Abstract
Endozoicomonas bacteria are emerging as extremely diverse and flexible symbionts of numerous marine hosts inhabiting oceans worldwide. Their hosts range from simple invertebrate species, such as sponges and corals, to complex vertebrates, such as fish. Although widely distributed, the functional role of Endozoicomonas within their host microenvironment is not well understood. In this review, we provide a summary of the currently recognized hosts of Endozoicomonas and their global distribution. Next, the potential functional roles of Endozoicomonas, particularly in light of recent microscopic, genomic, and genetic analyses, are discussed. These analyses suggest that Endozoicomonas typically reside in aggregates within host tissues, have a free-living stage due to their large genome sizes, show signs of host and local adaptation, participate in host-associated protein and carbohydrate transport and cycling, and harbour a high degree of genomic plasticity due to the large proportion of transposable elements residing in their genomes. This review will finish with a discussion on the methodological tools currently employed to study Endozoicomonas and host interactions and review future avenues for studying complex host-microbial symbioses.
Keywords: Endozoicomonas, Symbiosis, Marine, Coral reefs
Introduction
It is increasingly recognized that eukaryotic organisms rely on bacterial associates to provide a diversity of functions, from supplying nutrients and essential amino acids, to protection from pathogenic microbes and degradation of toxins (McFall-Ngai et al. 2013). Despite only being recently described (e.g. Kurahashi and Yokota 2007), the bacterial genus Endozoicomonas (Gammaproteobacteria; Oceanospirillales) has been reported to associate with a large diversity of marine organisms, including cnidarians, poriferans, molluscs, annelids, tunicates, and fish (Jensen et al. 2010; Morrow et al. 2012; Forget and Juniper 2013; Fiore et al. 2015; Katharios et al. 2015). They are also globally distributed and have been found living symbiotically with organisms in all major oceans of the world (Neave et al. 2016). However, although they are ubiquitously distributed, the functional role of Endozoicomonas is unclear. Their suggested roles have ranged from a beneficial symbiont required for healthy host functioning to a pathogen that can rapidly cause host death (Bourne et al. 2013; Katharios et al. 2015).
This mini-review provides an update on the ever-expanding range of known Endozoicomonas hosts and global distributions and addresses recent advancements in understanding the genetic potential and possible functions of bacteria in the genus Endozoicomonas. We conclude with a discussion on strategies for uncovering new insights into the lifestyle of this cryptic and enigmatic genus, including emerging tools for the study of microbial-animal symbioses, and provide recommendations for future work.
History and prevalence of Endozoicomonas in the scientific literature
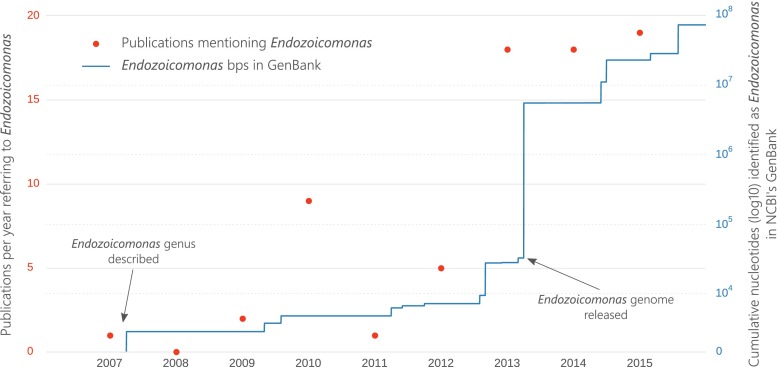
The genus Endozoicomonas was described less than a decade ago by Kurahashi and Yokota (2007) (Fig. 1), after they isolated an unknown gammaproteobacterial symbiont from the sea slug Elysia ornata. Physiological and phylogenetic analyses indicated that the creation of a new genus, Endozoicomonas (i.e. monad living inside an animal), was required (Kurahashi and Yokota 2007). After this initial description, references to Endozoicomonas remained relatively scarce in the literature until 2010, when Endozoicomonas montiporae (Yang et al. 2010) was described, and coral symbionts with similarity to Endozoicomonas were discovered (Kvennefors et al. 2010). Kvennefors et al. (2010) also noted that symbionts from earlier microbiome studies (before the creation of the genus) were closely related to Endozoicomonas. Examples include abundant gammaproteobacterial symbionts termed “PA1” from the coral Porites astreoides (Rohwer et al. 2002) and Gammaproteobacteria from the coral Pocillopora damicornis (Bourne and Munn 2005). These efforts to retroactively link past studies to the Endozoicomonas genus, in addition to the detection of Endozoicomonas in new hosts, resulted in a spike in citations of Endozoicomonas within the scientific literature. From 2013 onwards, more than 15 publications per year referred to the genus (Fig. 1). This relatively rapid rise in Endozoicomonas publications, and the novelty of the genus, has led to some taxonomic inconsistencies finding their way into journal articles and databases. For example, the Greengenes database (DeSantis et al. 2006) places the genus Endozoicomonas in an apparently newly created family called “Endozoicomonaceae”. However, despite not being explicitly stated, the type description of Endozoicomonas contained nucleotide information that placed the genus within the family Hahellaceae (Kurahashi and Yokota 2007) in the order Oceanospirillales and this taxonomy was maintained in subsequent type descriptions (Yang et al. 2010; Nishijima et al. 2013). The incorrect term Endozoicomonaceae has appeared in a number of scientific publications (e.g. Dishaw et al. 2014; Katharios et al. 2015; Lawler et al. 2016) paying tribute to just how rapid this genus became scientifically popular. Confusion surrounding the initial naming of the genus has also produced inconsistencies in the scientific literature. For example, in March 2005 (prior to the Endozoicomonas description), a sequence named “Spongiobacter nickelotolerans” was submitted to GenBank (#AB205011), which is very similar (>97 % SSU rRNA identity) to the Endozoicomonas type strains. The associated taxonomic paper describing the isolation of S. nickelotolerans from a marine sponge, however, was not published and the name appears to have been abandoned (e.g. Pike et al. 2013; McCauley et al. 2016). Nevertheless, several publications have referred to “Spongiobacter” or to the “Spongiobacter/Endozoicomonas” group (e.g. Costa et al. 2012; La Rivière et al. 2015).
Fig. 1.
Prevalence of Endozoicomonas in the scientific literature as shown by the number of publications referring to the genus per year (red markers) and the cumulative amount of Endozoicomonas nucleotide information uploaded to NCBI’s GenBank (blue line)
The volume of genetic data available for Endozoicomonas bacteria in NCBI’s GenBank (Benson et al. 2013) has also rapidly increased (Fig. 1). Since the initial deposition of the first 1437 bps SSU rRNA sequence of E. elysicola by Kurahashi and Yokota (2007), Endozoicomonas nucleotide information has steadily accumulated, reaching almost 100,000,000 bps by the start of 2016 (Fig. 1). Moreover, this number only takes into account data retrieved from GenBank; far more Endozoicomonas genetic information is available in other databases, such as NCBI’s Sequence Read Archive (SRA). Much of this rapid accumulation of genetic information can be attributed to the move towards whole genome sequencing, rather than marker gene sequencing. The first Endozoicomonas genome sequenced was E. elysicola in 2013 (Fig. 1) as part of the one thousand microbial genomes project (Kyrpides et al. 2014). The following year, Neave et al. (2014) released an updated version of the E. elysicola genome plus two new genomes, E. montiporae (Yang et al. 2010) and E. numazuensis (Nishijima et al. 2013). Since then, the E. montiporae genome has been re-sequenced (Ding et al. 2016), the genome of E. atrinae has been made available (Hyun et al. 2014), the genome of an undescribed pathogenic Endozoicomonas has been analysed (Katharios et al. 2015), and several other Endozoicomonas genome projects are underway (e.g. Appolinario et al. 2016). Most recently, Neave, Michell, Apprill and Voolstra (Endozoicomonas genomes reveals functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts, Submitted) applied single-cell genomics and metagenomic binning to recruit four additional Endozoicomonas genomes from native coral host assemblages.
Diversity and distribution of Endozoicomonas hosts
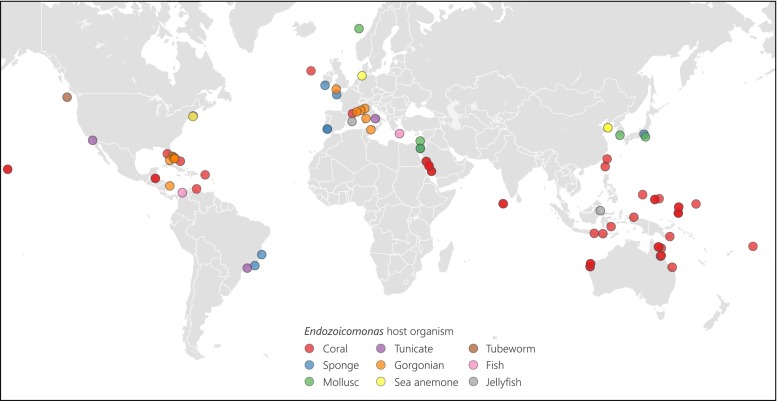
Endozoicomonas symbionts have a global distribution in numerous marine hosts, from abyssal depths to warm photic zones (Fig. 2). They are most frequently detected as coral symbionts and occur throughout the global distribution of coral reefs, from the Great Barrier Reef in Australia (Bourne et al. 2008; Lema et al. 2013), Papua New Guinea (Morrow et al. 2015), Indonesia and the Pacific (Yang et al. 2010; Neave et al. 2016) to the Red Sea (Bayer et al. 2013b; Jessen et al. 2013; Neave et al. 2016; Ziegler et al. 2016), Indian Ocean (Neave et al. 2016), and the Caribbean (Morrow et al. 2012; Rodriguez-Lanetty et al. 2013) (Fig. 2). Interestingly, different coral species appear to harbour specific Endozoicomonas genotypes (Neave et al. 2016). For example, the sympatric corals, Stylophora pistillata and Pocillopora verrucosa, each contain different Endozoicomonas genotypes, and these genotypes partition differently across large geographic scales (Neave et al. 2016). Moreover, patterns of symbiont specificity seem to co-align with differences in reproductive mode. For instance, the spawning coral P. verrucosa globally associates with the same Endozoicomonas symbionts, whereas the brooding coral S. pistillata harbours different Endozoicomonas genotypes in different regions (Neave et al. 2016).
Fig. 2.
Global distribution and diversity of Endozoicomonas host organisms
Although no quantitative methods have been applied to Endozoicomonas, trends in cell abundance have been inferred from SSU rRNA gene sequence abundances. These studies have linked the abundance of Endozoicomonas to the abundance of its coral host. For example, when the fungid coral Ctenactis echinata grew in its preferred Red Sea habitat, Endozoicomonas symbionts were more abundant than in habitats of degraded quality (Roder et al. 2015). Moreover, reduced abundances of the corals Acropora millepora and Porites cylindrica near carbon dioxide seeps in Papua New Guinea coincided with a 50 % reduction in Endozoicomonas symbionts (Morrow et al. 2015). Anthropogenic pollution can similarly decrease the abundance of Endozoicomonas bacteria. Near the large Red Sea city of Jeddah, the corals P. verrucosa and Acropora hemprichii contained a lower proportion of Endozoicomonas compared to corals further afield (Ziegler et al. 2016). In addition, bleaching of the coral A. millepora on the Great Barrier Reef induced a shift from Endozoicomonas-like symbionts to a Vibrio-dominated community (Bourne et al. 2008). Lesioned P. astreoides colonies also contained reduced Endozoicomonas sequence abundances, compared to non-lesioned colonies (Meyer et al. 2014). These studies suggest that Endozoicomonas bacteria are part of a healthy coral microbiome and reductions in their abundance may indicate unfavourable environmental conditions.
Gorgonians, commonly known as sea fans, are closely related to corals and also have a microbiome frequently dominated by Endozoicomonas. In the Mediterranean, the main gorgonian species (Eunicella cavolini, E. singularis, E. verrucosa, Leptogorgia sarmentosa, and Paramuricea clavata) harbour a microbiome dominated by Endozoicomonas symbionts (van de Water et al. In revision; Bayer et al. 2013a; La Rivière et al. 2015). Interestingly, patterns of host-specificity and possibly co-evolution between the gorgonian species and their specific Endozoicomonas genotype have also been observed (van de Water et al. In revision; La Rivière et al. 2015). In addition, another octocoral species, the red coral Corallium rubrum, also harbour Endozoicomonas, although it is much less abundant compared to the other gorgonians and is dominated by Spirochaetales (van de Water et al. 2016). Outside of the Mediterranean, Endozoicomonas symbionts were found in the gorgonian E. verrucosa living off the south-west coast of England (Ransome et al. 2014), as well as in the Caribbean gorgonian Pseudopterogorgia elisabethae (Correa et al. 2013). Although corals and gorgonians dominate the Endozoicomonas literature, these symbionts have also been found in a range of other cnidarian species. Early work by Schuett et al. (2007) detected an Endozoicomonas strain with 98 % similarity to E. elysicola and observed bacterial cell aggregates in the tentacles of the sea anemone Metridium senile from Helgoland in the North Sea. Although these aggregates were not confirmed as Endozoicomonas, they used scanning electron microscopy to obtain detailed images of the bacterial cells forming large aggregates within host tissues. More recently, an Endozoicomonas strain with 99 % similarity to E. elysicola was found in another sea anemone, Nematostella vectensis, from a marsh of varying salinity conditions in Massachusetts, USA (Har et al. 2015). Endozoicomonas symbionts also dominated two jellyfish species, Mastigias cf. papua and Tripedalia cf. cystophora, in several Indonesian lakes (Cleary et al. 2016). These examples show that Endozoicomonas bacteria are symbiotic with a large diversity of cnidarian species and are often abundant and host species-specific, suggesting an important and ancient evolutionary relationship with lineage-specific evolution.
Endozoicomonas are also known to associate with a wide range of other marine organisms (Fig. 2). A number of particularly interesting examples of the adaptability of Endozoicomonas come from deep-sea hydrothermal vent communities. Forget and Juniper (2013) collected the tubeworm, Ridgeia piscesae, from the Juan de Fuca Ridge in the north-east Pacific, which has high hydrogen sulphide concentrations (~40 μmol/L), temperatures up to 41.9 °C, and depths greater than 2000 m. Even in this extreme environment, R. piscesae contained abundant Endozoicomonas symbionts (Forget and Juniper 2013). Moreover, Endozoicomonas bacteria have been recovered from the gills of the hydrothermal vent snail Alviniconcha (Beinart et al. 2014), the gills of the deep-water bivalve Acesta excavata (Jensen et al. 2010), and from the tissues of deep-water corals (Meistertzheim et al. 2016). Endozoicomonas have also been associated with a number of sponge species, which are one of the oldest groups of metazoan invertebrates and often harbour a rich diversity of microbial symbionts (Rua et al. 2014). In the Caribbean, the giant barrel sponge (Xestospongia muta) not only contained Endozoicomonas bacteria, but evidence of their transcriptional activity was also detected (Fiore et al. 2015). Sponges in Brazil (Rua et al. 2014), Japan (Nishijima et al. 2013), and several other European seas also contain Endozoicomonas symbionts (Esteves et al. 2013; Gardères et al. 2015). In addition, tunicates, which are basal chordates, have a microbiome that consistently contains a substantial proportion of Endozoicomonas, suggesting that these symbionts are core members of the tunicate microbiome (Dishaw et al. 2014). Although reports to date mostly associate Endozoicomonas with marine invertebrates, several examples of associations with fish have also emerged in the recent literature. In these cases, fish were kept in artificial aquaculture environments and Endozoicomonas bacteria are suspected to have caused disease (Mendoza et al. 2013; Katharios et al. 2015). In both examples, Endozoicomonas formed cyst-like aggregations on the gills of the fish, resulting in epitheliocystis (Mendoza et al. 2013; Katharios et al. 2015).
The central picture that emerges is the remarkable ability of Endozoicomonas bacteria to adapt to a wide range of hosts and environments, from warm coral reefs to cold deep-sea mussels, and their apparent ability to transition from beneficial core microbiome members of corals and tunicates to disease-causing pathogens in fish.
Function and genetic potential of Endozoicomonas
The proposed functions of Endozoicomonas can be summarized into three categories: nutrient acquisition and provision, structuring of the host microbiome, and roles in host health or disease (Table 1). Nutrient acquisition spans from nitrogen and carbon recycling (Nishijima et al. 2013; Forget and Juniper 2013; Correa et al. 2013; Morrow et al. 2015), or methane and sulphur cycling (Bourne et al. 2013; Forget and Juniper 2013; Correa et al. 2013; Dishaw et al. 2014; Morrow et al. 2015), to the synthesis of amino acids and other essential molecules (Neave, Michell, Apprill and Voolstra, Endozoicomonas genomes reveals functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts, Submitted). Bourne et al. (2013) found that the abundance of Endozoicomonas-related sequences (referred to as Oceanospirillales sp. 1, 3, 5, and 6) in invertebrate microbial communities correlated with the presence of photosymbionts, such as Symbiodinium algae in coral tissues. They suggested that the photosymbionts provide carbon and sulphur to the bacteria from the large quantities of dimethylsulfopropionate (DMSP) produced (Bourne et al. 2013; Correa et al. 2013). On the other hand, Endozoicomonas bacteria are also found in hosts without photosymbionts (Bourne et al. 2013). In addition to nutrient cycling, Endozoicomonas-related members may also play a role in regulating bacterial colonization of the animal host via the production of bioactive secondary metabolites or probiotic mechanisms, such as competitive exclusion of pathogenic bacteria (Bayer et al. 2013b; Jessen et al. 2013; Rua et al. 2014; Morrow et al. 2015). Moreover, the loss of Endozoicomonas is often characteristic of corals with lesions, signs of disease, or if they are living in eutrophicated, warm, or acidic environments. Therefore, the abundance of Endozoicomonas seems to be linked with healthy colonies of diverse coral species (Morrow et al. 2012; Bayer et al. 2013b; Roder et al. 2015; Morrow et al. 2015; Ziegler et al. 2016).
Table 1.
Suggested functions of Endozoicomonas bacteria
| Host organism | Suggested function | Reference |
|---|---|---|
| Fish | Fish disease | (Mendoza et al. 2013; Katharios et al. 2015) |
| Sponge | Sponge health | (Gardères et al. 2015) |
| Bromopyrrole production for feeding deterence | (Haber and Ilan 2014) | |
| Carbohydrate fermentation/nitrate reduction | (Nishijima et al. 2013) | |
| Antibiotic production | (Rua et al. 2014) | |
| Tunicate | Sulphur cycling/nutrient metabolism | (Dishaw et al. 2014) |
| Hydrothermal vent snail | Host nutrition/sulphur cycling or breakdown of organic compounds | (Beinart et al. 2014) |
| Hydrothermal polychaete | Methane cycling/food degradation | (Forget and Juniper 2013) |
| Scleractinian corals | Quorum-sensing molecules | (Bayer et al. 2013b) |
| Microbiome structuring | (Jessen et al. 2013) | |
| Antimicrobial activity/N-acyl homoserine lactones | (Morrow et al. 2015) | |
| Coral health | (Meyer et al. 2014; Roder et al. 2015; Webster et al. 2016) | |
| Coral health and/or disease | (Ziegler et al. 2016) | |
| Protection from bleaching | (Pantos et al. 2015) | |
| Dimethylsulfoniopropionate (DMSP) metabolism/sulphur cycling | (Raina et al. 2009; Bourne et al. 2013; Correa et al. 2013) | |
| Carbohydrate metabolism/nutrient acquisition | (Correa et al. 2013; Morrow et al. 2015) | |
| Octocoral (gorgonians) | Host health | (Vezzulli et al. 2013; Ransome et al. 2014) |
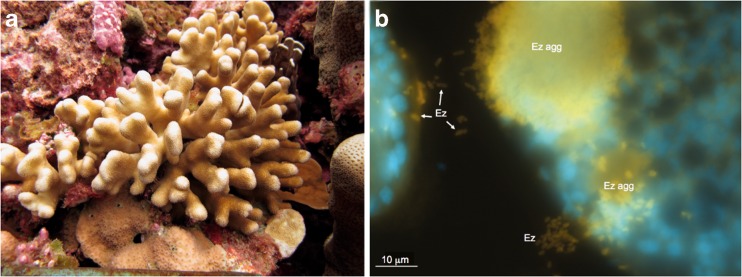
The spatial location of Endozoicomonas bacteria within host tissues may also have functional implications. For example, Endozoicomonas frequently form aggregations in various host habitats (Bayer et al. 2013b; Mendoza et al. 2013; Katharios et al. 2015; Schreiber et al. 2016). In corals, fluorescent oligonucleotides have been designed for Endozoicomonas and used to hybridize to Endozoicomonas cells, confirming their residence within host tissues (Bayer et al. 2013b), where they formed similar structures to cell-associated microbial aggregates or CAMAs as previously described using histological staining in corals (Work and Aeby 2014). More recently, using catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH) with this same probe, bacterial cells have been better resolved within the autofluorescent coral tissues and it was found that Endozoicomonas cells form dense aggregations and can reside within the tentacles of corals (Fig. 3; Neave et al. 2016). Aggregations filled with thousands of bacterial cells have also been found in anemone tentacles, and although SSU rRNA gene sequence data demonstrates that Endozoicomonas are in residence with these anemones, the tentacle-associated cells have not yet been confirmed as Endozoicomonas (Schuett et al. 2007). Another probe developed for Endozoicomonas also identified cells residing in extracellular aggregations in sea squirts (Schreiber et al. 2016). In fish, Endozoicomonas also form extremely dense aggregations containing thousands of individuals surrounded by a thin tightly enveloping membrane (Mendoza et al. 2013; Katharios et al. 2015). These formations, particularly the membrane barrier, may provide protection from host immune cells or other host responses to bacterial infection. Functionally, the aggregations may act as centres of protein transformation and production that could be beneficial for the host. Moreover, several genotypes of Endozoicomonas are known to inhabit individual hosts (Neave et al. 2016), and the aggregations could be comprised of several complementary genotypes, or even different bacterial species, that work together to passage nutrients and proteins. The formation of aggregations also suggests that some form of cell-to-cell communication is required, such as quorum-sensing molecules (Waters and Bassler 2005). To date, all Endozoicomonas microscopy studies have found aggregations in host tissues, suggesting that these formations are an important part of Endozoicomonas function and colonization.
Fig. 3.
Photograph of S. pistillata colony from Nukuoro atoll, Federated States of Micronesia (a), and microscopic photo of Endozoicomonas (Ez) probed cells (yellow) within the tentacles of S. pistillata residing in aggregates (Ez agg) as well as just outside the aggregate (b). Endozoicomonas cells are hybridized with the horseradish peroxidase-labelled probe targeting the Endozoicomonas bacteria, and the blue staining is a general nucleic acid stain (DAPI) (methodology detailed in Neave et al. (2016))
The recent availability of Endozoicomonas whole genome sequences has significantly improved our understanding of their genetic potential, possible functional roles, and life cycle. These data have revealed that Endozoicomonas genomes are relatively large, ranging from 5.60 Mbs for E. montiporae (Neave et al. 2014) to 6.83 Mbs for E. atrinae (Hyun et al. 2014). The genomes support a correspondingly large number of protein-coding genes, suggesting that genome streamlining, which often occurs in symbiotic bacteria (e.g. Kwan et al. 2012), is not occurring or at least not predominantly. This may indicate that Endozoicomonas have periods during their lifecycle that require a full complement of genetic pathways, e.g. a free-living stage. In a recent study of the E. montiporae genome, Ding et al. (2016) found genes coding for an N-deglycosylation enzyme that was hypothesized to help with penetration of the mucus layer of their coral host. They also found genes that may be involved in initiating internalization and evasion of the host immune response and hypothesized that Endozoicomonas was a beneficial symbiont that could help increase the efficiency of host gluconeogenesis (Ding et al. 2016). Interestingly, a high proportion of repeat sequences was found in the E. montiporae genome (Ding et al. 2016), which is similar to what was reported for a pathogenic Endozoicomonas strain (Katharios et al. 2015). This suggests that repeat and insertion sequences may help Endozoicomonas strains to adapt to new hosts or to transition between mutualistic and parasitic lifestyles. Importantly, Neave, Michell, Apprill and Voolstra (Endozoicomonas genomes reveals functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts, Submitted) comparatively analysed the genomes of E. elysicola, E. montiporae, E. numazuensis, and four newly sequenced Endozoicomonas strains from the Red Sea corals S. pistillata, P. verrucosa, and Acropora humilis and found a high proportion of transposable elements in the Endozoicomonas genomes, further implying that Endozoicomonas use these elements to rapidly evolve to new hosts or niches. In addition, the Endozoicomonas genomes were enriched for carbon sugar transport and protein secretion, suggesting that they contribute to carbohydrate cycling and delivery to their host organism (Neave, Michell, Apprill and Voolstra, Endozoicomonas genomes reveals functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts, Submitted). The common denominator among Endozoicomonas genome projects is the high incidence of transposable elements incorporated into their genomes, possibly allowing for rapid adaptation. These genomic findings are corroborated by field evidence showing that Endozoicomonas are associated with numerous hosts in numerous marine habitats. The genomes also tend to show the ability to transport and transform protein products that may then be used by the host. Although these studies provide important advances, future studies may be improved by obtaining in situ gene expression data or other forms of functional information, in order to refine our understanding of the specific role of Endozoicomonas with a particular host.
Future prospects—novel techniques to examine microbial functions
The functional role of Endozoicomonas bacteria within their many hosts is poorly understood. One fundamental but unknown aspect of many Endozoicomonas species is their genomic repertoire and metabolic potential. This question has traditionally been difficult to answer; however, new and emerging methods for determining bacterial function are likely to provide fascinating insights. Currently, many techniques that aim to discover bacterial function rely on the availability of cultured isolates. For example, bacterial cultures allow for efficient whole genome sequencing, for infectivity trials, and for traditional bacteriological tests of physiological and nutritional requirements. Although a number of Endozoicomonas species have been successfully cultured, many species are not readily amenable to isolation from host tissues (e.g. Katharios et al. 2015). In these cases, culture-independent methods can be used to obtain valuable data, such as whole genome sequences. For example, Katharios et al. (2015) micro-manipulated Endozoicomonas aggregates from sections of fish tissue, sequenced the DNA using high throughput technologies, and obtained a novel draft genome that provided valuable insights. Moreover, in cases where Endozoicomonas aggregates are not easily micro-manipulated, methods such as metagenomic binning and single-cell genomics can be used to acquire whole genome sequences. Recently, the genomes of Endozoicomonas associated with different species of Red Sea corals were obtained by differential coverage binning of coral metagenomes, as outlined by Albertsen et al. (2013), and by sequencing single-cell isolates (Neave, Michell, Apprill and Voolstra, Endozoicomonas genomes reveals functional adaptation and plasticity in bacterial strains symbiotically associated with diverse marine hosts, Submitted). With further improvements in metagenomic binning software (e.g. Imelfort et al. 2014; Alneberg et al. 2014; Eren et al. 2015) and in single-cell genomic methods, these may become useful for rapidly obtaining multiple genomes at a relatively small cost. A major improvement to these pipelines would be the addition of techniques that obtain gene expression data. One possible avenue that is already being developed is the use of RNA-Seq on isolated single cells (e.g. Wang and Navin 2015). Another is RNA subtraction technology, where total RNA is collected from host tissues, ribosomal RNA and eukaryotic messenger RNA are removed, and the remaining bacterial mRNA is sequenced (e.g. Stewart et al. 2010; Daniels et al. 2015). When combined, these techniques could provide fascinating insights into the intricate associations of Endozoicomonas-animal symbioses.
Although whole genome sequences and gene expression data are valuable, they cannot completely uncover the metabolic and physiological dynamics of Endozoicomonas bacteria in situ. For example, questions of how Endozoicomonas aggregates share metabolites with the host, or which Endozoicomonas genotypes are represented in aggregates, are difficult to answer. These questions may become tractable with the development of secondary ion mass spectrometry (SIMS), which has opened a new frontier in microbial ecology. While previously measurements of bacterial metabolism and physiology were restricted to community-based culture or whole-animal measurements, these novel technologies allow for the quantification of single-cell metabolisms at the spatial resolution of electron microscopy (Pernice and Levy 2014). Thereby, these technologies not only enable one to localize and quantify metabolic activity of selected tissues or microorganisms, but can also account for single-cell heterogeneity, a previously overlooked phenomenon, and of particular importance when hosts harbour multiple strains of Endozoicomonas (e.g. Neave et al. 2016). There are several SIMS technologies used to investigate metabolic functioning of bacteria in culture or within a host-symbiont framework. Nanoscale SIMS (NanoSIMS) is used to quantify stable isotope ratios at high spatial resolution. Coupled with pulse-chase isotope labelling, this technology can be used to detect the uptake, incorporation, and transfer of metabolites (Kopp et al. 2013). This technique may be particularly useful in detecting molecules that are consumed and produced by Endozoicomonas aggregations, providing data on molecule transformation within aggregations and allowing fascinating insights into potential benefits for the host. A recently developed modification of the traditional NanoSIMS approach is halogen in situ hybridization SIMS (HISH-SIMS) (Alonso et al. 2012), where specific probes are used to label targeted cells, which allows identification of the precise location of microbes within host tissue. This modification would permit Endozoicomonas aggregations to be interrogated for single-cell heterogeneity, including different genotypes or different bacterial species, and assess the relative contributions of these to aggregate functioning. Another technique called Time-of-Flight SIMS (ToF-SIMS) uses high-resolution mass spectrometry to identify molecules within a given cell or tissue (Colliver et al. 1997). This technology can be used to identify metabolic processes within microorganisms or the compounds they release, which would provide metabolic data at the level of individual Endozoicomonas cells and genotypes. While all of these technologies have proven to be powerful tools in understanding microbial function, they are even more powerful when combined. For instance, using Nano-, HISH- and ToF-SIMS on the same biological sample would make it possible to highlight specific Endozoicomonas genotypes in a host organism background and to characterize and quantify the nature of translocated metabolites, which should provide quantum leaps in our understanding of the function of host-associated Endozoicomonas bacteria.
Conclusions and outlook
Endozoicomonas bacteria are rapidly being recognized as globally important marine symbionts. Few other bacterial genera are symbiotic with such a large diversity and distribution of host species, suggesting that the function and adaptability of Endozoicomonas bacteria are particularly exceptional. Indeed, several Endozoicomonas species have unusually flexible genomes and a great diversity of metabolic pathways. Studies on the exact functional roles of Endozoicomonas within their many hosts are only in their infancy, and many exciting and novel discoveries await. Moreover, with new technologies allowing for in situ genome sequencing, gene expression analysis, and the measurement of metabolites, future functional discoveries will be greatly accelerated.
Acknowledgments
The authors would like to thank Nils Rädecker for providing input on SIMS technologies.
Compliance with ethical standards
Funding
We would like to acknowledge the following sources for funding: this work was supported by a KAUST-WHOI Post-doctoral Partnership Award to MJN and a KAUST-WHOI Special Academic Partnership Funding Reserve Award to CRV and AA. Research in this study was further supported by baseline research funds to CRV by KAUST and NSF award OCE-1233612 to AA.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors. Referenced studies adhere to applicable international, national and/or institutional guidelines for the care and use of animals.
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- Alneberg J, Bjarnason BS, de Bruijn I, Schirmer M, Quick J, Ijaz UZ, Lahti L, Loman NJ, Andersson AF, Quince C. Binning metagenomic contigs by coverage and composition. Nat Meth. 2014;11:1144–1146. doi: 10.1038/nmeth.3103. [DOI] [PubMed] [Google Scholar]
- Alonso C, Musat N, Adam B, Kuypers M, Amann R. HISH-SIMS analysis of bacterial uptake of algal-derived carbon in the Rio de la Plata estuary. Syst Appl Microbiol. 2012;35:541–548. doi: 10.1016/j.syapm.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Appolinario LR, Tschoeke DA, Rua CPJ, Venas T, Campeão ME, Amaral GRS, Leomil L, de Oliveira L, Vieira VV, Otsuki K, Swings J, Thompson FL, Thompson CC. Description of Endozoicomonas arenosclerae sp. nov. using a genomic taxonomy approach. Antonie Van Leeuwenhoek. 2016;109:431–438. doi: 10.1007/s10482-016-0649-x. [DOI] [PubMed] [Google Scholar]
- Bayer T, Arif C, Ferrier-Pagès C, Zoccola D, Aranda M, Voolstra CR. Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini. Mar Ecol Prog Ser. 2013;479:75–84. doi: 10.3354/meps10197. [DOI] [Google Scholar]
- Bayer T, Neave MJ, Alsheikh-Hussain A, Aranda M, Yum LK, Mincer T, Hughen K, Apprill A, Voolstra CR. The microbiome of the Red Sea coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79:4759–4762. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinart R, Nyholm S, Dubilier N, Girguis P. Intracellular Oceanospirillales inhabit the gills of the hydrothermal vent snail Alviniconcha with chemosynthetic, γ-proteobacterial symbionts. Environ Microbiol Rep. 2014;6:656–664. doi: 10.1111/1758-2229.12183. [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne D, Iida Y, Uthicke S, Smith-Keune C. Changes in coral-associated microbial communities during a bleaching event. ISME J. 2008;2:350–363. doi: 10.1038/ismej.2007.112. [DOI] [PubMed] [Google Scholar]
- Bourne DG, Dennis PG, Uthicke S, Soo RM, Tyson GW, Webster N (2013) Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J:1–7. doi:10.1038/ismej.2012.172 [DOI] [PMC free article] [PubMed]
- Bourne DG, Munn CB. Diversity of bacteria associated with the coral Pocillopora damicornis from the Great Barrier Reef. Environ Microbiol. 2005;7:1162–1174. doi: 10.1111/j.1462-2920.2005.00793.x. [DOI] [PubMed] [Google Scholar]
- Cleary DFR, Becking LE, Polónia ARM, Freitas RM, Gomes NCM. Jellyfish-associated bacterial communities and bacterioplankton in Indonesian Marine lakes. FEMS Microbiol Ecol. 2016 doi: 10.1093/femsec/fiw064. [DOI] [PubMed] [Google Scholar]
- Colliver TL, Brummel CL, Pacholski ML, Swanek FD, Ewing AG, Winograd N. Atomic and molecular imaging at the single-cell level with TOF-SIMS. Anal Chem. 1997;69:2225–2231. doi: 10.1021/ac9701748. [DOI] [PubMed] [Google Scholar]
- Correa H, Haltli B, Duque C, Kerr R. Bacterial communities of the gorgonian octocoral Pseudopterogorgia elisabethae. Microb Ecol. 2013;66:972–985. doi: 10.1007/s00248-013-0267-3. [DOI] [PubMed] [Google Scholar]
- Costa PM, Carreira S, Lobo J, Costa MH. Molecular detection of prokaryote and protozoan parasites in the commercial bivalve Ruditapes decussatus from southern Portugal. Aquaculture. 2012;370–371:61–67. doi: 10.1016/j.aquaculture.2012.10.006. [DOI] [Google Scholar]
- Daniels CA, Baumgarten S, Yum LK, Michell CT, Bayer T, Arif C, Roder C, Weil E, Voolstra CR. Metatranscriptome analysis of the reef-building coral Orbicella faveolata indicates holobiont response to coral disease. Front Mar Sci. 2015;2:62. doi: 10.3389/fmars.2015.00062. [DOI] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J-Y, Shiu J-H, Chen W-M, Chiang Y-R, Tang S-L. Genomic insight into the host–endosymbiont relationship of Endozoicomonas montiporae CL-33(T) with its coral host. Front Microbiol. 2016;7:251. doi: 10.3389/fmicb.2016.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishaw LJ, Flores-Torres J, Lax S, Gemayel K, Leigh B, Melillo D, Mueller MG, Natale L, Zucchetti I, De Santis R, Pinto MR, Litman GW, Gilbert JA. The gut of geographically disparate Ciona intestinalis harbors a core microbiota. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Esen C, Quince C, Vineis JH, Sogin ML, Delmont O. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3 doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves AIS, Hardoim CCP, Xavier JR, Gonçalves JMS, Costa R. Molecular richness and biotechnological potential of bacteria cultured from Irciniidae sponges in the north-east Atlantic. FEMS Microbiol Ecol. 2013;85:519–536. doi: 10.1111/1574-6941.12140. [DOI] [PubMed] [Google Scholar]
- Fiore CL, Labrie M, Jarett JK, Lesser MP. Transcriptional activity of the giant barrel sponge, Xestospongia muta holobiont: molecular evidence for metabolic interchange. Front Microbiol. 2015;6:364. doi: 10.3389/fmicb.2015.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget NL, Juniper KS. Free-living bacterial communities associated with tubeworm (Ridgeia piscesae) aggregations in contrasting diffuse flow hydrothermal vent habitats at the Main Endeavour Field, Juan de Fuca Ridge. Microbiologyopen. 2013;2:259–275. doi: 10.1002/mbo3.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardères J, Bedoux G, Koutsouveli V, Crequer S, Desriac F, Le Pennec G. Lipopolysaccharides from commensal and opportunistic bacteria: characterization and response of the immune system of the host sponge Suberites domuncula. Mar Drugs. 2015;13:4985–5006. doi: 10.3390/md13084985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M, Ilan M (2014) Diversity and antibacterial activity of bacteria cultured from Mediterranean Axinella spp. sponges. J Appl Microbiol 116(3):519–32. doi:10.1111/jam.12401 [DOI] [PubMed]
- Har JY, Helbig T, Lim JH, Fernando SC, Penn K, Reitzel AM, Renee J, Har JY, Helbig T, Lim JH, Fernando SC, Reitzel AM, Penn K, Thompson JR. Microbial diversity and activity in the Nematostella vectensis holobiont: insights from 16S rRNA gene sequencing, isolate genomes, and a pilot-scale survey of gene expression. Front Mar Sci. 2015;6:818. doi: 10.3389/fmicb.2015.00818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun D-W, Shin N-R, Kim M-S, Oh SJ, Kim PS, Whon TW, Bae J-W. Endozoicomonas atrinae sp. nov., a bacterium isolated from the intestine of a comb pen shell Atrina pectinata. Int J Syst Evol Microbiol. 2014;64:2312–2318. doi: 10.1099/ijs.0.060780-0. [DOI] [PubMed] [Google Scholar]
- Imelfort M, Parks D, Woodcroft BJ, Dennis P, Hugenholtz P, Tyson GW. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ. 2014;2 doi: 10.7717/peerj.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Duperron S, Birkeland N-K, Hovland M. Intracellular Oceanospirillales bacteria inhabit gills of Acesta bivalves. FEMS Microbiol Ecol. 2010;74:523–533. doi: 10.1111/j.1574-6941.2010.00981.x. [DOI] [PubMed] [Google Scholar]
- Jessen C, Villa Lizcano JF, Bayer T, Roder C, Aranda M, Wild C, Voolstra CR. In-situ effects of eutrophication and overfishing on physiology and bacterial diversity of the Red Sea coral Acropora hemprichii. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katharios P, Seth-Smith HMB, Fehr A, Mateos JM, Qi W, Richter D, Nufer L, Ruetten M, Guevara Soto M, Ziegler U, Thomson NR, Schlapbach R, Vaughan L. Environmental marine pathogen isolation using mesocosm culture of sharpsnout seabream: striking genomic and morphological features of novel Endozoicomonas sp. Sci Rep. 2015;5:17609. doi: 10.1038/srep17609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp C, Pernice M, Domart-Coulon I, Djediat C, Spangenberg JE, Alexander DTL, Hignette M, Meziane T, Meibom A. Highly dynamic cellular-level response of symbiotic coral to a sudden increase in environmental nitrogen. MBio. 2013;4:e00052–e00013. doi: 10.1128/mBio.00052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi M, Yokota A. Endozoicomonas elysicola gen. nov., sp. nov., a gamma-proteobacterium isolated from the sea slug Elysia ornata. Syst Appl Microbiol. 2007;30:202–206. doi: 10.1016/j.syapm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Kvennefors ECE, Sampayo E, Ridgway T, Barnes AC, Hoegh-Guldberg O. Bacterial communities of two ubiquitous Great Barrier Reef corals reveals both site- and species-specificity of common bacterial associates. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JC, Donia MS, Han AW, Hirose E, Haygood MG, Schmidt EW, Eric W. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci U S A. 2012;109:20655–20660. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Woyke T, Eisen JA, Garrity G, Lilburn TG, Beck BJ, Whitman WB, Hugenholtz P, Klenk H-P. Genomic encyclopedia of type strains, phase I: the one thousand microbial genomes (KMG-I) project. Stand Genomic Sci. 2014;9:1278–1284. doi: 10.4056/sigs.5068949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rivière M, Garrabou J, Bally M. Evidence for host specificity among dominant bacterial symbionts in temperate gorgonian corals. Coral Reefs. 2015;34:1087–1098. doi: 10.1007/s00338-015-1334-7. [DOI] [Google Scholar]
- Lawler SN, Kellogg CA, France SC, Clostio RW, Brooke SD, Ross SW. Coral-associated bacterial diversity is conserved across two deep-sea Anthothela species. Front Microbiol. 2016;7:1–18. doi: 10.3389/fmicb.2016.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lema KA, Willis BL, Bourne DG. Amplicon pyrosequencing reveals spatial and temporal consistency in diazotroph assemblages of the Acropora millepora microbiome. Environ Microbiol. 2013;16:3345–3359. doi: 10.1111/1462-2920.12366. [DOI] [PubMed] [Google Scholar]
- McCauley E, Haltli B, Correa H, Kerr RG. Spatial and temporal investigation of the microbiome of the Caribbean octocoral Erythropodium caribaeorum. FEMS Microbiol Ecol. 2016 doi: 10.1093/femsec/fiw147. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistertzheim AL, Lartaud F, Arnaud-Haond S, Kalenitchenko D, Bessalam M, Le Bris N, Galand PE. Patterns of bacteria-host associations suggest different ecological strategies between two reef building cold-water coral species. Deep Sea Res Part I Ocean Res Pap. 2016;114:12–22. doi: 10.1016/j.dsr.2016.04.013. [DOI] [Google Scholar]
- Mendoza M, Güiza L, Martinez X, Caraballo X, Rojas J, Aranguren LF, Salazar M. A novel agent (Endozoicomonas elysicola) responsible for epitheliocystis in cobia Rachycentrum canadum larvae. Dis Aquat Org. 2013;106:31–37. doi: 10.3354/dao02636. [DOI] [PubMed] [Google Scholar]
- Meyer JL, Paul VJ, Teplitski M. Community shifts in the surface microbiomes of the coral Porites astreoides with unusual lesions. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow KM, Bourne DG, Humphrey C, Botté ES, Laffy P, Zaneveld J, Uthicke S, Fabricius KE, Webster NS. Natural volcanic CO2 seeps reveal future trajectories for host-microbial associations in corals and sponges. ISME J. 2015;9:894–908. doi: 10.1038/ismej.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow KM, Moss AG, Chadwick NE, Liles MR. Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability. Appl Environ Microbiol. 2012;78:6438–6449. doi: 10.1128/AEM.01162-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave MJ, Michell CT, Apprill A, Voolstra R. Whole-genome sequences of three symbiotic Endozoicomonas strains. Genome Announc. 2014;2:e00802–e00814. doi: 10.1128/genomeA.00802-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neave MJ, Rachmawati R, Xun L, Michell CT, Bourne DG, Apprill A, Voolstra CR, (2016) Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. The ISME Journal. doi:10.1038/ismej.2016.95 [DOI] [PMC free article] [PubMed]
- Nishijima M, Adachi K, Katsuta A, Shizuri Y, Yamasato K. Endozoicomonas numazuensis sp. nov., a gammaproteobacterium isolated from marine sponges, and emended description of the genus Endozoicomonas Kurahashi and Yokota 2007. Int J Syst Evol Microbiol. 2013;63:709–714. doi: 10.1099/ijs.0.042077-0. [DOI] [PubMed] [Google Scholar]
- Pernice M, Levy O. Novel tools integrating metabolic and gene function to study the impact of the environment on coral symbiosis. Front Microbiol. 2014;5:448. doi: 10.3389/fmicb.2014.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike RE, Haltli B, Kerr RG. Endozoicomonas euniceicola sp. nov. and Endozoicomonas gorgoniicola sp. nov., bacteria isolated from the octocorals, Eunicea fusca and Plexaura sp. Int J Syst Evol Microbiol. 2013;63:4294–4302. doi: 10.1099/ijs.0.051490-0. [DOI] [PubMed] [Google Scholar]
- Raina JB, Tapiolas D, Willis BL, Bourne DG (2009) Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75(11):3492–501. doi:10.1128/AEM.02567-08 [DOI] [PMC free article] [PubMed]
- Ransome E, Rowley SJ, Thomas S, Tait K, Munn CB. Disturbance to conserved bacterial communities in the cold water gorgonian coral Eunicella verrucosa. FEMS Microbiol Ecol. 2014;90:404–416. doi: 10.1111/1574-6941.12398. [DOI] [PubMed] [Google Scholar]
- Roder C, Bayer T, Aranda M, Kruse M, Voolstra CR. Microbiome structure of the fungid coral Ctenactis echinata aligns with environmental differences. Mol Ecol. 2015;24:3501–3511. doi: 10.1111/mec.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lanetty M, Granados-Cifuentes C, Barberan A, Bellantuono AJ, Bastidas C. Ecological inferences from a deep screening of the complex bacterial consortia associated with the coral, Porites astreoides. Mol Ecol. 2013;22:4349–4362. doi: 10.1111/mec.12392. [DOI] [PubMed] [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- Rua CPJ, Trindade-Silva AE, Appolinario LR, Venas TM, Garcia GD, Carvalho LS, Lima A, Kruger R, Pereira RC, Berlinck RGS, Valle RAB, Thompson CC, Thompson F. Diversity and antimicrobial potential of culturable heterotrophic bacteria associated with the endemic marine sponge Arenosclera brasiliensis. PeerJ. 2014;2 doi: 10.7717/peerj.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber L, Kjeldsen KU, Funch P, Jensen J, Obst M, López-legentil S, Schramm A. Endozoicomonas are specific, facultative symbionts of sea squirts. Front Microbiol. 2016 doi: 10.3389/fmicb.2016.01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuett C, Doepke H, Grathoff A, Gedde M. Bacterial aggregates in the tentacles of the sea anemone Metridium senile. Helgol Mar Res. 2007;61:211–216. doi: 10.1007/s10152-007-0069-4. [DOI] [Google Scholar]
- Stewart FJ, Ottesen EA, Delong EF. Development and quantitative analyses of a universal rRNA-subtraction protocol for microbial metatranscriptomics. ISME J. 2010;4:896–907. doi: 10.1038/ismej.2010.18. [DOI] [PubMed] [Google Scholar]
- van de Water J, Melkonian R, Voolstra CR, Junca H, Beraud E, Allemand D, Ferrier-Pagès C Comparative assessment of Mediterranean gorgonian-associated microbial communities reveals local impacts on spatially conserved core microbiomes. In revision [DOI] [PubMed]
- van de Water JAJM, Melkonian R, Junca H, Voolstra CR, Reynaud S, Allemand D, Ferrier-Pagès C. Spirochaetes dominate the microbial community associated with the red coral Corallium rubrum on a broad geographic scale. Sci Rep. 2016;6:27277. doi: 10.1038/srep27277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzulli L, Pezzati E, Huete-Stauffer C, Pruzzo C, Cerrano C (2013) 16SrDNA pyrosequencing of the Mediterranean gorgonian paramuricea clavata reveals a link among alterations in bacterial holobiont members, anthropogenic influence and disease outbreaks. PloS one 8(6):e67745. doi:10.1371/journal.pone.0067745 [DOI] [PMC free article] [PubMed]
- Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Work TM, Aeby GS. Microbial aggregates within tissues infect a diversity of corals throughout the Indo-Pacific. Mar Ecol Prog Ser. 2014;500:1–9. doi: 10.3354/meps10698. [DOI] [Google Scholar]
- Webster NS, Negri AP, Botte ES, Laffy PW, Flores F, Noonan S, Schmidt C, Uthicke S (2016) Hostassociated coral reef microbes respond to the cumulative pressures of ocean warming and ocean acidification. Sci Rep 6:19324. doi:10.1038/srep19324 [DOI] [PMC free article] [PubMed]
- Yang C-S, Chen M-H, Arun AB, Chen CA, Wang JT, Chen WM. Endozoicomonas montiporae sp. nov., isolated from the encrusting pore coral Montipora aequituberculata. Int J Syst Evol Microbiol. 2010;60:1158–1162. doi: 10.1099/ijs.0.014357-0. [DOI] [PubMed] [Google Scholar]
- Ziegler M, Roik A, Porter A, Zubier K, Mudarris MS, Ormond R, Voolstra CR. Coral microbial community dynamics in response to anthropogenic impacts near a major city in the central Red Sea. Mar Pollut Bull. 2016;105:629–640. doi: 10.1016/j.marpolbul.2015.12.045. [DOI] [PubMed] [Google Scholar]