulator of G protein signaling 4 in the prefrontal cortex in schizophrenia. Am J Psychiatry. 2010;167(12):1489–1498. doi: 10.1176/appi.ajp.2010.10030318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Lewis DA. Prenatal ontogeny as a susceptibility period for cortical GABA neuron disturbances in schizophrenia. Neuroscience. 2013;248C:154–164. doi: 10.1016/j.neuroscience.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Lewis DA. Early developmental disturbances of cortical inhibitory neurons: contribution to cognitive deficits in schizophrenia. Schizophr Bull. 2014;40(5):952–957. doi: 10.1093/schbul/sbu111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Matsubara T, Li S, Sengupta EJ, Georgiev D, Minabe Y, Sampson A, Hashimoto T, Lewis DA. Deficits in transcriptional regulators of cortical parvalbumin neurons in schizophrenia. Am J Psychiatry. 2012;169(10):1082–1091. doi: 10.1176/appi.ajp.2012.12030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Radchenkova PV, Walker EM, Sengupta EJ, Lewis DA. Cortical opioid markers in schizophrenia and across postnatal development. Cereb Cortex. 2011;22(5):1215–1223. doi: 10.1093/cercor/bhr202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Siegel BI, Verrico CD, Lewis DA. Endocannabinoid metabolism in the prefrontal cortex in schizophrenia. Schizophr Res. 2013;147(1):53–57. doi: 10.1016/j.schres.2013.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dye CA, Sohal V, Long JE, Estrada RC, Roztocil T, Lufkin T, Deisseroth K, Baraban SC, Rubenstein JL. Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J Neurosci. 2010;30(15):5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li G, Stanco A, Long JE, Crawford D, Potter GB, Pleasure SJ, Behrens T, Rubenstein JL. CXCR4 and CXCR7 have distinct functions in regulating interneuron migration. Neuron. 2011;69(1):61–76. doi: 10.1016/j.neuron.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecevic N, Hu F, Jakovcevski I. Interneurons in the developing human neocortex. Dev Neurobiol. 2011;71(1):18–33. doi: 10.1002/dneu.20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510(1):79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.