Abstract
Motivation: The identification of microRNA (miRNA) target sites is fundamentally important for studying gene regulation. There are dozens of computational methods available for miRNA target site prediction. Despite their existence, we still cannot reliably identify miRNA target sites, partially due to our limited understanding of the characteristics of miRNA target sites. The recently published CLASH (crosslinking ligation and sequencing of hybrids) data provide an unprecedented opportunity to study the characteristics of miRNA target sites and improve miRNA target site prediction methods.
Results: Applying four different machine learning approaches to the CLASH data, we identified seven new features of miRNA target sites. Combining these new features with those commonly used by existing miRNA target prediction algorithms, we developed an approach called TarPmiR for miRNA target site prediction. Testing on two human and one mouse non-CLASH datasets, we showed that TarPmiR predicted more than 74.2% of true miRNA target sites in each dataset. Compared with three existing approaches, we demonstrated that TarPmiR is superior to these existing approaches in terms of better recall and better precision.
Availability and Implementation: The TarPmiR software is freely available at http://hulab.ucf.edu/research/projects/miRNA/TarPmiR/.
Contacts: haihu@cs.ucf.edu or xiaoman@mail.ucf.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The prediction of microRNA (miRNA) target sites is critical in understanding miRNA function and their involvement in various biological processes (Lewis et al., 2003). MiRNAs are short noncoding RNAs that bind and regulate their target mRNAs in a variety of biological processes, such as cell development, differentiation, proliferation and apoptosis pathways (Sassen et al., 2008; Schanen and Li, 2011). The binding of miRNAs to their target mRNAs degrades the target mRNAs and/or prevents the target mRNAs from being translated into proteins, and thus modulates gene expression at the post-transcriptional level (Axtell et al., 2011; Bartel, 2009; Muljo et al., 2010; Wang et al., 2011). By identifying miRNA target sites, the target mRNAs and the potential functional roles of miRNAs may thus be discovered.
Several features are commonly believed to be important for predicting miRNA target sites. Among them, seed match, the exact sequence matching between the positions 2–7 of an miRNA and a segment of 6 nucleotides (nt) long in target mRNAs, has been reported to be essential for miRNA–mRNA binding (Brennecke et al., 2005). Accessibility, which measures how likely a region in an mRNA sequence is ‘open’ or accessible for an miRNA to bind, is well known to be important for functional miRNA–mRNA binding (Kertesz et al., 2007). In addition, other features such as AU content (Grimson et al., 2007), folding energy (Enright et al., 2004; Grimson et al., 2007; Yousef et al., 2007) and conservation (Helwak et al., 2013) are also regarded as informative indicators of functional miRNA–mRNA bindings.
Dozens of tools for miRNA target site prediction have been developed in the past decade, based on different subsets of the aforementioned features (Peterson et al., 2014). For instance, miRanda (Enright et al., 2004) utilizes the features of seed match, conservation and free energy for target site prediction. TargetScan (Friedman et al., 2009; Grimson et al., 2007) uses seed match, pairing of mRNAs with 3′ of miRNAs, local AU content, etc., for target site identification. In addition to these traditional miRNA target site prediction tools, recently, several tools based on next-generation sequencing technologies have been developed (Chou et al., 2013; Vejnar and Zdobnov, 2012; Wang et al., 2014). For instance, miRTarCLIP (Chou et al., 2013) identifies miRNA target sites from the data generated by high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) experiments (Chi et al., 2009; Licatalosi et al., 2008) and photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) experiments (Hafner et al., 2010).
Despite the existence of dozens of computational methods, computational identification of miRNA target sites remains a challenging problem partially due to our limited understanding of the characteristics of miRNA target sites. For instance, although matching seed is not always sufficient for a functional miRNA–mRNA interaction (Brennecke et al., 2005; Didiano and Hobert, 2006), it has been thought to be necessary for most animal miRNA–mRNA binding. However, studies have shown non-canonical pairings that allow G:U wobbles and even mismatches can be functional (Brennecke et al., 2005; Didiano and Hobert, 2006). Recent crosslinking ligation and sequencing of hybrids (CLASH) experiments (Helwak et al., 2013) have further shown that seed match, including canonical and non-canonical seed-matching, is not required for certain miRNA–mRNA interactions.
The CLASH experiments (Helwak et al., 2013) provide an unprecedented opportunity to advance our understanding of miRNA target sites and to develop better computational methods for miRNA target site prediction. Compared with other high-throughput experimental approaches such as HITS-CLIP (Chi et al., 2009; Licatalosi et al., 2008) and PAR-CLIP (Hafner et al., 2010) that identify miRNA target sequences only, CLASH experiments provide both miRNAs and their corresponding target sequences. With thousands of target sequences for dozens of miRNAs in one CLASH experiment, new features of miRNA target sites may be inferred and better computational methods for miRNA target site prediction may be developed.
In this study, we developed a new approach for miRNA target site prediction called Target Prediction for miRNAs (TarPmiR). TarPmiR applies a random-forest-based approach to integrate six conventional features and seven new features to predict miRNA target sites. These features were learned from the only CLASH dataset in mammal that is made publically available by Helwak et al. (2013). By cross-validation, we showed that TarPmiR had an average recall of 0.543 and an average precision of 0.181. Tested on three independent datasets, including two human PAR-CLIP datasets and one mouse HITS-CLIP dataset, we demonstrated that TarPmiR identified more than 74.2% of known miRNA target sites in each dataset. Compared with three existing approaches, we found that TarPmiR is superior to existing approaches, in terms of both higher recall and higher precision. The TarPmiR method is implemented in a python package, which is freely available at http://hulab.ucf.edu/research/projects/miRNA/TarPmiR/.
2 Materials and Methods
2.1 Training and testing data
We downloaded 18 514 miRNA target sites of 399 miRNAs from CLASH experiments (Helwak et al., 2013). These target sites were considered as positive target sites. We also generated 18 514 corresponding negative or ‘false’ target sites in a manner similar to a previous study (Li et al., 2014), with the following criteria: (i) A positive site and its corresponding negative site are on the same mRNA; (ii) The positive and its corresponding negative site has similar CG dinucleotide frequency; (iii) The positive and its corresponding negative site has similar number of the nucleotide G; (iv) A negative site does not overlap with any positive site; and (v) With multiple candidate negative sites in an mRNA, select the one with the lowest folding energy.
We performed cross-validation to determine which machine learning method to be used in TarPmiR and to assess the accuracy of TarPmiR. To determine which method to be used, we randomly chose 10 000 positive sites and 10 000 negative sites for training and the remaining positive and negative sites for testing. We repeated this process five times and selected the method with the F2 scores. To test TarPmiR, we used the same five training datasets. For a corresponding testing dataset, we input the mRNA sequences that contain the corresponding remaining 8514 positive sites and the remaining 8514 negative sites for testing. The final model used to predict miRNA target sites by TarPmiR in this study was trained using the first set of randomly chosen 10 000 positive sites and 10 000 negative sites.
We also collected two independent PAR-CLIP datasets from the human HEK293 cell line for testing. PAR-CLIP datasets were used because a large number of potential miRNA target regions called crosslink-centered regions (CCRs) could be obtained from PAR-CLIP. CCRs were considered as positive target sites. One PAR-CLIP dataset with 17 310 CCRs was from Hafner et al. (2010). Only 16 041 of these CCRs were able to be mapped to mRNAs and resulted in 10 023 target mRNAs. In this dataset, 60 miRNAs accounted for more than 90% of total miRNA reads and 120 miRNAs accounted for 99% of total miRNA reads. In other words, depending on the cutoff to define active miRNAs, there were mainly 60 or 120 miRNAs related to these 17 310 CCRs. The other PAR-CLIP dataset with 44 497 CCRs was obtained from Kishore et al. (2011). Only 43 251 of the 44 497 CCRs were able to be mapped to mRNAs and resulted in 17 794 target mRNAs. Same as the first PAR-CLIP dataset, depending on the cutoff to define active miRNAs, there were mainly 60 or 120 related miRNAs in this dataset.
To test TarPmiR on general datasets, we compared the TarPmiR predictions with the experimentally validated miRNA targets by general methods in TarBase 7.0 (Vlachos et al., 2014). There are 421 086 POSITIVE TarBase 7.0 miRNA–mRNA interactions in human. We chose the top 100 and 50 miRNAs, which had the largest number of interactions in TabBase 7.0, for further analyses. The rationale to choose top miRNAs was that we had more experimentally validated target mRNAs of these miRNAs and thus could assess the accuracy of TarPmiR and other tools better. The top 100 and 50 miRNAs in TarBase 7.0 accounted for 100 608 (23.9%) and 60 818 (14.4%) of human TarBase 7.0 interactions, respectively. There were 9869 and 9823 mRNAs associated with these 100 and 50 top miRNAs, respectively. We ran TarPmiR and other tools with the 100 or 50 miRNAs and the corresponding mRNAs they interacted as input to predict miRNA target sites.
In addition to the human datasets, we collected an independent HITS-CLIP dataset from the mouse cortex cell (Chi et al., 2009). This dataset provided an Argo–miRNA–mRNA ternary interaction map related to 20 miRNA families, 2953 mRNAs and 11 080 miRNA–mRNA interactions. We further downloaded the corresponding 119 miRNAs from the 20 miRNA families from miRBase (Griffiths-Jones et al., 2006).
2.2 Potential features considered
We considered the following 18 features of miRNA target sites in miRNA–mRNA duplexes: (i) folding energy; (ii) seed match; (iii) accessibility; (iv) AU content; (v) stem conservation; (vi) flanking conservation; (vii) difference between stem and flanking conservation; (viii) m/e motif; (ix) the total number of paired positions; (x) the length of the target mRNA region; (xi) the length of the largest consecutive pairs; (xii) the position of the largest consecutive pairs relative to the miRNA 5′; (xiii) the length of the largest consecutive pairs allowing 2 mismatches; (xiv) the position of the largest consecutive pairs allowing 2 mismatches; (xv) the number of paired positions at the miRNA 3′ end, where 3′ miRNA end was defined as the last 7 positions of the miRNA; (xvi) the total number of paired positions in the seed region and the miRNA 3′ end; (xvii) the difference between the number of paired positions in the seed region and that in the miRNA 3′ end and (xviii) exon preference (Ding et al., 2015). The first seven features had been used in existing tools (Peterson et al., 2014), we thus considered them as conventional features. Remaining features that had not been commonly used by miRNA-target prediction tools were defined as ‘new’ features.
The detailed definition of all 18 features and how to calculate their values are provided in the Supplementary File S1. We briefly explain the m/e motif feature here, as it is not as self-evident as others. The m/e motif describes how different positions in miRNAs match the corresponding positions in target sites. Here two positions match means that nucleotides at the two positions are complement to each other. For instance, nucleotides at positions in miRNA seed regions tend to match the nucleotides at the corresponding positions in target sites and nucleotides at positions in other miRNA regions tend to form mismatches or bulges with the corresponding positions in target sites. We thus have a sequential pattern composed of two letters ‘m’ and ‘e’ to describe preferred matching and non-matching positions, respectively. To calculate the m/e scores, for each position in miRNAs, we calculate a probability that this position matches the corresponding position in target sites by using all positive target sites in the training dataset. The m/e motif score of a potential target site is calculated as , where x is the length of the miRNA and x is smaller than 24.
2.3 Four computational methods for feature selection
Not all of the aforementioned 18 features are effective for target site prediction. To select important features, we applied the following four machine learning methods: step-wise logistic regression (Ralston and Wilf, 1960), least absolute shrinkage and selection operator (LASSO) (Tibshirani, 1996), randomized logistic regression (Meinshausen and Bühlmann, 2010) and random forests (Svetnik et al., 2003). The step-wise logistic regression repeatedly eliminates the least significant feature until all significant features are found, which is performed by using the GLM package in R (http://data.princeton.edu/R/glms.html). LASSO constructs a linear model and shrinks the coefficients of non-important features to zero. All features with non-zero regression coefficients are 'selected' as important features. We used the glmnet package (http://cran.r-project.org/web/packages/glmnet/index.html) in R for the LASSO analysis. The randomized logistic regression randomly chooses a portion of the training samples and performs the logistic regression to select significant features. It repeats this procedure many times and counts the number of times each feature is selected, which is regarded as the importance of the features. The randomized logistic regression was performed with the scikit-learn package (http://scikit-learn.org/stable/) in python. The random forests method grows many classification trees and assigns a new object to the class most trees vote for this object. We used the random forest model from sklearn package (http://scikit-learn.org/stable/) in python. Each of the four methods has been applied to select features in previous studies (Chen and Lin, 2006; Chou et al., 2001; Díaz-Uriarte and De Andres, 2006; Kim and Kim, 2004; Kokaly and Clark, 1999; Ma and Huang, 2008; Saeys et al., 2007; Yeo et al., 1995) and demonstrated good performance in feature selection. We claim a feature as an important feature if at least two of the four methods consider this feature important. By applying the four methods to the training data, we selected 13 important features (Section 3.1).
2.4 TarPmiR, a random-forest-based approach for miRNA target site prediction
With the 13 selected features, we developed a random-forest-based approach called TarPmiR for miRNA target site prediction. We chose the random forests method because we applied the above four approaches to the aforementioned training and testing datasets and found that random forests gave the best performance (Section 3.2).
TarPmiR predicts miRNA target sites in three steps with the input of a set of miRNAs and a set of mRNAs. First, TarPmiR generates candidate target sites based on seed match or minimal folding energy (Enright et al., 2004; Grimson et al., 2007; Yousef et al., 2007). For a given miRNA, TarPmiR scans an mRNA sequence with the seed region of the miRNA (positions 2–7) to find perfect seed-matching sites. These sites are defined as the first set of candidate target sites. In addition, TarPmiR applies RNA-duplex from the Vienna RNA package (Hofacker, 2003) to obtain the top target sites with the lowest folding energy. These energy-based sites are defined as the second set of candidate target sites. The combination of seed match and folding energy helps TarPmiR to pick up almost all true target sites from the beginning. Second, for each candidate target sites, TarPmiR calculates the values of the 13 selected features (Supplementary File S1). Finally, TarPmiR applies the trained random-forest based predictor to predict target sites. The output of the random-forest model is the predicted probability that a candidate target site is a true target site. We have compared nine probability cutoffs to define target sites using the F2 score, since we put more emphasis on the recall than the precision. The cutoffs 0.5 and 0.6 have almost the similar F2 scores, while the cutoff 0.5 has the largest recall (Supplementary File S2). Therefore, we used 0.5 for the following analyses. We provide a parameter –p in TarPmiR, users can choose other cutoffs based on their own needs.
2.5 Comparisons with other methods
We compared TarPmiR with the following methods: targetScan V2010 (Friedman et al., 2009; Grimson et al., 2007), targetScan V2015 (Agarwal et al., 2015), miRanda (Enright et al., 2004) and miRmap (Vejnar et al., 2013; Vejnar and Zdobnov, 2012). The targetScan and miRanda are two of the most widely used miRNA target prediction tools. We used the following commands to run them: targetScan V2010-perl targetscan.pl <miRNA.> <mRNA> <targetscan_out>, perl targetscan_60_context_scores.pl <miRNA> <mRNA> <targetscan_out> <targetscan_context_score_out>; targetSan V2015- perl targetscan_70.pl <miRNA> <mRNA> <targetscan_out>, targetscan_70_BL_bins.pl <Mrna> > <BL_bins_out>, targetscan_70_BL_PCT.pl <miRNA> <targetscan_out> <BL_bins_out> > <PCT_out>, perl targetscan_count_8mers.pl <mir> <Mrna_ORF> >ORF_out, perl targetscan_70_context_scores.pl <miRNA> <Mrna> <PCT_out> <tar.lengths.txt> <ORF_out> <context+ score>; perl and miranda <miRNA> <mRNA> -sc 120 –en 1. MiRmap is a recently developed tool, which takes high-throughput sequencing data as input to predict miRNA target sites. MiRmap provides a python library and users can write a script to output miRmap predictions with the functions in the library. We used similar parameters as in Vejnar and Zdobnov (2012) when running miRmap.
3 Results
3.1 All but one conventional features and seven new features were selected by different approaches
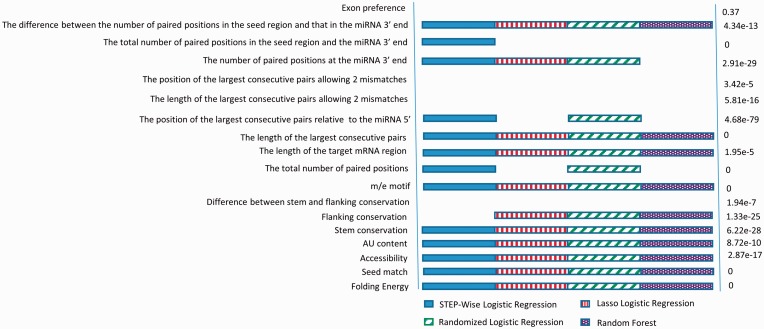
We applied four approaches to select important features from the 18 potential features. Each approach selected a similar but slightly different subset of features (Supplementary File S3). By defining features selected by at least two approaches as important features, we discovered 13 important features (Fig. 1) . They are: (i) folding energy; (ii) seed match; (iii) accessibility; (iv) AU content; (v) stem conservation; (vi) flanking conservation; (vii) m/e motif; (viii) the total number of paired positions; (ix) the length of the target mRNA region; (x) the length of the largest consecutive pairings; (xi) the position of the largest consecutive pairings relative to the 5′ end of miRNA; (xii) the number of paired positions at the miRNA 3′ end. Recall miRNA 3′ end meant the last 7 positions of a miRNA and (xiii) the difference between the number of paired positions in the seed region and that in the miRNA 3′ end.
Fig. 1.
Features selected by four different methods
An interesting observation from Figure 1 was the removal of one and only one conventional feature, the difference between stem and flanking conservation. This feature was used in previous studies (Helwak et al., 2013; Pollard et al., 2010). The removal of this feature may be explained by the fact that most positive target sites from CLASH experiments were from coding regions and there was not much difference in terms of conservation between the seed regions and the flanking regions of target sites in coding regions. Because true target sites were functional and conserved, two features related to the conservation in miRNA–mRNA stem regions and in flanking regions around the stems, respectively, were selected.
In addition to the six selected conventional features (folding energy, seed-matching, accessibility, AU content), four new features were selected by all four approaches (Fig. 1). These features were the m/e motif, the length of the target site, the length of the largest consecutive pairings and the difference between the number of paired positions in the seed region and that in the miRNA 3′ end. The inclusion of the m/e motif implied that there existed preferred matching positions shared by all miRNAs. The length of the target site was selected, showing the importance of the binding preference of miRNAs to mRNA regions with specific lengths. The length of the largest consecutive pairing positions mattered, which extended the concept of seed match, as seed match was just a simple case with a long consecutive pairing positions. The difference between the number of paired positions in the seed region and that in the miRNA 3′ end also suggested that the seed match may be unimportant, given a high-quality 3′ end region matching. This also supported the idea that a long consecutive matching region is critical for functional miRNA target sites.
We further investigated the importance of the 13 selected features by the rank-sum test (Mann and Whitney, 1947) (Fig. 1). In brief, for each selected feature, we calculated its value for all positive target sites and for their corresponding negative target sites. We then compared the two groups of numbers by the rank-sum test. The numbers on the right side of Figure 1 showed the P-values of the corresponding features. All 13 selected features had a significant difference between the positive target sites and negative sites (P-value < 1.95e−5). Some significant features based on the rank-sum test were not selected by the four machine learning methods, which may be due to the fact that the contribution from the combination of the selected 13 features can already replace that of these removed features. In fact, we calculated the correlation between every pair of the 18 features and found that the discarded significant features correlate well with certain important features (Supplementary File S3).
3.2 TarPmiR had a >55% recall and a >19.1% precision
With the 13 selected features, we developed the TarPmiR method to predict miRNA target sites in the entire regions of mRNAs. TarPmiR applied the random-forest-based approach for target site prediction. It applied the random forests approach instead of the other three approaches because when tested on five testing datasets, the random-forests-based approach always gave better recalls and precisions (Table 1).
Table 1.
Recall and precision of different methods on five testing datasets
| Lasso logistic |
Randomized logistic |
STEP-wise logistic |
Random forest |
TarPmiR |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Recall | Precision | Recall | Precision | Recall | Precision | Recall | Precision | Recall | Precision | |
| T1 | 0.8549 | 0.7765 | 0.8539 | 0.7785 | 0.8559 | 0.7795 | 0.8740 | 0.8283 | 0.5514 | 0.1905 |
| T2 | 0.8736 | 0.7713 | 0.8746 | 0.7730 | 0.8751 | 0.7736 | 0.8921 | 0.8296 | 0.5227 | 0.1626 |
| T3 | 0.8315 | 0.7626 | 0.8319 | 0.7898 | 0.8320 | 0.7904 | 0.8686 | 0.8253 | 0.5303 | 0.1661 |
| T4 | 0.836 | 0.7871 | 0.8411 | 0.7903 | 0.838 | 0.7894 | 0.8776 | 0.8266 | 0.5507 | 0.1902 |
| T5 | 0.8856 | 0.7639 | 0.8878 | 0.7662 | 0.8895 | 0.7649 | 0.8989 | 0.8173 | 0.5583 | 0.1909 |
To investigate the recall and precision of TarPmiR, we tested it on the five testing datasets described in Material and Methods . The precision and recall of TarPmiR in each set of test data were shown in Table 1. Since the TarPmiR predictors built on each of the five training datasets had similar precision and recall, we chose the first TarPmiR predictor in our developed tool and in the following analyses. TarPmiR had a 55.1% recall and a 19.1% precision, which were higher than the recall and precision of existing methods reviewed in Reczko et al. (2011). Note that TarPmiR had a much smaller precision and recall than the above four methods (columns 2–5 in Table 1), because it predicted target sites from the entire mRNA sequences instead of the 8514 sites that were not used for training.
3.3 TarPmiR predicted the majority of true target sites in independent datasets
To investigate whether TarPmiR was able to predict true target sites in non-CLASH datasets, we applied it to two PAR-CLIP datasets in the HEK293 cell (Material and Methods). There were 16 041 ‘true’ target sites in 10 023 mRNAs from the first dataset (dataset I). Moreover, the reads of the top 60 miRNAs and top 120 miRNAs accounted for more than 90% and 99%, respectively, of the total PAR-CLIP reads in this dataset. By inputting 60 miRNAs and 10 023 mRNAs, TarPmiR predicted 240 605 target sites, which included 74.2% of true target sites (Table 2). Similarly, by inputting 120 miRNAs and 10 023 mRNAs, TarPmiR predicted 481 135 target sites, which included 86.3% of true target sites (Table 2). The percentages of correctly predicted true target sites should be considered underestimated, as a portion of true target sites may not be target sites of the 60 or 120 miRNAs. By considering the 16 041 ‘true’ target sites as all target sites in these mRNAs, we found that TarPmiR had a >74% recall in this dataset (Table 2). For the second PAR-CLIP dataset (dataset II), there were 43 251 ‘true’ target sites in 17 794 mRNAs. Because the cell was the same as that in the first PAR-CLIP dataset, we assumed that mainly 60 or 120 miRNAs related to these target sites. Similarly, we found that TarPmiR was able to identify 79.3% and 89.8% of ‘true’ target sites, when inputting 60 miRNAs and 120 miRNAs, respectively, together with the 17 794 mRNAs (Table 2).
Table 2.
Comparison of four methods on independent datasets
| Dataset | # of miRNAs input | Performance measurement | TarPmiR | miRanda | TargetScan V2010 | miRmap | TargetScan V2015 |
|---|---|---|---|---|---|---|---|
| I | 60 | # of predictions | 240 605 | 246 311 | 219 304 | 504 447 | 215 885 |
| % of correct predictions | 11 904/16 041=74.2% | 7061/16 041=44.0% | 6248/16 041=39.0% | 7121/16 041 =44.4% | 7472/16 041=46.6% | ||
| Recall | 0.742 | 0.440 | 0.390 | 0.444 | 0.466 | ||
| Precision | 0.0495 | 0.0287 | 0.0285 | 0.014 | 0.0346 | ||
| 120 | # of predictions | 481 135 | 476 827 | 461 280 | 906 654 | 446 074 | |
| % of correct predictions | 13 846/16 041=86.3% | 9683/16 041=60.4% | 8969/16 041=55.9% | 10 342/16 041=64.5% | 10 614/16 041=66.2% | ||
| Recall | 0.863 | 0.604 | 0.559 | 0.645 | 0.662 | ||
| Precision | 0.0288 | 0.0203 | 0.0194 | 0.0114 | 0.0238 | ||
| II | 60 | # of predictions | 469 752 | 453 880 | 437 791 | 971 238 | 399 746 |
| % of correct predictions | 34 301/43 251 =79.3% | 20 378/43 251 =47.1% | 17 556/43 251 =40.6% | 20 543/43 251 =47.5% | 19 442/43 251=46.1% | ||
| Recall | 0.793 | 0.471 | 0.406 | 0.475 | 0.461 | ||
| Precision | 0.0730 | 0.0449 | 0.0401 | 0.0211 | 0.0486 | ||
| 120 | # of predictions | 961 112 | 902 611 | 922 373 | 1 952 258 | 832 842 | |
| % of correct predictions | 38 821/43 251= 89.8% | 23 762/43 251=54.9% | 24 578/43 251= 56.8% | 25 667/43 251= 59.3% | 27 980/43 251=64.7% | ||
| Recall | 0.898 | 0.549 | 0.568 | 0.593 | 0.647 | ||
| Precision | 0.0403 | 0.0263 | 0.0266 | 0.0131 | 0.0336 | ||
| III | 119 | # of predictions | 285 491 | 439 485 | 875 442 | 341 773 | 382 173 |
| % of correct predictions | 10 766/11 080=97.2% | 9069/11 080=81.8% | 10 084/11 080=91.0% | 7840/11 080=70.8% | 10 334/11 080=93.3% | ||
| Recall | 0.972 | 0.818 | 0.910 | 0.708 | 0.933 | ||
| Precision | 0.0377 | 0.0206 | 0.0115 | 0.0229 | 0.0270 | ||
| IV | 50 | # of predicted interactions | 184 842 | 172 256 | 141 717 | 173 378 | 149 142 |
| % of correct predictions | 31 779/60 818=52.3% | 25 326/60 818=41.6% | 19 873/60 818=32.7% | 19 785/60 818=32.5% | 23 757/60 818=39.1% | ||
| Recall | 0.523 | 0.416 | 0.327 | 0.325 | 0.391 | ||
| Precision | 0.172 | 0.147 | 0.140 | 0.114 | 0.159 | ||
| 100 | # of predicted interactions | 412 149 | 337 863 | 286 667 | 413 213 | 298 004 | |
| % of correct predictions | 52 955/100 608=52.6% | 41 722/100 608=41.5% | 32 649/100 608=32.5% | 33 412/100 608=33.2% | 37 616/100 608=37.4% | ||
| Recall | 0.526 | 0.415 | 0.325 | 0.332 | 0.374 | ||
| Precision | 0.128 | 0.123 | 0.114 | 0.081 | 0.126 |
The above analyses demonstrated the successful performance of TarPmiR in the human dataset in the same cell type. It was unclear how well TarPmiR performed in other species and in other cell types. We thus applied TarPmiR to a third independent dataset, the mouse HITS-CLIP dataset in the cortex cell (dataset III). There were 119 potential miRNAs and 2953 mRNAs involved in a total of 11 080 target sites. With the input of these 119 miRNAs and 2953 mRNAs, TarPmiR predicted 285 491 target sites in total. There were 10 766 of the 11 080 (97.2%) target sites predicted by TarPmiR (Table 2).
In addition to the above analyses on the crosslinking-based data, we tested TarPmiR using the annotated miRNA–mRNA interactions in TarBase 7.0 (dataset IV) (Table 2). For the top 50 miRNAs and the corresponding 9823 target mRNAs, TarPmiR predicted 52.3% of true target mRNAs (Methods). For the top 100 miRNAs and the corresponding 9869 target mRNAs, TarPmiR predicted 52.6% of true target mRNAs (Table 2) (Methods).
3.4 TarPmiR showed superior performance to existing approaches
We compared TarPmiR with two widely used tools miRanda (Enright et al., 2004), targetScan V2010 (Friedman et al., 2009; Grimson et al., 2007), targetScan V2015 (Agarwal et al., 2015) and a recently published tool, miRmap (Vejnar and Zdobnov, 2012; Vejnar et al., 2013). The comparison was made on the CLASH dataset, the three independent datasets and the two databases described above. Overall, TarPmiR with the default cutoff 0.5 had a much higher recall and precision than the three existing methods on the CLASH dataset (Table 3). For instance, TarPmiR had a recall of 55.1%, which was at least 10% higher than other approaches. TarPmiR had a precision of 19.1%, which was at least 0.2% higher than other approaches.
Table 3.
Comparison of different methods on the CLASH dataset
| Method | TP | FN | FP | Recall TP/(TP+FN) | Precision TP/(TP+FP) | F1-score |
|---|---|---|---|---|---|---|
| TarPmiR | 4695 | 3819 | 19 950 | 0.551 | 0.191 | 0.284 |
| miRanda | 3852 | 4662 | 51 849 | 0.452 | 0.069 | 0.120 |
| TargetScan V2010 | 1164 | 7350 | 10 281 | 0.136 | 0.101 | 0.116 |
| TargetScan V2015 | 2368 | 6146 | 10 182 | 0.278 | 0.189 | 0.225 |
| Mirmap | 1821 | 6693 | 30 746 | 0.214 | 0.056 | 0.089 |
On the three independent datasets, we compared TarPmiR with the other three methods, including two versions of TargetScan (Table 2). Overall, TarPmiR had a similar or much smaller number of predicted target sites, while it had much more known miRNA target sites predicted in each dataset. By assuming the CCRs from PAR-CLIP and target sites from HITS-CLIP were the only true miRNA target sites in the corresponding mRNAs in the corresponding datasets, we found that TarPmiR had a recall at least 3.9% higher than other methods, and a precision at least 0.5% higher than other methods. Note that the performance of all five methods was relatively high in the mouse dataset than other independent datasets, because miRNA–mRNA interactions in this dataset were mainly inferred and majorly based on seed regions (Chi et al., 2009).
For the known miRNA–mRNA interactions in TarBase 7.0, we also compared TarPmiR with other three methods (Table 2). TarPmiR had a similar or slightly larger number of predicted interactions, while it predicted much more known miRNA–mRNA interactions. Similar to the results on crosslinking-based datasets, TarPmiR had a much higher recall and a higher precision than other methods.
We also compared the running speed of the all methods. Because TarPmiR was a machine learning based method and it calculated more features, it was much slower than miRanda and TargetScan. The running speed was similar to that of Mirmap, which was also a machine learning-based method. It was worth pointing out that, although TarPmiR was relatively slow, its speed was reasonable. For instance, it took TarPmiR about 7940 CPU s to predict target sites of 20 miRNAs in 400 mRNA sequences, on average each 2000 nt long.
4 Discussion
In this study, we identified seven new features together with six conventional features of miRNA target sites. Based on these 13 selected features, we developed a new approach called TarPmiR to predict miRNA target sites. We tested TarPmiR on a human CLASH dataset, two human PAR-CLIP datasets, a mouse HITS-CLIP dataset and a general dataset from TarBase 7.0, and showed that TarPmiR performed at least the same or better than three existing approaches. TarPmiR is freely available at http://hulab.ucf.edu/research/projects/miRNA/TarPmiR/.
Not all new features were completely new. We claimed some features as new because they were not used by most of the existing tools, such as miRanda (Enright et al., 2004), TargetScan (Friedman et al., 2009; Grimson et al., 2007), DIANA-microT-CDS (Maragkakis et al., 2009; Paraskevopoulou et al., 2013), rna22-gui (Loher and Rigoutsos, 2012), TargetMiner (Bandyopadhyay and Mitra, 2009), PITA (Kertesz et al., 2007) and RNAhybrid (Krüger and Rehmsmeier, 2006). However, several new features were mentioned in previous studies directly or indirectly. For instance, Thomson et al. (2011) stated that ‘some validated miRNA target sites do not have a complete seed match but instead exhibit 11–12 continuous base pairs in the central region of the miRNA’. We observed similar target sites in the CLASH dataset and proposed the feature ‘The length and position of the longest consecutive pairs’.
The selected new features significantly improved the prediction accuracy of TarPmiR. To show the contribution of the new features to the accuracy of TarPmiR, we removed the seven new features and retrained random forests in TarPmiR. Compared with the original TarPmiR with 13 features, the recall and precision of the modified TarPmiR dropped 8.6% and 9.7%, respectively.
We also compared the predicted true target sites by different approaches (Supplementary File S4). TarPmiR had the largest number of predicted true sites shared by other tools. However, the percentage of shared true target sites predicted by TarPmiR was lower than that of other tools, suggesting that TarPmiR complements existing tools by predicting sites that cannot be predicted by other tools. In fact, there are 2090 ‘non-seed-matching’ sites in the first CLASH test dataset. TarPmiR was able to identify 1585 (75.8%) of those sites. On the other hand, miRanda and TargetScan were only able to predict 173 (8.28%) and 34 (1.6%) sites, respectively. This also suggested that the traditional tools like TargetScan and miRanda almost cannot predict non-seed-matching binding sites.
It is also worth mentioning that CLASH experiments may pick up direct and indirect miRNA target sites. The Argonaut proteins are guided by miRNAs to bind mRNAs, which is referred to as miRNA-dependent recruitment and results in direct miRNA target sites. There is also a miRNA-independent Argonaut protein recruitment mechanism, in which Argonaut proteins are recruited to target mRNAs by protein–protein interaction with RNA-binding proteins and thus miRNAs do not interact with the mRNAs directly (Meister, 2013). In the future, one may want to distinguish these two types of target sites from the CLASH experiments before training predictors for target site prediction. In this way, we may also obtain better features and improve the prediction accuracy.
Because of the existence of indirect target sites in CLASH data, the recall of TarPmiR on the CLASH testing datasets may be underestimated. In fact, TarPmiR had a much higher recall on the three independent human and mouse datasets, suggesting that TarPmiR may have a recall larger than 74%. On the other hand, TarPmiR had a much lower precision on the independent datasets, which may be underestimated as well. This was because we treated all segments other than the CCRs or identified miRNA target sites in these independent datasets as true negative target sites, which may not be the case.
By the time of this study, only one CLASH dataset was publicly available (Helwak et al., 2013). This human CLASH dataset was used to train TarPmiR. We applied TarPmiR to human and mouse datasets and demonstrated that it works well on these datasets. In the future, with more CLASH datasets available, more important miRNA target site features including tissue-specific features may be discovered and the accuracy of TarPmiR, especially its precision, may be further improved.
Supplementary Material
Acknowledgements
We appreciate the anonymous reviewers for their insightful comments and suggestions. We thank Vikram Dhillon and Amlan Talukder for the proofreading of the manuscript and their helpful comments.
Funding
This work is supported by the National Science Foundation [grants 1356524, 1149955 and 1218275] and the National Institute of Health [grant 2R01HL048044]. Funding for open access charge: The National Science Foundation grant 1356524.
Conflict of Interest: none declared.
References
- Agarwal V. et al. (2015) Predicting effective microRNA target sites in mammalian mRNAs. Elife, 4, e05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. et al. (2011) Vive la difference: biogenesis and evolution of microRNAs in plants and animals. Genome Biol., 12, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S., Mitra R. (2009) TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics, 25, 2625–2631. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. (2009) MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J. et al. (2005) Principles of microRNA-target recognition. PLoS Biol., 3, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.W., Lin C.J. (2006) Combining SVMs with Various Feature Selection Strategies In: Guyon I. et al. (eds) Feature Extraction. Springer, Berlin Heidelberg, pp. 315–324. [Google Scholar]
- Chi S.W. et al. (2009) Argonaute HITS-CLIP decodes microRNA–mRNA interaction maps. Nature, 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C.H. et al. (2013) A computational approach for identifying microRNA-target interactions using high-throughput CLIP and PAR-CLIP sequencing. BMC Genomics, 14, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.H. et al. (2001) Stepwise logistic regression analysis of tumor contour features for breast ultrasound diagnosis. Ultrasound Med. Biol., 27, 1493–1498. [DOI] [PubMed] [Google Scholar]
- Díaz-Uriarte R., De Andres S.A. (2006) Gene selection and classification of microarray data using random forest. BMC Bioinformatics, 7, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D., Hobert O. (2006) Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol., 13, 849–851. [DOI] [PubMed] [Google Scholar]
- Ding J. et al. (2015) MicroRNA modules prefer to bind weak and unconventional target sites. Bioinformatics, 31, 1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J. et al. (2004) MicroRNA targets in Drosophila. Genome Biol., 5, R1-R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.C. et al. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res., 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. et al. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res., 34, D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A. et al. (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell, 27, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M. et al. (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell, 141, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A. et al. (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell, 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L. (2003) Vienna RNA secondary structure server. Nucleic Acids Res., 31, 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M. et al. (2007) The role of site accessibility in microRNA target recognition. Nat. Genet., 39, 1278–1284. [DOI] [PubMed] [Google Scholar]
- Kim Y., Kim J. (2004) Gradient LASSO for feature selection. In: Proceedings of the twenty-first international conference on Machine learning. ACM, pp. 60.
- Kishore S. et al. (2011) A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Methods, 8, 559–564. [DOI] [PubMed] [Google Scholar]
- Kokaly R.F., Clark R.N. (1999) Spectroscopic determination of leaf biochemistry using band-depth analysis of absorption features and stepwise multiple linear regression. Remote Sens. Environ., 67, 267–287. [Google Scholar]
- Krüger J., Rehmsmeier M. (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res., 34, W451–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P. et al. (2003) Prediction of mammalian microRNA targets. Cell, 115, 787–798. [DOI] [PubMed] [Google Scholar]
- Li J. et al. (2014) Identifying mRNA sequence elements for target recognition by human Argonaute proteins. Genome research, 24, 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D. et al. (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature, 456, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loher P., Rigoutsos I. (2012) Interactive exploration of RNA22 microRNA target predictions. Bioinformatics, 28, 3322–3323. [DOI] [PubMed] [Google Scholar]
- Ma S., Huang J. (2008) Penalized feature selection and classification in bioinformatics. Brief. Bioinf., 9, 392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann H.B., Whitney D.R. (1947) On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat., 18, 50–60. [Google Scholar]
- Maragkakis M. et al. (2009) DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res., gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinshausen N., Bühlmann P. (2010) Stability selection. J. R. Stat. Soc. Ser. B (Statistical Methodology), 72, 417–473. [Google Scholar]
- Meister G. (2013) Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet., 14, 447–459. [DOI] [PubMed] [Google Scholar]
- Muljo S.A. et al. (2010) MicroRNA targeting in mammalian genomes: genes and mechanisms. Wiley Interdisc. Rev. Syst. Biol. Med., 2, 148–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou M.D. et al. (2013) DIANA-microT web server v5. 0: service integration into miRNA functional analysis workflows. Nucleic Acids Res., 41, W169–W173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.M. et al. (2014) Common features of microRNA target prediction tools. Front. Genet., 5, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S. et al. (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res., 20, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston A., Wilf H.S. eds. (1976) Mathematical methods for digital computers. Vol. 1, John Wiley & Sons, New York. [Google Scholar]
- Reczko M. et al. (2011) Accurate microRNA target prediction using detailed binding site accessibility and machine learning on proteomics data. Front. Genet., 2, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeys Y. et al. (2007) A review of feature selection techniques in bioinformatics. Bioinformatics, 23, 2507–2517. [DOI] [PubMed] [Google Scholar]
- Sassen S. et al. (2008) MicroRNA—implications for cancer. Virchows Arch., 452, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanen B.C., Li X. (2011) Transcriptional regulation of mammalian miRNA genes. Genomics, 97, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetnik V. et al. (2003) Random forest: a classification and regression tool for compound classification and QSAR modeling. J. Chem. Inf. Comput. Sci., 43, 1947–1958. [DOI] [PubMed] [Google Scholar]
- Thomson D.W. et al. (2011) Experimental strategies for microRNA target identification. Nucleic Acids Res., 39, 6845–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. (1996) Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B (Methodological), 267–288. [Google Scholar]
- Vejnar C.E. et al. (2013) miRmap web: comprehensive microRNA target prediction online. Nucleic Acids Res., 41, W165–W168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejnar C.E., Zdobnov E.M. (2012) miRmap: Comprehensive prediction of microRNA target repression strength. Nucleic Acids Res., 40, 11673–11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos I.S. et al. (2014) DIANA-TarBase v7. 0: indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res., gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. et al. (2014) dCLIP: a computational approach for comparative CLIP-seq analyses. Genome Biol., 15, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2011) Transcriptional regulation of co-expressed microRNA target genes. Genomics, 98, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C.J. et al. (1995) A prospective randomized trial of pancreaticogastrostomy versus pancreaticojejunostomy after pancreaticoduodenectomy. Ann. Surg., 222, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef M. et al. (2007) Naïve Bayes for microRNA target predictions—machine learning for microRNA targets. Bioinformatics, 23, 2987–2992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.