Abstract
Long noncoding RNAs (lncRNA) have been recognized as important regulators in diverse biological processes, such as transcriptional regulation, stem cell proliferation, and differentiation. Previous study has demonstrated that lncRNA-ANCR (antidifferentiation ncRNA) plays a key role in regulating the proliferation and osteogenic differentiation of periodontal ligament stem cells (PDLSCs). However, little is known about the role of ANCR in regulating other types of dental tissue-derived stem cells (DTSCs) behaviours (including proliferation and multiple-potential of differentiation). In this study, we investigated the regulatory effects of lncRNA-ANCR on the proliferation and differentiation (including osteogenic, adipogenic, and neurogenic differentiation) of DTSCs, including dental pulp stem cells (DPSCs), PDLSCs, and stem cells from the apical papilla (SCAP) by downregulation of lncRNA-ANCR. We found that downregulation of ANCR exerted little effect on proliferation of DPSCs and SCAP but promoted the osteogenic, adipogenic, and neurogenic differentiation of DTSCs. These data provide an insight into the regulatory effects of long noncoding RNA-ANCR on DTSCs and indicate that ANCR is a very important regulatory factor in stem cell differentiation.
1. Introduction
Dental tissue-derived stem cells (DTSCs) are stem cells separated from the dental tissue that have self-proliferation and multidirectional differentiation potential. Dental pulp stem cells (DPSCs), periodontal ligament stem cells (PDLSCs), and stem cells from the apical papilla (SCAP) are three important DTSCs which are used in tissue engineering research [1–4]. Hilkens et al. found that DPSCs are capable of osteogenic, adipogenic, and chondrogenic differentiation via different induction methods [5]. Kim et al. induced differentiation of DPSCs into cementoblast and neurocytes-like cells [6]. Dissanayaka and Janebodin found that DPSCs could be differentiated into vascular endothelial cells and had angiopoietic abilities after stimulation with an angiopoietic factor [7, 8]. Seo et al. combined PDLSCs with HA/TCP and implanted the complex into a nude mouse where the PDLSCs formed periodontium and cementum-like tissues [2]. Sonoyama et al. seeded PDLSCs and SCAP mixed cells into HA/TCP and implanted the complex into young swine where the mixed cells formed functional bioroot and periodontium [9].
Many factors, including growth factors, inflammatory factors, and microRNAs, play important roles in regulating the proliferation and differentiation of DTSCs (Table 1) [7, 10–25]. After lucubration on the regulatory effects of microRNAs on DTSCs, scholars discovered that many noncoding RNAs were key regulating factors in the proliferation and differentiation of DTSCs [14–16].
Table 1.
The effects of different regulating factors on DTSC.
| Research team | Cell | Regulating factors | Regulatory effect |
|---|---|---|---|
| Osathanon et al. [10] | DPSC | BFGF | Inhibit osteogenic differentiation |
| Dissanayaka et al. [7] | DPSC | VEGF | Promote angiopoietic differentiation |
| Feng et al. [11] | DPSC | IGF | Promote osteogenic differentiation |
| He et al. [12] | DPSC | LPS | Promote osteogenic differentiation |
| Yang et al. [13] | DPSC | IL-1, TNF-α | Promote osteogenic differentiation |
| Gay et al. [14] | DPSC | MicroRNA-218 | Inhibit osteogenic differentiation |
| Hara et al. [15] | DPSC | MicroRNA-720 | Promote osteogenic differentiation |
| Wei et al. [16] | PDLSC | MicroRNA-21 | Promote osteogenic differentiation |
| Ye et al. [17] | PDLSC | BMP9 | Promote osteogenic differentiation |
| Osathanon et al. [18] | PDLSC | BFGF | Inhibit osteogenic differentiation |
| Chen et al. [19] | PDLSC | NFκ-B | Promote osteogenic differentiation |
| Song et al. [20] | PDLSC | BMP2 | Promote adipogenic differentiation |
| Osathanon et al. [21] | PDLSC | Notch signaling | Promote neurogenic differentiation |
| Wu et al. [22] | SCAP | BFGF | Inhibit osteogenic differentiation |
| Wang et al. [23] | SCAP | Canonical Wnt pathway | Promote osteogenic differentiation |
| Zhang et al. [24] | SCAP | BMP2 | Promote osteogenic differentiation |
| Li et al. [25] | SCAP | 17β estradiol | Promote osteogenic differentiation |
While 93% of human genomic DNA is translated into RNA, only 2% of RNA is transcribed into protein. Most of the DNA is translated into noncoding RNAs [26]. Long noncoding RNA (lncRNA) used to be recognized as useless transcriptional noise [27]. In recent years, scientists discovered that they were very important in regulating epigenetic, transcriptional, and posttranscriptional gene expression [26–31]. Antidifferentiation noncoding RNA (ANCR) is a newly found lncRNA that is downregulated during stem cell differentiation and required to keep epidermal stem cells or osteoblast cells in an undifferentiated cell state. Depleting ANCR in progenitor-containing populations, without any other stimuli, resulted in rapid differentiation and gene induction [32]. Zhu and Xu found that downregulating ANCR promoted osteoblast differentiation by targeting EZH2 and regulating Runx2 expression [33]. Additionally, our previous research also showed that ANCR promoted the osteogenic differentiation of PDLSCs [34]. In this experiment, we used RNA interference to downregulate ANCR expression to further analyze the regulatory function of ANCR on the proliferation and differentiation of DTSCs, which would give a laterally comparative view on the regulatory function of ANCR on DTSCs.
2. Materials and Methods
2.1. Sample Collection and Cell Culture
Healthy impacted third molars were collected from adult humans between 19 and 29 years old from the dental hospital of the Fourth Military Medical University. All tooth extractions were conducted under the approval of the Ethical Committee of School of Stomatology, Fourth Military Medical University (permission number IRB-REV-2014-018). Informed consent was obtained from all subjects and the methods were carried out in accordance with the approved guidelines. Tissues from the dental pulp, periodontal ligament, and apical papilla were isolated from each molar, as previously described [1–3, 35]. Briefly, the tissues were gently separated and digested in a solution composed of 3 mg/mL of collagenase type I (Invitrogen, USA) and 4 mg/mL of dispase (Invitrogen, USA) for 40 min at 37°C. The cell suspension was obtained by passing the solution through a 70-μm strainer. Then, 1 × 104 cells were seeded into each well of a 6-well plate. α-modification of Eagle's Medium (α-MEM, Invitrogen, USA) supplemented with 10% fetal bovine serum (HyClone, USA), 100 mol/L of L-ascorbic acid 2-phosphate (Sigma, USA), 2 mmol/L of L-glutamine (Sigma, USA), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (Gibco, USA) was used as the standard culturing medium for cells at 37°C with 5% CO2. The medium was changed every 2-3 days. A limited dilution technique was applied and single cells were seeded into a 96-well plate to obtain single cell–derived colonies. Upon reaching 80% confluency, a number of these single-colony-derived lines were passaged at a 1 : 3 ratio for further culturing.
2.2. Immunophenotype Analysis
DTSCs were stained with stem cell surface markers and analyzed by flow cytometry as described previously [16]. Briefly, to identify the phenotypes of DTSCs, 5 × 105 cells at the 3rd passage were incubated with phycoerythrin (PE) conjugated monoclonal antibodies for human CD29, CD34, CD45, CD90, and CD146 or Allophycocyanin (APC) conjugated monoclonal antibody against human STRO-1, based on the manufacturer's instructions. The incubation procedure was carried out at 4°C away from light for 1 h. After washing with PBS, the cells were subjected to flow cytometric analysis.
2.3. Lentivirus Infection
The third-passage self-inactivating lentivirus vector was purchased from Neuron Biotech (Shanghai Neuron Biotech Co., Ltd., Shanghai, China). The vector contained a puromycin marker and U6 PolIII promoter, which allowed the introduction of oligonucleotides encoding short hairpin RNAs (shRNAs) and synthetic oligonucleotides containing the human ANCR (NCBI accession number NR_024031.1) splice variant target sequence (GCTGACCCTTACCCTGAATAC). The sequences for cloning were synthesized, annealed, and ligated into the pLKD-CMV-G lentiviral vector between the AgeI and EcoRI enzyme sites after the U6 promoter. The oligonucleotide sequences were 5′-CCGGGCTGACCCTTACCCTGAATACCTCGAGGTATTCAGGGTAAGGGTCAGCTTTTTT-3′ (sense); and 5′- AATTCAAAAAAGCTGACCCTTACCCTGAATACCTCGAGGTATTCAGGGTAAGGGTCAGC-3′ (antisense). DTSCs at passage 3 were plated at a density of 5 × 104 cells/well into 6-well plates. The cells were cultured for 3 days along with recombinant lentivirus encoding shRNA against ANCR at a multiplicity of infection (MOI) of 10, in α-MEM supplemented with 10% FBS containing 5 mg/mL of polybrene at 37°C and 5% CO2. The cells were then replated in 25-cm2 flasks in 90% α-MEM, 10% FBS, and 5 μg/mL of puromycin and cultured at 37°C with 5% CO2 and constant humidity. The ANCR expression after ANCR-RNA interference (RNAi) was examined by qRT-PCR analysis. Cells successfully infected with specific ANCR-RNAi were designated as DPSC/ANCR-RNAi, PDLSC/ANCR-RNAi, and SCAP/ANCR-RNAi, while the wild-type DTSCs were designated as DPSC/wt, PDLSC/wt, and SCAP/wt and cells infected by control lentivirus vector were designated as DPSC/vector, PDLSC/vector, and SCAP/vector.
2.4. Cell Viability Assay (CCK8 Kit)
Cells were seeded into two 96-well plates at a density of approximately 1 × 104 cells per well. The assay was performed 24 h after seeding and lasted for 7 days; data were collected at the same time point each day. A Cell Counting Kit-8 (CCK8) was used to perform this experiment. The number of cells per well was tested by the absorbance (450 nm) of reduced WST-8 at the indicated time points.
2.5. Cell Viability Assay (Edu Kit)
Cells were seeded into two 96-well plates at a density of approximately 1 × 104 cells per well and cultivated to logarithmic phase. Then, the cell viability assay was performed with Edu kit (RiboBio, China) as the manufacturers' instruction. Briefly, 50 μM 5-ethynyl-2′-deoxyuridine (Edu) was added into the culture medium and cells were cultured for 2 hours. After 2-time PBS washing and paraformaldehyde fixation, the cell nucleus were stained with Apollo 567 and Hoechst 33342. Then, the cells were imaged with a laser scanning confocal microscope (Zeiss, Germany) at 550 nm and 350 nm in the same vision. Then, the images of the same vision were overlapped in one image. Cell counting was done by Image J.
2.6. Alkaline Phosphatase (ALP) Activity Quantification
Cells were seeded into 96-well plates at approximately 1 × 105 cells per well. 24 h after seeding, the medium was changed into standard osteogenic differentiation induction medium (50 mg/mL of ascorbic acid (Sigma, USA)), 10 mmol/L of beta-glycerophosphate (BGP, Sigma, USA) and 10 ng/mL of dexamethasone diluted in 10% FBS α-MEM. ALP quantification assay was performed at 3, 6, 9, 12, 15, 18, and 21 days using an alkaline phosphatase assay quantification kit (JianCheng, China). The results were measured spectrophotometrically at a wavelength of 520 nm, based upon the manufacturer's instructions.
2.7. Alizarin Red Staining and Quantification
Cells were seeded into 24-well plates at a density of approximately 1 × 105 cells per well separately. After cells reached 80% confluence, the medium was changed into standard osteogenic differentiation induction medium and then cultured for another 3 weeks. The induction medium was changed every 3 days. Finally, cells were stained with Alizarin red (pH = 4.1) staining solution and were quantified according to previously described methods [10].
2.8. Oil Red O Staining
Cells were seeded into 24-well plates at a density of approximately 1 × 105 cells per well. After the cells reached 80% confluence, the medium was changed into standard adipogenic differentiation induction medium (0.5 mmol/L of methylisobutylxanthine (Sigma, USA), 1 mmol/L of dexamethasone, 10 μg/mL of insulin (Sigma, USA), and 200 μmol/L of indomethacin (Sigma, USA)) and then cultured for another 3 weeks. The induction medium was changed every 3 days. Finally, cells were stained with Oil red O staining solution and microscopically observed under phase contrast (Zeiss, Germany).
2.9. Immunofluorescence
Cells were seeded into confocal dishes at approximately 1 × 105 cells per well. After adherence, the culturing medium was changed into neurogenic differentiation induction medium (Gibco, USA) containing 20 ng/mL of EGF (Calbiochem, USA) and 20 ng/mL of bFGF (Calbiochem, USA) and then cultured for another 2 weeks. The induction medium was changed every 3 days. Finally, cells were stained with mouse anti-human βIII-TUBULIN primary antibody (Cell Signaling Technology, USA) at a concentration of 1 : 100. After overnight incubation, the cells were stained with DyLight 405 (ThermoFisher, USA) at a concentration of 1 : 50 and Hoechst dye (RiboBio, China) at a concentration of 1 : 100. Then, the cells were imaged with a laser scanning confocal microscope (Zeiss, Germany).
2.10. Quantitative Real-Time Polymerase Chain Reaction (Q-PCR)
Cells were seeded into 60-mm dishes. At 80% confluence, cells were induced by osteogenic/adipogenic/neurogenic differentiation medium for 2 weeks. Then, the total RNA was extracted from cells using Trizol reagent (TAKARA, Japan), and a reverse-transcription reaction was performed using a TAKARA reverse transcriptase kit (TAKARA, Japan). Real-time PCR was performed using a standard SYBR Green PCR kit (TAKARA, Japan) protocol on an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, USA), according to the manufacturer's instructions. β-ACTIN was used as an internal reference. The mRNA expression of osteogenic, adipogenic, and neurogenic marker genes was analyzed. Each sample was analyzed in triplicate. The 2−ΔΔCt value was used to determine the relative expression levels. The results were expressed as log10(2−ΔΔCt). The gene names and primer sequences are shown in Table 2.
Table 2.
Primer sequences.
| Primer | Sequence (5′-3′) | |
|---|---|---|
| β-ACTIN-forward | TGGCACCCAGCACAATGAA | |
| β-ACTIN-reverse | CTAAGTCATAGTCCGCCTAGAAGCA | |
| ANCR-forward | GCCACTATGTAGCGGGTTTC | |
| ANCR-reverse | ACCTGCGCTAAGAACTGAGG | |
|
| ||
| Osteogenic differentiation marker genes | ALP-forward | CCACGTCTTCACATTTGGTG |
| ALP-reverse | AGACTGCGCCTGGTAGTTGT | |
| OCN-forward | GGCAGCGAGGTAGTGAAGAG | |
| OCN-reverse | CTGGAGAGGAGCAGAACTGG | |
| BSP-forward | AAAGTGAGAACGGGGAACCT | |
| BSP-reverse | GATGCAAAGCCAGAATGGAT | |
|
| ||
| Adipogenic differentiation marker genes | PPAR-γ2-forward | CATTCTGGCCCACCAACTT |
| PPAR-γ2-reverse | CCTTGCATCCTTCACAAGCA | |
| C/EBPα-forward | TGGACAAGAACAGCAACGAG | |
| C/EBPα-reverse | TTGTCACTGGTCAGCTCCAG | |
|
| ||
| Neurogenic differentiation marker genes | βIII-TUBULIN-forward | GGCCTCTTCTCACAAGTACG |
| βIII-TUBULIN-reverse | CCACTCTGACCAAAGATGAAA | |
| GAP43-forward | CCTGTGGGAGTCCACTTTCC | |
| GAP43-reverse | CGATATTTTGGACTCCTCAGATGA | |
| NEFL-forward | TGATGGTAATGGATTGGAACTATGA | |
| NEFL-reverse | TCAACCCAGGTCTAGTAAGCAGAA | |
| NESTIN-forward | GTAGCTCCCAGAGAGGGGAA | |
| NESTIN-reverse | CTCTAGAGGGCCAGGGACTT | |
2.11. Statistical Analysis
Each experiment was performed at least three times, unless otherwise indicated. Data are reported as the mean ± SD (Standard Deviation). The significance of the differences between the experimental and the control groups was determined by using post hoc analysis for ANOVA. A value of P < 0.05 was considered to be statistically significant.
3. Results
3.1. The Characterization and ANCR Interference of DTSCs
Immunophenotype analysis showed that the separated DPSCs (Figure 1(a)), PDLSCs (Figure 1(b)), and SCAP (Figure 1(c)) were CD29, CD90, CD146, and STRO-1 positive and CD34 and CD45 negative. Q-PCR analysis was performed 3 days after infection. The results showed that ANCR was significantly knocked down by the ANCR-specific lentivirus-delivered shRNA in the ANCR-RNAi groups compared with control groups (wt and vector) in DPSCs (Figure 1(d)), PDLSCs (Figure 1(e)), and SCAP (Figure 1(f)).
Figure 1.
The characterization and ANCR interference of DTSCs. The DPSCs (a), PDLSCs (b), and SCAP (c) that we isolated expressed mesenchymal stem cell markers, including CD29, CD90, CD146, and STRO-1, but were negative for the hematopoietic cell marker CD34 and leukocyte marker CD45. Three days after lentivirus infection, we evaluated the effects of ANCR-RNAi by Q-PCR. The results showed that ANCR expression in DPSC/ANCR-RNAi (d), PDLSC/ANCR-RNAi (e), and SCAP/ANCR-RNAi (f) cells was remarkably downregulated when compared with control cells. ∗ P < 0.05 when compared with the control group.
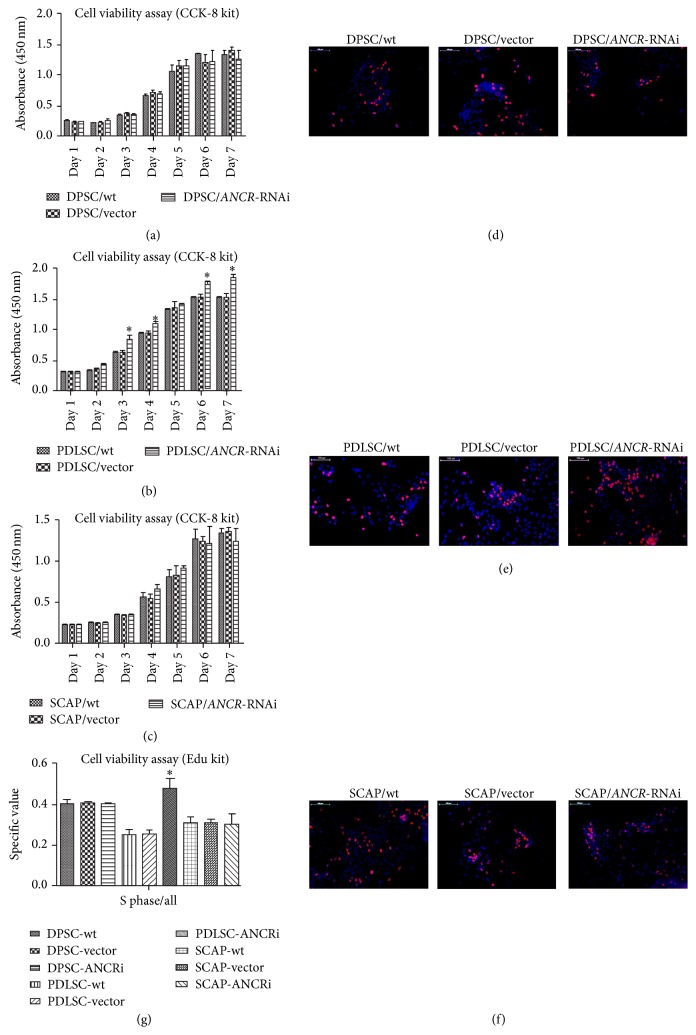
3.2. The Effect of ANCR-RNAi on DTSC Proliferation
A cell viability assay using CCK8 kit was performed to quantify the proliferation of DTSCs. The results showed that the growth rate of cells from PDLSC/ANCR-RNAi group was increased compared with the control groups (Figure 2(b)). However, downregulating ANCR had little impact on DPSCs (Figure 2(a)) and SCAP (Figure 2(c)) proliferation. To confirm this result, we used Edu kit to analyze the cell viability. The results showed that there were more PDLSC/ANCR-RNAi cells (Figure 2(e)) in S phase (purple) than control cells. However, there was little difference between ANCR-RNAi cells and control cells in DPSCs (Figure 2(d)) and SACP (Figure 2(f)). Cell counting by Image J confirmed this result (Figure 2(g)). In summary, downregulating ANCR promoted the cell viability of PDLSCs but had little effect on the cell viability of DPSCs and SCAP.
Figure 2.
The effects of ANCR-RNAi on DTSCs proliferation. From day 1 to day 7, the cell viability of PDLSC/ANCR-RNAi cells (b) was greater than that of the control cells; however, there were no statistically significant differences in DPSCs (a) and SCAP (c). Edu assay showed that there were more PDLSC/ANCR-RNAi cells (e) in S phase (purple) than control cells. However, there was little difference between ANCR-RNAi cells and control cells in DPSCs (d) and SACP (f). The cell counting result confirmed this result (g). The specific value of cells in S phase was higher in PDLSC/ANCR-RNAi cells compared to the control group; however, there were no statistically significant differences in DPSCs and SCAP. ∗ P < 0.05 when compared with the control group. Bars 100 μm.
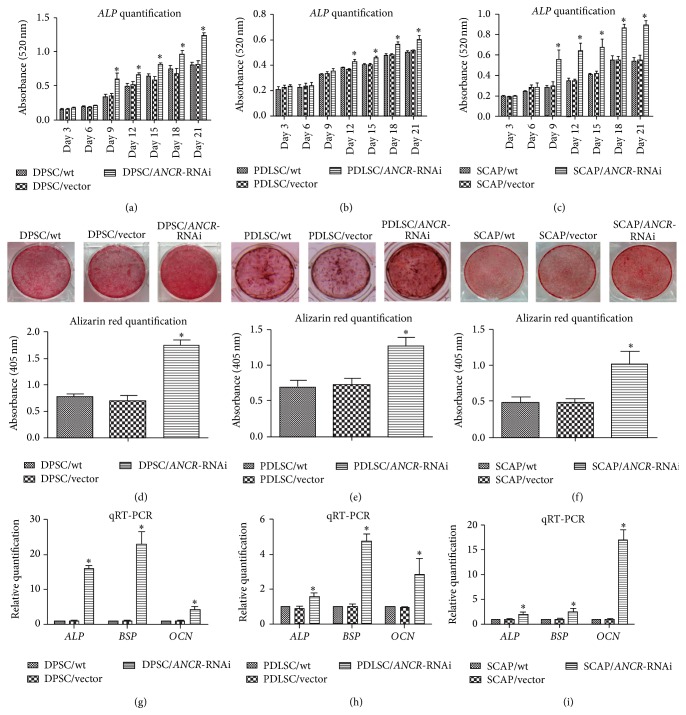
3.3. Downregulation of ANCR Promoted the Osteogenic Differentiation of DTSCs
ALP activity quantification showed that downregulating ANCR increased ALP secretion in DTSCs. A significant difference appeared at day 9 in DPSCs (Figure 3(a)) and SCAP (Figure 3(c)) but not until day 12 in PDLSCs (Figure 3(b)). The Alizarin red staining showed that ANCR-RNAi cells secreted more mineralizing matrix than the control groups. Quantification showed that the absorbance of DPSC/ANCR-RNAi cells (Figure 3(d)) was twofold higher than the control groups and the absorbance of PDLSC/ANCR-RNAi (Figure 3(e)) and SCAP/ANCR-RNAi (Figure 3(f)) cells was onefold higher than the control groups. Q-PCR showed that all of the osteogenic marker genes (ALP, BSP, and OCN) were upregulated in ANCR-RNAi cells (Figures 3(g)–3(I)).
Figure 3.
The effect of ANCR-RNAi on osteogenic differentiation in DTSCs. ALP activity was significantly increased in DPSC/ANCR-RNAi (a), PDLSC/ANCR-RNAi (b), and SCAP/ANCR-RNAi (c) cells when compared with control cells. Three weeks after induction, the cells were stained with Alizarin red (pH = 4.1), and DPSC/ANCR-RNAi (d), PDLSC/ANCR-RNAi (e), and SCAP/ANCR-RNAi (f) cells formed more mineralized nodules than the control groups. The Alizarin red quantification confirmed this result. The mRNA expression of ALP, BSP, and OCN was upregulated in DPSC/ANCR-RNAi (g), PDLSC/ANCR-RNAi (h), and SCAP/ANCR-RNAi (i) cells compared with control cells after two weeks of induction. ∗ P < 0.05 when compared with the control group.
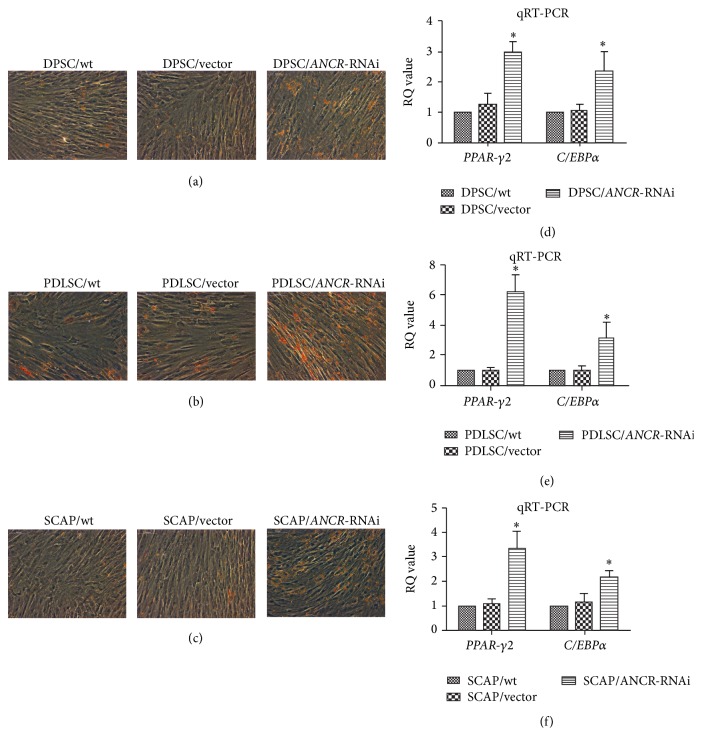
3.4. Downregulation of ANCR Promoted the Adipogenic Differentiation of DTSCs
Oil red O staining showed that downregulating ANCR promoted adipogenic differentiation of DPSCs (Figure 4(a)), PDLSCs (Figure 4(b)), and SCAP (Figure 4(c)). Q-PCR showed that all of the adipogenic marker genes (PPARγ-2 and C/EBPα) were upregulated in DPSCs (Figure 4(d)), PDLSCs (Figure 4(e)), and SCAP (Figure 4(f)).
Figure 4.
The effect of ANCR-RNAi on adipogenic differentiation in DTSCs. Oil red O staining showed that DPSC/ANCR-RNAi (a), PDLSC/ANCR-RNAi (b), and SCAP/ANCR-RNAi (c) cells formed more lipid droplets than the control groups. The mRNA expression of PPAR-γ2 and C/EBPα was upregulated in DPSC/ANCR-RNAi (d), PDLSC/ANCR-RNAi (e), and SCAP/ANCR-RNAi (f) cells compared with control cells after 2 weeks of induction. ∗ P < 0.05 when compared with the control group.
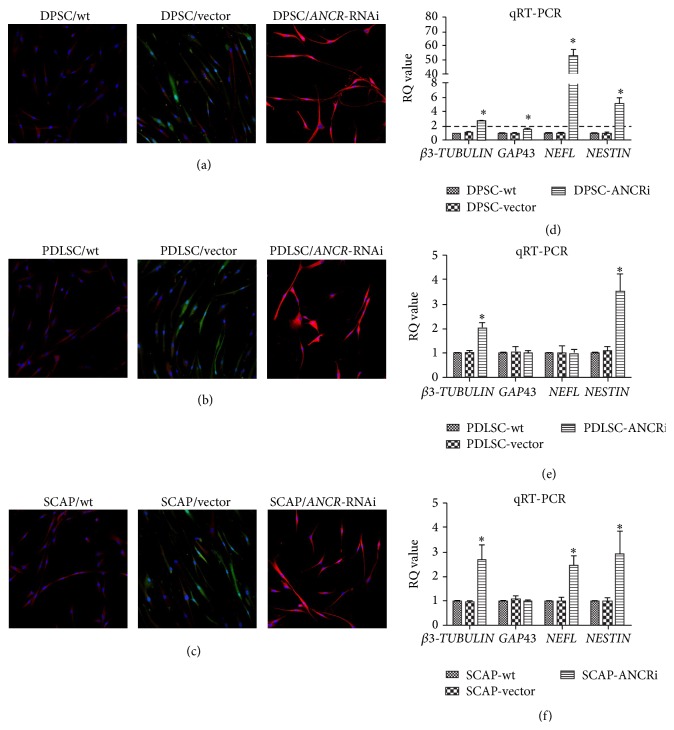
3.5. Downregulation of ANCR Promoted the Neurogenic Differentiation of DTSCs
Immunofluorescence showed stronger βIII-TUBULIN expression in DPSC/ANCR-RNAi (Figure 5(a)), PDLSC/ANCR-RNAi (Figure 5(b)), and SCAP/ANCR-RNAi (Figure 5(c)) cells than in the control cells. After two weeks of induction, the cellular morphology of DPSC/ANCR-RNAi and PDLSC/ANCR-RNAi cells was markedly changed compared with control groups. DPSC/ANCR-RNAi cells were connected by an axon/dendron-like structure that extended from the cell body. The cellular morphology of the PDLSC/ANCR-RNAi cells became irregular triangles or oval. However, there was little change in the cellular morphology of SCAP/ANCR-RNAi cells. Q-PCR showed that βIII-TUBULIN, GAP43, NEFL, and NESTIN were upregulated in DPSC/ANCR-RNAi cells (Figure 5(d)) compared with the control groups. In PDLSC/ANCR-RNAi cells (Figure 5(e)), βIII-TUBULIN and NESTIN were upregulated, while GAP43 and NEFL had little change compared with the control groups. In SCAP/ANCR-RNAi cells (Figure 5(f)), βIII-TUBULIN, NEFL, and NESTIN were upregulated while GAP43 had little change compared with the control groups.
Figure 5.
The effect of ANCR-RNAi on neurogenic differentiation in DTSCs. Immunofluorescence results showed that the expression of βIII-TUBULIN on DPSC/ANCR-RNAi (a), PDLSC/ANCR-RNAi (b), and SCAP/ANCR-RNAi (c) cells was stronger than control groups. The green is the color of GFP (Green Fluorescent Protein) transfected by the vector. The red is the color of βIII-TUBULIN stained by DyLight 405. The morphology of DPSC/ANCR-RNAi and PDLSC/ANCR-RNAi cells was remarkably changed. The mRNA expressions of βIII-TUBULIN, GAP43, NEFL, and NESTIN were upregulated in DPSC/ANCR-RNAi cells (d) after 2 weeks of induction. In PDLSC/ANCR-RNAi cells (e), βIII-TUBULIN and NESTIN were upregulated, while GAP43 and NEFL had little change compared with control groups. In SCAP/ANCR-RNAi cells (f), βIII-TUBULIN, NEFL, and NESTIN were upregulated while GAP43 had little change compared with control groups. ∗ P < 0.05 when compared with the control group.
4. Discussion
Compared with other mesenchymal stem cells (MSCs) derived from bone marrow, adipose tissue, peripheral blood, and umbilical cord blood, MSCs derived from dental tissues have marked advantages of easy access with least invasive procedures without any ethical issues [36]. The ability to undergo self-renewal and multidifferentiation is the most remarkable characteristic of DTSCs, which make them particularly suited for tissue engineering and gene therapy application. Before using DTSCs for clinical therapy, the ability of in vitro expansion and differentiation should be taken into account. A range of factors participate in the regulation of DTSCs differentiation and proliferation [37]. We herein used a laterally comparative analysis to investigate the role of long noncoding RNA-ANCR in regulating the ex vivo expansion and differentiation of three types of DTSCs, including DPSCs, PDLSCs, and SCAP.
Under osteogenic conditions, the ANCR-RNAi cells highly mineralized compared with the control groups demonstrated by the quantification of the Alizarin red staining, as well as the ALP activity. These results indicate that downregulation of ANCR may promote osteogenic differentiation of DTSCs. Thus to further confirm osteogenic differentiation thoroughly, we chose to evaluate the expression of several osteogenic related genes (including OCN, BSP, and ALP). OCN is secreted solely by osteoblasts and often used as a marker for the bone formation process [38]. Its gene expression is recognized to increase at a late stage of osteoblastic differentiation [39]. BSP, a component of mineralized tissues such as bone, dentin, and calcified cartilage, is considered as a specific marker for osteoblasts [40]. The gene expression of OCN, BSP, and ALP was upregulated in the ANCR-RNAi cells compared with control groups. Taken together, we speculate that the osteogenic differentiation of DTSCs that is promoted by downregulated ANCR is implemented by upregulating OCN, BSP, and ALP.
Downregulation of ANCR promoted the adipogenic differentiation of DTSCs demonstrated by Oil red O staining and confirmed by PPAR-γ2 and C/EBPα expression. More lipid droplets were form in DTSCs/ANCR-RNAi cells compared with the control groups. And the gene expression of PPAR-γ2 and C/EBPα was upregulated after the downregulation of ANCR. PPARγ-2 is known as a transcription factor in the directed differentiation of preadipocytes that is specifically expressed at the early stage of adipocyte differentiation. It is a key factor in adipogenic differentiation. C/EBPα works with PPARγ-2 to promote adipogenic differentiation [41]. We speculate that promotion of adipogenic differentiation caused by downregulating ANCR is implemented by upregulating PPARγ-2 and C/EBPα.
βIII-TUBULIN is often identified as the structural protein of neuronal cells. It plays a vital role in the process of neurogenic differentiation [42]. After two weeks of neurogenic differentiation induction, we examined the immunofluorescence staining of βIII-TUBULIN. Compared with the control groups, the morphology of DPSC/ANCR-RNAi cells and PDLSC/ANCR-RNAi cells was obviously changed. The morphology of DPSC/ANCR-RNAi cells was similar to neuronal-like cells. There were several axon-like structures extending from the cell body that connected with adjacent cells. The morphology of the PDLSC/ANCR-RNAi cells was changed from long fusiform cells into irregular triangles or ovals, with no axon-like structures, while the morphology of the control cells became long and thin. This same long, thin morphology was observed in SCAP/ANCR-RNAi cells and the control groups. There was little difference between the SCAP/ANCR-RNAi cells and control groups. The red fluorescence of βIII-TUBULIN became more obvious after ANCR downregulation in DPSCs, PDLSCs, and SCAP.
We used Q-PCR to evaluate the gene expression of βIII-TUBULIN, GAP43, neurofilament-light polypeptide (NEFL), and NESTIN. The βIII-TUBULIN expression was upregulated after downregulation of ANCR in DTSCs. These results could explain the increased red fluorescence of βIII-TUBULIN in DTSC/ANCR-RNAi cells. NESTIN, an intermediate filament protein, is an important surface marker on neural progenitor cells [43]. Q-PCR showed that NESTIN expression was upregulated after downregulating ANCR in DTSCs. GAP43 or neuromodulin is a neural specific axon membrane protein, which plays a vital role in axonogenesis [44]. The expression of GAP43 was upregulated in DPSCs but was not significantly different in PDLSCs and SCAP. NEFL is a key protein in axon intermediate filament formation that also plays an important role in axonogenesis [21]. After ANCR downregulation in DPSCs, the expression of NEFL was increased more than 50-fold compared with the control groups, indicating that DPSC/ANCR-RNAi cells have very strong axonogenesis. However, the expression of NEFL was approximately twofold higher in SCAP/ANCR-RNAi but largely unchanged in PDLSCs compared with the control groups. We found that the changes in the expression of GAP43 and NEFL were in accordance with the morphology changes of DTSCs/ANCR-RNAi cells. From these data, we concluded that downregulating ANCR promoted neurogenic differentiation of DTSCs and that the neurogenic differentiation potential of DPSCs was more obviously enhanced by downregulated ANCR compared with PDLSCs and SCAP.
As for the proliferation aspect, the regulation effect of ANCR on three types of DTSCs was different. Downregulation of ANCR promoted the proliferation of PDLSCs but had little effect on DPSCs and SCAP proliferation. We used cck-8 and Edu assay to confirm this results. Previous studies demonstrate that stem cells derived from dental tissues show a difference in terms of cell proliferation [3, 45]. Although most of stem cells from dental tissues exhibit the similar fibroblast-like morphology and clonogenic abilities after in vitro expansion, they orient from different niches of postnatal stem cells. The stem cells niches refer to an in vivo or in vitro stem cells microenvironment which maintain stem cells in a quiescent state [46, 47]. However, following tissue injury, the surrounding microenvironment interacts with stem cells to regulate cell fate. Stem cells behaviours (self-renewal or differentiation for tissue regeneration) are mainly governed by these distinct microenvironment which includes growth factors, cytokines, and extracellular matrix components, as well as cell signaling pathways [48]. A recent study in rats demonstrates that Notch signaling pathway plays a vital role in controlling DTSCs fate during the tooth development [49]. Additionally, our previous research found that ANCR promotes the proliferation and osteogenic differentiation of PDLSCs by activating the canonical Wnt signaling pathway [34]. The canonical Wnt signaling pathway which is an ancient and evolutionarily conserved pathway extensively participates in osteogenic, adipogenic, and neurogenic differentiation of stem cells [50]. It has been demonstrated that downregulating ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression [33]. EZH2 is known to be a key cellular homeostasis and differentiation regulator which interacts with canonical Wnt signaling pathway in stem cells [51]. Based on these researches, we speculate that ANCR may interact with EZH2 via canonical Wnt signaling pathway, thus regulating the DTSCs differentiation. Belonging to lncRNA, ANCR acts as the upstream of the regulatory network. It is a novel factor in regulating stem cells behaviours. Therefore, further in-depth investigation shall be performed to fully evaluate the control mechanism of lncRNA-ANCR during the proliferation and differentiation of DTSCs, as well as crosstalk between multiple pathways regulated by ANCR.
5. Conclusion
This study demonstrates that lncRNA-ANCR acts as a key regulator in terms of DTSCs proliferation and multiple-potential of differentiation. Downregulation of ANCR promoted the proliferation of PDLSCs, as well as osteogenic, adipogenic, and neurogenic differentiation of DTSCs (DPSCs, PDLSCs, and SCAP). More work is required to investigate the related genes and signaling pathways via regulation of ANCR controlling the DTSCs behaviours. This study represents a step forward in providing a novel regulator factor for further research in better understanding the behaviours of stem cells from dental tissues.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (no. 81371139 and no. 81470754) and the National Key Technologies R&D Program of the Twelve-Five Year Plan, the Ministry of Science and Technology of China (2012BAI07B03).
Competing Interests
The authors deny any competing interests.
Authors' Contributions
Qian Jia, Xiaolin Chen, and Wenkai Jiang contributed equally to this work.
References
- 1.Gronthos S., Mankani M., Brahim J., Robey P. G., Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(25):13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seo B.-M., Miura M., Gronthos S., et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. The Lancet. 2004;364(9429):149–155. doi: 10.1016/s0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 3.Sonoyama W., Liu Y., Yamaza T., et al. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. Journal of Endodontics. 2008;34(2):166–171. doi: 10.1016/j.joen.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song B., Jiang W., Alraies A., et al. Bladder smooth muscle cells differentiation from dental pulp stem cells: future potential for bladder tissue engineering. Stem Cells International. 2016;2016:11. doi: 10.1155/2016/6979368.6979368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilkens P., Gervois P., Fanton Y., et al. Effect of isolation methodology on stem cell properties and multilineage differentiation potential of human dental pulp stem cells. Cell and Tissue Research. 2013;353(1):65–78. doi: 10.1007/s00441-013-1630-x. [DOI] [PubMed] [Google Scholar]
- 6.Kim B.-C., Bae H., Kwon I.-K., et al. Osteoblastic/cementoblastic and neural differentiation of dental stem cells and their applications to tissue engineering and regenerative medicine. Tissue Engineering Part B: Reviews. 2012;18(3):235–244. doi: 10.1089/ten.teb.2011.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dissanayaka W. L., Zhan X., Zhang C., Hargreaves K. M., Jin L., Tong E. H. Y. Coculture of dental pulp stem cells with endothelial cells enhances osteo-/odontogenic and angiogenic potential in vitro . Journal of Endodontics. 2012;38(4):454–463. doi: 10.1016/j.joen.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 8.Janebodin K., Zeng Y., Buranaphatthana W., Ieronimakis N., Reyes M. VEGFR2-dependent angiogenic capacity of pericyte-like dental pulp stem cells. Journal of Dental Research. 2013;92(6):524–531. doi: 10.1177/0022034513485599. [DOI] [PubMed] [Google Scholar]
- 9.Sonoyama W., Liu Y., Fang D., et al. Mesenchymal stem cell-mediated functional tooth regeneration in swine. PLoS ONE. 2006;1(1, article e79) doi: 10.1371/journal.pone.0000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osathanon T., Nowwarote N., Pavasant P. Basic fibroblast growth factor inhibits mineralization but induces neuronal differentiation by human dental pulp stem cells through a FGFR and PLCγ signaling pathway. Journal of Cellular Biochemistry. 2011;112(7):1807–1816. doi: 10.1002/jcb.23097. [DOI] [PubMed] [Google Scholar]
- 11.Feng X., Huang D., Lu X., et al. Insulin-like growth factor 1 can promote proliferation and osteogenic differentiation of human dental pulp stem cells via mTOR pathway. Development Growth and Differentiation. 2014;56(9):615–624. doi: 10.1111/dgd.12179. [DOI] [PubMed] [Google Scholar]
- 12.He W., Wang Z., Luo Z., et al. LPS promote the odontoblastic differentiation of human dental pulp stem cells via MAPK signaling pathway. Journal of Cellular Physiology. 2015;230(3):554–561. doi: 10.1002/jcp.24732. [DOI] [PubMed] [Google Scholar]
- 13.Yang X., Zhang S., Pang X., Fan M. Pro-inflammatory cytokines induce odontogenic differentiation of dental pulp-derived stem cells. Journal of Cellular Biochemistry. 2012;113(2):669–677. doi: 10.1002/jcb.23396. [DOI] [PubMed] [Google Scholar]
- 14.Gay I., Cavender A., Peto D., et al. Differentiation of human dental stem cells reveals a role for microRNA-218. Journal of Periodontal Research. 2014;49(1):110–120. doi: 10.1111/jre.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara E. S., Ono M., Eguchi T., et al. MiRNA-720 controls stem cell phenotype, proliferation and differentiation of human dental pulp cells. PLoS ONE. 2013;8(12, article e83545) doi: 10.1371/journal.pone.0083545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei F., Liu D., Feng C., et al. MicroRNA-21 mediates stretch-induced osteogenic differentiation in human periodontal ligament stem cells. Stem Cells and Development. 2015;24(3):312–319. doi: 10.1089/scd.2014.0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye G., Li C., Xiang X., et al. Bone morphogenetic protein-9 induces PDLSCs osteogenic differentiation through the ERK and p38 signal pathways. International Journal of Medical Sciences. 2014;11(10):1065–1072. doi: 10.7150/ijms.8473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osathanon T., Nowwarote N., Manokawinchoke J., Pavasant P. BFGF and JAGGED1 regulate alkaline phosphatase expression and mineralization in dental tissue-derived mesenchymal stem cells. Journal of Cellular Biochemistry. 2013;114(11):2551–2561. doi: 10.1002/jcb.24602. [DOI] [PubMed] [Google Scholar]
- 19.Chen X., Hu C., Wang G., et al. Nuclear factor-κB modulates osteogenesis of periodontal ligament stem cells through competition with β-catenin signaling in inflammatory microenvironments. Cell Death & Disease. 2013;4(2, article e510) doi: 10.1038/cddis.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song D.-S., Park J.-C., Jung I.-H., et al. Enhanced adipogenic differentiation and reduced collagen synthesis induced by human periodontal ligament stem cells might underlie the negative effect of recombinant human bone morphogenetic protein-2 on periodontal regeneration. Journal of Periodontal Research. 2011;46(2):193–203. doi: 10.1111/j.1600-0765.2010.01328.x. [DOI] [PubMed] [Google Scholar]
- 21.Osathanon T., Manokawinchoke J., Nowwarote N., Aguilar P., Palaga T., Pavasant P. Notch signaling is involved in neurogenic commitment of human periodontal ligament-derived mesenchymal stem cells. Stem Cells and Development. 2013;22(8):1220–1231. doi: 10.1089/scd.2012.0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J., Huang G. T.-J., He W., et al. Basic fibroblast growth factor enhances stemness of human stem cells from the apical papilla. Journal of Endodontics. 2012;38(5):614–622. doi: 10.1016/j.joen.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Liu B., Gu S., Liang J. Effects of Wnt/β-catenin signalling on proliferation and differentiation of apical papilla stem cells. Cell Proliferation. 2012;45(2):121–131. doi: 10.1111/j.1365-2184.2012.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W., Zhang X., Ling J., et al. Proliferation and odontogenic differentiation of BMP2 gene-transfected stem cells from human tooth apical papilla: An In Vitro Study. International Journal of Molecular Medicine. 2014;34(4):1004–1012. doi: 10.3892/ijmm.2014.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Yan M., Wang Z., et al. 17β-estradiol promotes the odonto/osteogenic differentiation of stem cells from apical papilla via mitogen-activated protein kinase pathway. Stem Cell Research & Therapy. 2014;5(6):p. 125. doi: 10.1186/scrt515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipovich L., Johnson R., Lin C.-Y. MacroRNA underdogs in a microRNA world: evolutionary, regulatory, and biomedical significance of mammalian long non-protein-coding RNA. Biochimica et Biophysica Acta (BBA)—Gene Regulatory Mechanisms. 2010;1799(9):597–615. doi: 10.1016/j.bbagrm.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Ponjavic J., Ponting C. P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Research. 2007;17(5):556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Bakel H., Nislow C., Blencowe B. J., Hughes T. R. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biology. 2010;8(5, article e1000371) doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dinger M. E., Gascoigne D. K., Mattick J. S. The evolution of RNAs with multiple functions. Biochimie. 2011;93(11):2013–2018. doi: 10.1016/j.biochi.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Ponting C. P., Oliver P. L., Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Guttman M., Amit I., Garber M., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kretz M., Webster D. E., Flockhart R. J., et al. Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes and Development. 2012;26(4):338–343. doi: 10.1101/gad.182121.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu L., Xu P.-C. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochemical and Biophysical Research Communications. 2013;432(4):612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 34.Jia Q., Jiang W., Ni L. Down-regulated non-coding RNA (lncRNA-ANCR) promotes osteogenic differentiation of periodontal ligament stem cells. Archives of Oral Biology. 2015;60(2):234–241. doi: 10.1016/j.archoralbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Jiang W., Lv H., Wang H., et al. Activation of the NLRP3/caspase-1 inflammasome in human dental pulp tissue and human dental pulp fibroblasts. Cell and Tissue Research. 2015;361(2):541–555. doi: 10.1007/s00441-015-2118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang W., Ni L., Sloan A., Song B. Tissue engineering and regenerative medicine, from and beyond the dentistry. Dentistry. 2015;5(6, article 306) doi: 10.4172/2161-1122.1000306. [DOI] [Google Scholar]
- 37.Potdar P. D., Jethmalani Y. D. Human dental pulp stem cells: applications in future regenerative medicine. World Journal of Stem Cells. 2015;7(5):839–851. doi: 10.4252/wjsc.v7.i5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee N. K., Sowa H., Hinoi E., et al. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130(3):456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizuno M., Kuboki Y. Osteoblast-related gene expression of bone marrow cells during the osteoblastic differentiation induced by type I collagen. Journal of Biochemistry. 2001;129(1):133–138. doi: 10.1093/oxfordjournals.jbchem.a002824. [DOI] [PubMed] [Google Scholar]
- 40.Ogata Y. Bone sialoprotein and its transcriptional regulatory mechanism. Journal of Periodontal Research. 2008;43(2):127–135. doi: 10.1111/j.1600-0765.2007.01014.x. [DOI] [PubMed] [Google Scholar]
- 41.Siersbæk R., Nielsen R., Mandrup S. PPARγ in adipocyte differentiation and metabolism—novel insights from genome-wide studies. FEBS Letters. 2010;584(15):3242–3249. doi: 10.1016/j.febslet.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 42.Mead B., Logan A., Berry M., Leadbeater W., Scheven B. A. Intravitreally transplanted dental pulp stem cells promote neuroprotection and axon regeneration of retinal ganglion cells after optic nerve injury. Investigative Ophthalmology and Visual Science. 2013;54(12):7544–7556. doi: 10.1167/iovs.13-13045. [DOI] [PubMed] [Google Scholar]
- 43.Ryu J.-S., Ko K., Lee J.-W., et al. Gangliosides are involved in neural differentiation of human dental pulp-derived stem cells. Biochemical and Biophysical Research Communications. 2009;387(2):266–271. doi: 10.1016/j.bbrc.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Bressan R. B., Melo F. R., Almeida P. A., et al. EGF-FGF2 stimulates the proliferation and improves the neuronal commitment of mouse epidermal neural crest stem cells (EPI-NCSCs) Experimental Cell Research. 2014;327(1):37–47. doi: 10.1016/j.yexcr.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 45.Eleuterio E., Trubiani O., Sulpizio M., et al. Proteome of human stem cells from periodontal ligament and dental pulp. PLoS ONE. 2013;8(8) doi: 10.1371/journal.pone.0071101.e71101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bianco P., Robey P. G. Stem cells in tissue engineering. Nature. 2001;414(6859):118–121. doi: 10.1038/35102181. [DOI] [PubMed] [Google Scholar]
- 47.Fuchs E., Segre J. A. Stem cells: a new lease on life. Cell. 2000;100(1):143–155. doi: 10.1016/s0092-8674(00)81691-8. [DOI] [PubMed] [Google Scholar]
- 48.Moore K. A., Lemischka I. R. Stem cells and their niches. Science. 2006;311(5769):1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 49.Løvschall H., Tummers M., Thesleff I., Füchtbauer E.-M., Poulsen K. Activation of the Notch signaling pathway in response to pulp capping of rat molars. European Journal of Oral Sciences. 2005;113(4):312–317. doi: 10.1111/j.1600-0722.2005.00221.x. [DOI] [PubMed] [Google Scholar]
- 50.Logan C. Y., Nusse R. The Wnt signaling pathway in development and disease. Annual Review of Cell and Developmental Biology. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y.-H., Hung M.-C., Li L.-Y. EZH2: a pivotal regulator in controlling cell differentiation. American Journal of Translational Research. 2012;4(4):364–375. [PMC free article] [PubMed] [Google Scholar]