Abstract
Summary: MIA detects and visualizes isotopic enrichment in gas chromatography electron ionization mass spectrometry (GC–EI-MS) datasets in a non-targeted manner. It provides an easy-to-use graphical user interface that allows for visual mass isotopomer distribution analysis across multiple datasets. MIA helps to reveal changes in metabolic fluxes, visualizes metabolic proximity of isotopically enriched compounds and shows the fate of the applied stable isotope labeled tracer.
Availability and Implementation: Linux and Windows binaries, documentation, and sample data are freely available for download at http://massisotopolomeanalyzer.lu. MIA is a stand-alone application implemented in C ++ and based on Qt5, NTFD and the MetaboliteDetector framework.
Contact: karsten.hiller@uni.lu
1 Introduction
Stable isotope assisted metabolomics is widely applied to analyze metabolism and to determine reaction mechanisms (Buescher et al., 2015). However, current approaches are usually highly targeted and only take into account a small set of metabolites. We recently demonstrated the potential of non-targeted stable isotope labeling in the analysis of hypoxic cancer cells to detect hypoxia-induced metabolic flux changes in a non-targeted manner (Weindl et al., 2016).
There have been tools available for the non-targeted and quantitative detection of isotopic enrichment in complex samples analyzed by GC-MS (Hiller et al., 2013) or LC-MS (Bueschl et al., 2012; Capellades et al., 2016; Chokkathukalam et al., 2013; Creek et al., 2012; Huang et al., 2014). These tools provide the isotopic enrichment of all detected compounds in the form of mass isotopomer distributions (MIDs), which represent the relative abundances of the different isotopic isomers (isotopologues) grouped by their masses. However, such metabolome-wide stable isotope labeling data has barely been analyzed to extract biological information in a truly non-targeted manner. One reason is, that there is still a lack of adequate software to facilitate the analysis of such datasets. Without proper data analysis tools, a global mass isotopolome analysis and its biological interpretation is tedious.
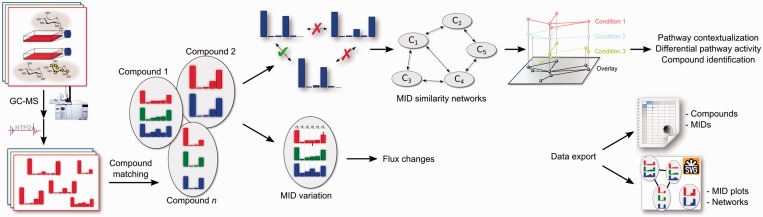
Until recently, there was neither free nor commercial software available that sufficiently supported the biological analysis of global stable isotope labeling datasets beyond the mere detection of isotopic enrichment. To this end, we developed MIA, an easy-to-use software tool to determine, visualize and analyze mass isotopomer distributions across multiple GC–EI-MS datasets in a non-targeted manner (Fig. 1).
Fig. 1.
MIA workflow. After performing a stable isotope labeling experiment, isotopic enrichment is detected and quantified in a non-targeted manner. Labeled compounds are matched across all datasets and MIDs are visualized. Data can be filtered and analyzed by changes in MIDs which indicate metabolic flux changes. MID-similarity can be visualized to reveal metabolic proximity of the isotopically enriched compounds to aid compound identification or to reveal their biosynthetic pathways. Finally, data can be exported either as spreadsheet or vector graphics for further use (Color version of this figure is available at Bioinformatics online.)
2 Features
MIA, the Mass Isotopolome Analyzer, provides the following features:
Non-targeted determination of mass isotopomer distributions
Matching detected compounds across different datasets
Visualization of MIDs of all mass-spectrometric fragments
Analysis of MID changes
Visualization of compound networks based on MID similarity
Compound identification using reference libraries
Export of graphics or spreadsheet data
Integration with MetaboliteDetector software for further data analysis (http://metabolitedetector.tu-bs.de/)
2.1 Data visualization
MIA visualizes the MIDs of the detected fragments as barplots. All detected isotopically enriched mass spectrometric fragments can be analyzed. If replicate measurements are provided, confidence intervals and quality measures of MID determination such as coefficient of determination are provided (Weindl et al., 2015b).
2.2 Mass isotopomer abundance variation analysis
MIDs from different datasets can be analyzed for changes in relative mass isotopomer abundance. Such variations are caused by, and are indicative of, altered metabolic fluxes. Therefore, metabolic flux changes are detected in a non-targeted manner (Weindl et al., 2015a, 2016).
2.3 MID similarity networks
Usually compounds closely connected within the metabolic network show very similar MIDs (Weindl et al., 2016). MIA exploits this fact for the analysis of stable isotope labeling data: Isotopically enriched compounds can be visualized in form of a network with connectivity based on their pairwise similarity. Due to this network visualization, closely related compounds can easily be detected. Knowledge on metabolic similarity can be valuable for compound identification, addressing a common bottleneck of non-targeted metabolomics.
3 Experimental requirements
MIA operates on low resolution GC–EI-MS data in the MetaboliteDetector or the commonly used netCDF format. MIDs are determined from the difference between mass spectra of an isotopically enriched compound and those of the non-enriched compound (Weindl et al., 2015b). Thus, two metabolite extracts need to be measured: One from a stable isotope labeling experiment and one from a label-free experiment.
4 Implementation
MIA is implemented in C ++ and Qt5. The MetaboliteDetector (Hiller et al., 2009) library is used for GC–EI-MS data handling, the NTFD library (Hiller et al., 2010, 2013) for non-targeted detection and quantification of stable isotope labeling and the GraphViz library for graph layouts (Gansner and North, 2000). Binaries are available for Linux and Windows operating systems.
5 Conclusion
Non-targeted stable isotope labeling analysis is a versatile tool for metabolic research. However, it has not yet reached its full potential due to the lack of appropriate data analysis software. MIA helps the user to quickly visualize and analyze this complex data and to gain new biological insights eventually. A more in-depth description of the implemented workflows and a case study applying the described workflows are available in Weindl et al. (2016).
Funding
This project was supported by the Fonds National de la Recherche (FNR) Luxembourg (ATTRACT A10/03, THActivity CORE).
Conflict of Interest: none declared.
References
- Buescher J.M. et al. (2015) A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol., 34, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueschl C. et al. (2012) MetExtract: a new software tool for the automated comprehensive extraction of metabolite-derived LC/MS signals in metabolomics research. Bioinformatics, 28, 736–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capellades J. et al. (2016) geoRge: A computational tool to detect the presence of stable isotope labeling in LC/MS-based untargeted metabolomics. Anal. Chem., 88, 621–628. [DOI] [PubMed] [Google Scholar]
- Chokkathukalam A. et al. (2013) mzMatch-ISO: an R tool for the annotation and relative quantification of isotope-labelled mass spectrometry data. Bioinformatics, 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D.J. et al. (2012) Stable isotope-assisted metabolomics for network-wide metabolic pathway elucidation. Anal. Chem., 84, 8442–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gansner E.R., North S.C. (2000) An open graph visualization system and its applications to software engineering. Softw. Pract. Exp., 30, 1203–1233. [Google Scholar]
- Hiller K. et al. (2009) MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal. Chem., 81, 3429–3439. [DOI] [PubMed] [Google Scholar]
- Hiller K. et al. (2010) Nontargeted elucidation of metabolic pathways using stable-isotope tracers and mass spectrometry. Anal. Chem., 82, 6621–6628. [DOI] [PubMed] [Google Scholar]
- Hiller K. et al. (2013) NTFD–a stand-alone application for the non-targeted detection of stable isotope-labeled compounds in GC/MS data. Bioinformatics, 29, 1226–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. et al. (2014) X13cms: global tracking of isotopic labels in untargeted metabolomics. Anal. Chem., 86, 1632–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindl D. et al. (2015a) Metabolome-wide analysis of stable isotope labeling — is it worth the effort? Front. Physiol., 6, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindl D. et al. (2015b). Non-targeted tracer fate detection In: Metallo C.M. (ed.), Metabolic Analysis Using Stable Isotopes, Volume 561 of Methods in Enzymology. Academic Press, Waltham, MA, pp. 277–302. [DOI] [PubMed] [Google Scholar]
- Weindl D. et al. (2016) Bridging the gap between non-targeted stable isotope labeling and metabolic flux analysis. Cancer Metab., 4, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]