Abstract
Motivation: Many drug combinations are routinely assessed to identify synergistic interactions in the attempt to develop novel treatment strategies. Appropriate software is required to analyze the results of these studies.
Results: We present Combenefit, new free software tool that enables the visualization, analysis and quantification of drug combination effects in terms of synergy and/or antagonism. Data from combinations assays can be processed using classical Synergy models (Loewe, Bliss, HSA), as single experiments or in batch for High Throughput Screens. This user-friendly tool provides laboratory scientists with an easy and systematic way to analyze their data. The companion package provides bioinformaticians with critical implementations of routines enabling the processing of combination data.
Availability and Implementation: Combenefit is provided as a Matlab package but also as standalone software for Windows (http://sourceforge.net/projects/combenefit/).
Contact: Giovanni.DiVeroli@cruk.cam.ac.uk
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The combination of drugs is an important strategy to treat various diseases. The goal is to increase efficacy and, through the avoidance of overlapping toxicity, improve the therapeutic index of treatment (Long et al., 2014). At the discovery stage, research often focuses on the identification of synergistic target effects using in vitro systems (Lehár et al., 2009; Roemer and Boone, 2013; Sun et al., 2013). Large scale combination screens testing multiple pairwise combinations of drugs across different concentration ranges and cell lines are being performed with increasing frequency (He et al., 2015; Held et al., 2013; Mathews Griner et al., 2014). Typically, the dose–response data generated during these experiments are then analyzed in terms of synergistic or antagonistic effects. Free software were developed in the past but these are limited in terms of data handling and available options, particularly given high throughput screens (HTS) requirements (http://www.combosyn.com/; Dressler et al., 1999; Prichard and Shipman, 1990). Two recent commercial software packages offer advanced features such as automated analyses, choice from a variety of models, quality curve-fitting, variety of graphical displays and metrics quantification (Chalice http://cwr.horizondiscovery.com; Genedata https://www.genedata.com). There is a lack of free, advanced, scalable, software for the analysis of drug combinations. As part of the growing effort in the search of effective combinations, we present here Combenefit (‘Combinations Benefit’), a new software tool that enables the visualization, analysis and quantification of drug combination effects, in terms of synergy and/or antagonism, for single combination experiments but also HTS as per commercial packages (Table 1).
Table 1.
Features of available software for drugs combinations analyses. L: Loewe, B: Bliss, H: HSA, DR: Dose–response curve, M: Matrix view, C: contours, S: surfaces, I: Isobolograms, SM: synergy mapped to dose–response surfaces, F: Fa related curves, see (Chou, 2010). Note: we have not included CombiTool (Dressler et al., 1999) because it does not appear to be available anymore
| Combenefit | ChaliceTM | Genendata Screener® | CompuSyn | MacSynergy™ II | |
|---|---|---|---|---|---|
| Available | Free | Commercial | Commercial | Free | Free |
| Source code | Yes | No | No | No | Spreadsheet |
| Replicates | Yes | Yes | Yes | No | Yes |
| Fitting (Hill model) | 3 params | 3 params | 3 params + baseline | 2 params (no max) | No |
| Synergy models | L,B,H (+planned) | L,B,H and 2 others | L,B,H | L based | B |
| Scores/metrics | Several | Several | Several | No | No |
| Graphics | DR,M,C,S,SM | DR,M,C,I | DR,M,C,I | DR,I,F | C,S |
| Data input | File | File | File | Manual | Manual |
| HTS suitable | Yes | Yes | Yes | No | No |
2 General overview
Combenefit implements a surface approach, where in vitro experimental data is compared to mathematical models of dose–responses for non-synergistic combinations (also termed additive or independent combinations; Greco and Bravo, 1995). The three classical models, namely the Loewe (Loewe and Muchnick, 1926; Loewe, 1953), the Bliss (Bliss, 1939; Webb, 1963) and the Highest Single Agent (HSA; Mathews Griner et al., 2014; Tan et al., 2012) models have been incorporated. These models have been used extensively in the literature, with different fields having their preference and sometimes alternative names for these same models (Greco and Bravo, 1995; Odds, 2003). It is important to use these models in a consistent way and to provide results in the context of the model used. Additionally, our own group is currently developing a new general model which, together with any other future feature, will be incorporated into Combenefit.
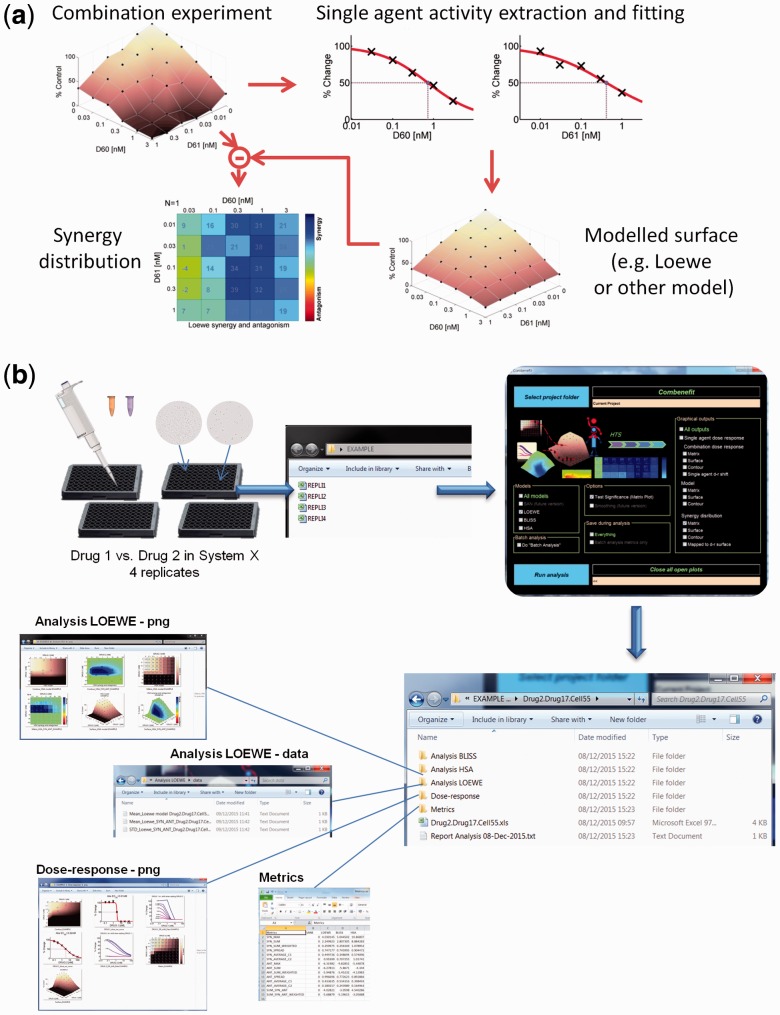
The approach implemented in Combenefit can be summarized as follows (Fig. 1a). The experimental dose–response surface that delineates combination effects in concentration space, is first read by the software as a matrix of % of the control value across concentrations. Single agent effects are extracted from this data and fitted with a dose response curve. Based on the two single agent dose response curves, a model-based combination dose–response surface is derived. This surface provides a ‘reference’ dose–response surface for a non-synergistic (additive/independent) combination, whose characteristics are determined by the selected model (Loewe, Bliss, etc.). The experimental combination dose response surface is then compared to the model-generated one, resulting in a synergy distribution in concentration space. This synergy distribution can be further summarized via various metrics as described below.
Fig. 1.
Analysis and visualization of drug combinations with Combenefit. (a) Illustration of analysis principle. (b) Illustration of automated processing (Color version of this figure is available at Bioinformatics online.)
3 Using Combenefit
The software is available as a standalone application for Windows or as a Matlab package. The standalone application can be installed automatically and is then directly accessed through its own icon. The Matlab package needs to be saved in a dedicated folder which is then accessed via Matlab by calling the Combenefit routine contained in the package. Laboratory scientists can quickly and easily analyze their data using either version. Bioinformaticians can use the code provided in the Matlab package to facilitate their own projects involving drugs combinations.
3.1 Typical analysis
A typical analysis is performed as follows. Upon completion of experiments, the results of a specific combination (including replicates if available) are saved in .xls files (the data is ordered with the first drug in columns and the second in rows; a template is provided). Once Combenefit is launched, the project folder containing all experimental .xls files is selected (Fig. 1b). Then, one or more reference models can be selected. Other options and graphical outputs can be chosen as per requirements/preferences. The graphical outputs consist of: (i) the single agent dose response data and its fitting, (ii) the combination dose response (Four different displays), (iii) the model-generated reference combination dose response, i.e. the prediction of effect if the drugs are not synergistic (Three different displays) and (iv) the resulting synergy distribution (Three different displays). Additionally, a fourth graphic mapping the synergy distribution onto the dose–response surface can also be displayed to facilitate interpretation. All figures, as well as corresponding data, fitting parameters and synergy metrics (see below) can be saved in the project folder. Figures are saved as high quality .png or .pdf files which can be directly used in publications. A user’s guide with step-by-step guidance for installing and running the software is available online.
3.2 Metrics and batch analysis
The analyses described above result in a synergy distribution for the combination being processed. To facilitate comparison across experiments, or for HTS applications, it is useful to summarize the resulting synergy and/or antagonism distribution using one or more metrics. Combenefit provides a set of metrics (or scores) which captures information about the synergy distribution. These include metrics such as the maximum synergy, the integrated and the weighted integrated synergy and concentration value at which synergy is most dense (for a full list of metrics and their formulation, please see the Supplementary Material).
During HTS, large volumes of combinations and cell lines are investigated using automated technology. Combenefit can be used to analyze the large amount of data derived with these screens. Using Combenefit, the user selects the folder containing all experiments (each in an individual sub-folder) and then selects the ‘Batch Analysis’ option. Upon running the analysis, Combenefit will batch process in a sequential way all the folders. The results of the analysis are summarized in a table containing all the experiments in rows and the metrics in columns. Typically, the highest hits are then visually inspected using Combenefit’s graphical outputs to improve understanding of the data and prioritize drug combinations. Combenefit has been recently used to generate the AstraZeneca dataset (∼11 500 combinations) provided for the current AstraZeneca-Sanger Drug Combination Prediction DREAM challenge (http://dreamchallenges.org/).
In summary, we have described the overall implementation and functioning of Combenefit, new software which offers advanced graphical capabilities and allows for model-based quantification of drug combinations in single and high-throughput settings. We hope that this new tool will continue to improve and will help many scientists in a variety of fields involving combination data analyses.
Supplementary Material
Acknowledgements
We acknowledge all support and feedback from Combenefit users.
Funding
This work has been supported by the Cancer Research UK grant C14303/A17197.
Conflict of Interest: none declared.
References
- Bliss C.I. (1939) The toxicity of poisons applied jointly. Ann. Appl. Biol., 26, 585–615 [Google Scholar]
- Chou T.C. (2010) Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res., 70, 440–446. [DOI] [PubMed] [Google Scholar]
- Dressler V. et al. (1999) CombiTool–a new computer program for analyzing combination experiments with biologically active agents. Comput. Biomed. Res., 32, 145–160. [DOI] [PubMed] [Google Scholar]
- Greco W.R., Bravo G.,P.J.C., (1995) The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev., 47, 331–385. [PubMed] [Google Scholar]
- He S. et al. (2015) Repurposing of the antihistamine chlorcyclizine and related compounds for treatment of hepatitis C virus infection. Sci. Transl. Med., 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M. a. et al. (2013) Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer Discov., 3, 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehár J. et al. (2009) Synergistic drug combinations tend to improve therapeutically relevant selectivity. Nat. Biotechnol., 27, 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe S. (1953) The problem of synergism and antagonism of combined drugs. ArzneimiettelForschung, 3, 286–290. [PubMed] [Google Scholar]
- Loewe S., Muchnick H. (1926) Effect of combinations: mathematical basis of problem. Arch. Exp. Pathol. Pharmacol., 114, 313–326. [Google Scholar]
- Long G.V. et al. (2014) Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun., 5, 5694. [DOI] [PubMed] [Google Scholar]
- Mathews Griner L.A. et al. (2014) High-throughput combinatorial screening identifies drugs that cooperate with ibrutinib to kill activated B-cell-like diffuse large B-cell lymphoma cells. Proc. Natl. Acad. Sci. USA, 111, 2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds F.C. (2003) Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother., 52, 1.. [DOI] [PubMed] [Google Scholar]
- Prichard M.N., Shipman C. (1990) A three-dimensional model to analyze drug-drug interactions. Antiviral Res., 14, 181–205. [DOI] [PubMed] [Google Scholar]
- Roemer T., Boone C. (2013) Systems-level antimicrobial drug and drug synergy discovery. Nat. Chem. Biol., 9, 222–231. [DOI] [PubMed] [Google Scholar]
- Sun X. et al. (2013) High-throughput methods for combinatorial drug discovery. Sci. Trans. Med., 5, 205rv1. [DOI] [PubMed] [Google Scholar]
- Tan X. et al. (2012) Systematic identification of synergistic drug pairs targeting HIV. Nat. Biotechnol., 30, 1125–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb J.L. (1963) Effect of more than one inhibitor. Enzyme Metab. Inhib, 1, 66–79. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.