Abstract
Motivation: Recent advances of next-generation sequence technologies have made it possible to rapidly and inexpensively identify gene variations. Knowing the disease association of these gene variations is important for early intervention to treat deadly diseases and provide possible targets to cure these diseases. Genome-wide association studies (GWAS) have identified many individual genes associated with common diseases. To exploit the large amount of data obtained from GWAS studies and leverage our understanding of common as well as rare diseases, we have developed a knowledge-based approach to predict gene–disease associations. We first derive gene–gene mutual information by utilizing the cooccurrence of genes in known gene–disease association data. Subsequently, the mutual information is combined with known protein–protein interaction networks by a boosted tree regression method.
Results: The method called Know-GENE is compared with the method of random walking on the heterogeneous network using the same input data. For a set of 960 diseases, using the same training data in testing in 3-fold cross-validation, the average recall rate within the top ranked 100 genes by Know-GENE is 65.0% compared with 37.9% by the state of the art random walking on heterogeneous network. This significant improvement is mostly due to the inclusion of knowledge-based mutual information.
Availability and Implementation: Predictions for genes associated with the 960 diseases are available at http://cssb2.biology.gatech.edu/knowgene.
Contact: skolnick@gatech.edu
1 Introduction
Complex diseases such as Parkinson’s Disease (PD) are attributed to both genetic and/or environmental causes (Goldman, 2014). Environmental toxins cause disease through their effects on genes (Qi et al., 2014). Knowing the genes associated with a disease is useful for preventing and curing the disease. It is also fundamentally important for understanding the biological functions of genes. Genome-wide association studies (GWAS) posit that multiple, common small-risk variants interact to cause common diseases (Reich and Lander, 2001; TA et al., 2010). GWAS relies on testing several hundred thousand common genetic variants found throughout the human genome in large-control cohorts. Over the past few years, many gene loci have been implicated as associated with various human diseases by GWAS (www.genome.gov/gwastudies/) as well as linkage studies (Morton, 1955).
Despite the fruitful insights provided by GWAS, heritability estimates have shown that large proportions of genetic risk underlying complex disease have not yet been explained (Manolio et al., 2009). This is due to lack of the ability to detect ‘common disease by rare variants’ using a GWAS approach. The introduction of next-generation sequencing technologies allows cost-effective sequencing of entire genomes and has led to the discovery of numerous rare variants in the human genome. The disease associations of these rare variants cannot be inferred by GWAS for further experimental verification. Thus, alternative computational methods that predict the association of gene with a given disease have been developed (Köhler et al., 2008; Li and Patra, 2010; Natarajan and Dhillon, 2014; Qian et al., 2014; Singh-Blom et al., 2013; van Driel et al., 2006; Vanunu et al., 2010). The general idea of prediction methods is the ‘guilt by association’ principle (Wolfe et al., 2005) with respect to a set of known genes related to the given disease. The most frequently used types of evidence for inference of association are (Piro and Di Cunto, 2012): (i) text mining of the biomedical literature; (ii) phenotype relationships such as disease–disease similarity; (iii) protein–protein interactions; (iv) regulatory information and (v) gene expression information. For example, Van Driel et al. (2006) used a text-mining approach to associate genes with human phenotypes found in the Online Mendelian Inheritance in Man (OMIM) database (Hamosh et al., 2002). Köhler et al. found gene–disease associations by using a global network distance measure—a random walk analysis—for the definition of similarities in protein–protein interaction networks (the ‘interactome’). Encouragingly, they find that this approach significantly outperforms previous methods based on local distance measures in the interactome (Köhler et al., 2008). A slightly modified approach called ‘network propagation’ that differs from a random walk only in the normalization of the adjacency matrix W representing the interactome (Qian et al., 2014; Vanunu et al., 2010) has also been developed. The random walk method normalizes the adjacency matrix W by columns (the summation of each column equals to one), whereas the network propagation normalizes W by the diagonal matrix: , where . An extension of the random walk approach is walking on a heterogeneous network that includes protein–protein interaction, disease–disease and gene–disease networks (Li and Patra, 2010). Singh-Blom et al. (2013) have developed a truncated version of the random walking on a heterogeneous network by using a limited steps for the walks but it also includes phenotypes from multiple species. The method uses simple dampening coefficients for longer walks and learns the coefficients for longer walks using a support vector machine (SVM) (Cortes and Vapnik, 1995). Natarajan and Dhillon (2014)developed an inductive method that uses a machine learning approach to incorporate different biological sources of evidence such as microarray expression data, gene functional interaction data and disease-related textual data from human as well as other species. The best performing of the aforementioned methods for prioritizing genes associated with a given disease is the inductive matrix completion developed by Natarajan and Dhillon (2014). It has an average recall rate of 25% within the top 100 ranked genes. There are also many disease specific methods that focus on a single or group of diseases to prioritize genes for further experimental validation. For a survey of methods for predicting gene–disease association, please see Piro and Di Cunto (2012).
Here, we develop a new type of approach for prioritizing candidate genes associated with a given disease. Our approach applies the idea of word association in the context of texts (Church and Hanks, 1990) to gene–gene association in the context of diseases and then employs gene–gene association to infer gene–disease association. This is a knowledge-based approach that learns gene–gene association propensity in diseases from known gene–disease association. It is also analogous to methods of knowledge-based statistical potentials for protein structure prediction that learn residue–residue or atom–atom pairwise interaction potentials from experimental protein structures (Lu and Skolnick, 2001; Zhou and Zhou, 2002). Mutual information (Fano, 1961) is used to measure the strength of gene–gene association in a given disease. Owing to the increasing amount of data for known gene–disease associations, the mutual information of gene–gene pairs can be derived from these known associations. Subsequently, mutual information is combined with the properties of the protein–protein physical interaction network by means of boosted tree regression (Roe et al., 2006). The resulting method called Know-GENE is then benchmarked in 3-fold cross-validation (training on 2/3 of the known gene–disease associations and testing on the remaining 1/3) and compared with network propagation, random walking on the interactome as well as on the heterogeneous network-based methods. For 960 diseases defined by the medical subject headings (MeSHs) ontology with at least two known seed genes associated with a given disease [as provided by the OMIM and GWAS databases (Mottaz et al., 2008; Ramos et al., 2014; Zhang et al., 2010)], using the same input evidence (known gene–disease association and interactome), Know-GENE achieves a significantly better recall rate (65.0%) within the top 100 ranked of 15 948 total screened genes compared with the network propagation (19.2%), the random walk on interactome (18.1%) and the random walk on heterogeneous network (37.9%) methods. Thus, Know-GENE is a promising method for prioritizing candidate genes associated with a given disease. Then, we apply Know-GENE in prediction mode (training on all known gene–disease associations) to predict and rank genes in the human exome for each of the 960 diseases. Likewise, our predictions can rank diseases for a given gene, a useful feature for diagnosing diseases in a given mutated gene. It is also useful for predicting and understanding possible side effects of drug targets. Predictions are available for academic users at http://cssb2.biology.gatech.edu/knowgene.
2 Materials and methods
This work is based on the assumption that gene–disease association can be inferred from gene–gene functional interactions and protein–protein physical interactions given a set of genes known to be associated with a given disease. We will utilize the known protein–protein physical interaction network and known gene–disease associations as provided by GWAS (Reich and Lander, 2001; TA et al., 2010) and linkage studies (Morton, 1955).
2.1 Datasets and sources
Protein–protein physical interactions (interactome) are compiled from the HIPPIE database (http://cbdm.mdc-berlin.de/tools/hippie/hippie_current.txt) (Schaefer et al., 2012) and the work of Menche et. al. (2015) (http://www.sciencemag.org/content/347/6224/1257601/suppl/DC1). In total, there are 246 502 human protein–protein interactions involving 15 948 genes/proteins that are experimentally documented to involve regulatory, metabolic pathway and kinase–substrate interactions. Among these, 3179 genes are known to be associated with at least one disease. The interactome data can be found at http://cssb2.biology.gatech.edu/knowgene.
We then compiled 960 diseases defined by the MeSH ontology that have at least two associated genes (proteins) in the interactome from two sources as in: (i) Ref. (Zhang et al., 2010) (http://www.biomedcentral.com/1755-8794/3/1) that also uses the MeSH disease definition and (ii) Ref. (Menche et al., 2015) (http://www.sciencemag.org/content/347/6224/1257601/suppl/DC1). The primary sources of these data are from the genetic association databases (Becker et al., 2004), the OMIM database (Hamosh et al., 2002) and the GWAS databases (Mottaz et al., 2008; Ramos et al., 2014). This provides 31 993 gene–disease associations for 3171 genes. Of these, 283 genes are known to be associated with only one disease, and the disease association of the remaining 15 948 genes is unknown. The known association data can also be found at http://cssb2.biology.gatech.edu/knowgene.
2.2 Core genes of a disease
It was found that even with an incomplete interactome, genes associated with given disease tend to interact with each other and form connected clusters (Menche et al., 2015). A connected cluster consists of genes directly connected (that is interact) to one another in interactome space. Genes in the largest interacting cluster of a disease are called core genes. To measure if a cluster is formed by chance or due to intrinsic properties of the disease, we conduct the same statistical test as in Ref. (Menche et al., 2015). For the size S (number of genes) of the cluster, we calculate the z-score:
| (1) |
where <Srand> and σ(Srand) are the average largest cluster size value and SD of randomly picked Ng number of genes in the interaction network. Here, Ng is the number of known genes of the considered disease. To calculate the z-score, we simulate the random process 10 000 times. Each time, Ng genes were randomly selected from the 15 948 screened genes. Then, the size Srand of the largest connected cluster from these Ng genes was obtained. The process was repeated 10 000 times. A cluster with z-score > 1.65 (P-value < 0.05) is considered significant. This leads to 537 of the 960 diseases having a statistically significant cluster. The largest significant cluster of each disease gives the core set of genes: i.e. core genes of the disease. The average number of known genes for the 537 diseases having core genes is 25.5; whereas for the 423 diseases not having core genes, it is 9.4. Thus, a disease having core genes usually has more known genes than diseases lacking core genes.
2.3 Gene–gene pairwise mutual information
Borrowing the idea of measuring word association strength (Church and Hanks, 1990), to measure the strength of functional association of two genes in diseases that might include the effect of direct physical interactions, we use their mutual information (Fano, 1961) defined as:
| (2) |
where P(gx), P(gy) are the probabilities of observing genes gx and gy, independently in a given disease, and P(gx,gy) is the probability of observing genes gx and gy, together in a given disease. If there is a genuine association between genes gx and gy, then the joint probability P(gx,gy) will be larger than that by chance, P(gx)P(gy). Thus, I(gx,gy) will be > 0.
In Know-GENE (knowledge-based approach for predicting gene-disease association), the probabilities P(gx), P(gy) are estimated by counting the number of genes gx, gy associated with diseases N(gx) and N(gy), normalized by the number of diseases, Nd (here, Nd= 960) and P(gx,gy) is estimated by counting the number of cooccurrences of genes gx, gy associated with the disease N(gx, gy) normalized by Nd.
2.4 Gene–disease association measures
We first consider the network distance of a gene to a given disease. A unit network distance is defined as a path from one protein to another with a direct connection in the interactome. We bin the shortest distances of an unknown gene to all the known genes of a given disease from 1 to 10 and fill each bin with the number of genes known to be associated with the given disease, i.e. we have a vector (n1, n2,…, n10) with Σni = Ng, Ng is total number of known genes and ni is the number of genes in the known set with shortest distance i to the unknown gene. A similar histogram is also done for the core genes (if there is no core gene, all 10 histogram values are zero).
We then consider the functional association strength of an unknown gene gx to a given disease D that is defined as:
| (3) |
where the summation is over all known genes of the disease. P(gx,gy) and I(gx,gy) are the probabilities of observing two genes associated with a disease together and the resulting mutual information [Equation (2)], respectively.
To combine the above network distances and functional association strength, we employ the boosted tree regression machine learning method. Boosted tree regression has been employed in many applications (Friedman and Meulman, 2003; Roe et al., 2006) and has been shown to be much better than SVMs (Cortes and Vapnik, 1995) and random forests (Breiman, 2001) in predicting genomic breeding values (Ogutu et al., 2011). It involves generating a sequence of decision trees; each grows on the basis of the residuals of all previous trees (Roe et al., 2006; Thusberg et al., 2011). Here, a decision tree regression is implemented with a maximal depth of eight. The scoring function is represented as a boosted decision tree (Roe et al., 2006):
| (4) |
where Tm is a decision tree, ε is the shrinkage factor or learning rate, Ntree is the number of trees and x represents a set of features. In this application, Ntree is set to 500 and ε = 0.01. The following 23 features are derived from the above network distances and mutual information:
(a) 11 features: histogram of network distances to all known genes (n1, n2,…,n10) plus the mean value Σ (ni × i)/Ng;
(b) 11 features: histogram of network distances to all core genes (n1c, n2c,…,n10c) plus the mean value Σ(nic × i)/Σnic; these values are set to zero for diseases without core genes;
(c) 1 feature for the functional association strength S(D,gx) by Equation (3).
2.5 Training and testing
We considered 960 diseases and 15 948 genes having protein–protein interactions. There are 15 310 080 gene–disease pairs with 31 993 known associations. To test our method, we perform 3-fold cross-validation. All gene–disease pairs are randomly partitioned into three approximately equal size sets. For each set, we use the other two sets for training the boosted tree regression model and predict scores for the third, testing set. In the training process, all known associations are assigned an objective function value of 1 and unknown pairs assigned 0. This might introduce a few false negatives because unknown ones could be true associations. However, since the number of unknown associations is much larger than possible true associations, the chance of a false negative is small. In the 2/3 of training data pairs, only 0.2% of total gene–disease pairs are positives (assigned a value of 1). If all pairs are used for training, the model will be overwhelmingly dominated by negative samples. Thus, we shall use only a partial set (∼4%) of the negative pairs in the training data for actual training. This results in a ratio of 1:20 positives versus negatives in the actual training data. In either training or testing, when calculating the network distances, the gene is removed from known set of genes for the given disease if it is a known gene of the disease. Any known association is not counted in mutual information derivation by Equation (2) if it is being tested. The predicted scores are then used to rank genes for each disease. Similarly, one can also use the score to rank diseases of a given gene.
2.6 Comparison to other methods
We shall compare Know-GENE to methods that utilize the same input data (interactome and known set of gene–disease associations) to tease out the effects of different methods. We will not compare methods that use multiple other sources of data (Natarajan and Dhillon, 2014; Singh-Blom et al., 2013) since they used more data, and the effects of the methods themselves are unclear. We implemented three other methods as described in the corresponding references for comparison: random walk (Köhler et al., 2008), network propagation (Vanunu et al., 2010) and random walk on a heterogeneous network (Li and Patra, 2010). Random walk on a heterogeneous network requires a disease–disease similarity matrix calculated using MimMiner (van Driel et al., 2006). In order not to introduce additional data, we used the overlap score to measure disease–disease similarity between disease d1 and d2 without using additional input data (Goh et al., 2007):
| (5) |
2.7 Evaluation
For each of the 960 diseases, 15 948 genes are ranked according to their tree regression scores. We evaluated the performance of the methods using the following three criteria: AUC (area under the Receiver Operating Characteristics (ROC) curve–true positive (TP) rate/false positive (FP) rate curve), area under the precision/recall curve (AUPR) and recall rate [the ratio (predicted TPs)/(total TPs)] within the top 100 ranked genes. AUC measures discrimination, that is, the ability of the test to correctly classify those with and without the association to the given disease. Of the 15 948 genes, the majority are true negatives (TNs) for a given disease, a large number change in the number of FPs can lead to a small change in the FP rate [FP/(FP + TN)] used in the ROC analysis (Davis and Goadrich, 2006). On the other hand, AUPR, by comparing FPs to TPs rather than TNs, captures the effect of the large number of negative examples on the algorithm’s performance [precision = TP/(TP + FP)]. Thus, we include AUPR as an additional performance measure. AUPR is sensitive to FPs among the top ranked genes (the region where FP rate has the smallest value and precision has the largest value). The recall rate gives the experimentalist’s expectation of TPs. Based on our earlier work for ligand virtual screening for the human proteome, the recall rate does not depend on whether the known set of genes for a given disease is complete or partial whereas precision, AUC and AUPR will (Zhou et al., 2015). Thus, recall rate can be considered as having absolute meaning whereas precision, AUC and AUPR are meaningful only for comparing different methods (and are not good for assessing the absolute performance of a method).
3 Results
3.1 Overall performance
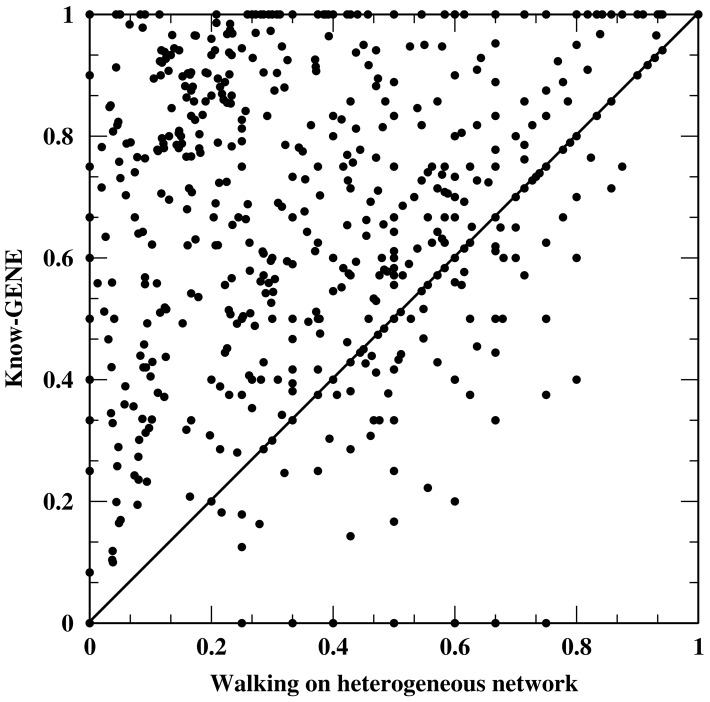
The 3-fold cross-validation for the 960 diseases is shown in Table 1 in comparison with the other three methods. Know-GENE with AUC = 0.967, AUPR = 0.405 and recall rate 65.0% is the best for all three measures. Random walk and network propagation methods have very similar results whereas method by walking on the heterogeneous network has much better performance than both the random walk and network propagation. This performance boost is due to the inclusion of information from the gene–disease network. The AUPR and recall rate within the top 100 genes by Know-GENE are almost twice those by walking on a heterogeneous network. We also see that network propagation is slightly better than a random walk. A scatter plot of recall rates for 960 diseases by Know-GENE versus method of walking on a heterogeneous network is given in Figure 1. Know-GENE wins for 651 diseases whereas walking on the heterogeneous network wins for 129 diseases.
Table 1.
Performance of methods for 960 diseases against 15 948 genes
| Method | Average AUC | Average AUPR | Average recall rate within the top 100 genes (%) |
|---|---|---|---|
| Random walk | 0.780 | 0.053 | 18.1 |
| Network propagation | 0.790 | 0.055 | 19.2 |
| Random walk on heterogeneous network | 0.930 | 0.138 | 37.9 |
| Know-GENE | 0.967 | 0.405 | 65.0 |
| Know-GENE without using mutual information | 0.700 | 0.167 | 24.0 |
| Know-GENE without using core gene | 0.967 | 0.352 | 63.8 |
Fig. 1.
Scatter plot of recall rates within the top 100 genes by Know-GENE versus the walking on heterogeneous network approach
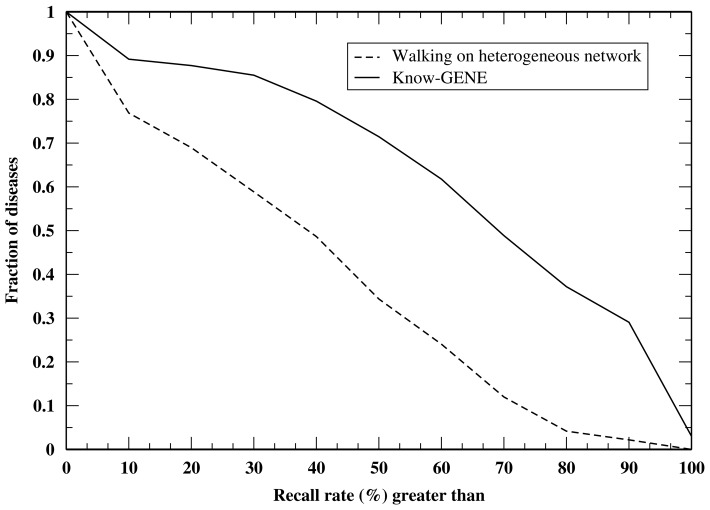
The histograms of recall rates by those two methods are given in Figure 2. For the 960 diseases, Know-GENE has 71.4% of diseases with a recall rate ≥ 50%, whereas walking on heterogeneous network has 34.3% of diseases with a recall rate ≥ 50%. The P-value for the difference between recall rates of Know-GENE and those of walking on heterogeneous network is 2.8 × 10−92. Thus, Know-GENE gives significant improvement over extant methods using exactly the same input data.
Fig. 2.
Fractions of diseases having recall rates within the top 100 genes ≥ given value by Know-GENE and the walking on heterogeneous network approach, respectively
3.2 Effects of mutual information and core genes
We next examine the effects of some factors in Know-GENE by removing from the features: (i) the mutual information term or (ii) the network distances to core genes. The results are also given in Table 1, and clearly indicate that without the mutual information term, Know-GENE performs significantly worse. Thus, the mutual information term derived from the gene–disease network contributes the most to the good performance of Know-GENE. This is consistent with the method by walking on the heterogeneous network that gets boosted performance by including the gene–disease network. The fact that Know-GENE performs significantly better than walking on the heterogeneous network indicates that mutual information captures more important information than the random walk approach. The terms related to core genes slightly affect the AUPR (from 0.405 to 0.352) and recall rate (from 65.0 to 63.8%).
In Table 2, we separately examined the performance of the methods for the 537 diseases having core genes and 423 diseases with no core genes. All methods have better performance for diseases having core genes. The performance differences between the two datasets are relatively larger for random walk and network propagation methods where known gene–disease associations are not utilized. Know-GENE has the best performance for all criteria for both datasets. It has a 70.7% average recall rate for the 537 diseases having core genes.
Table 2.
Performance of methods to predict disease association of the 15 948 genes for diseases with or without core genes
| Method | Average AUC | Average AUPR | Average recall rate within the top 100 genes (%) |
|---|---|---|---|
| 537 diseases having core genes | |||
| Random walk | 0.810 | 0.082 | 25.7 |
| Network propagation | 0.823 | 0.086 | 27.1 |
| Random walk on heterogeneous network | 0.957 | 0.179 | 41.2 |
| Know-GENE | 0.980 | 0.533 | 70.7 |
| 423 diseases not having core genes | |||
| Random walk | 0.741 | 0.016 | 8.4 |
| Network propagation | 0.748 | 0.015 | 9.0 |
| Random walk on heterogeneous network | 0.896 | 0.086 | 33.7 |
| Know-GENE | 0.951 | 0.243 | 57.7 |
3.3 Test on singleton genes
Next, we analyze the performance of methods for singleton genes (Natarajan and Dhillon, 2014; Singh-Blom et al., 2013) defined as having only one known disease association in the data. In this case, the gene will have no mutual information contribution at training and testing times because it is removed from the known association with a given disease. Therefore, the test on singleton genes tests the ability of the method to predict associations for genes that have no known association to any disease. The test results are compiled in Table 3 using measures of the average rank of genes and number of genes ranked within the top 100 of 15 948 genes for a given disease. Table 3 shows that all methods perform much poorer (the best recall rate by network propagation method is 32/283 = 11%) for singleton genes than that for genes known to be associated with diseases. Especially for Know-GENE and walking on heterogeneous network that utilized known gene–disease associations, the recall rate is around 1%. This is because these methods favor genes that have known gene–disease associations and thus drag down the relative ranks of those genes have no known gene–disease associations. Therefore, in practice, if a gene has no known disease association, one should use methods like network propagation, random walk or Know-GENE without mutual information whose recall rate is 26/283 or 9%
Table 3.
Performance of different methods to assign diseases for 283 singleton genes
| Method | Average rank | # of genes ranked within the top 100 genes |
|---|---|---|
| Random walk | 5485 | 24 |
| Network propagation | 5014 | 32 |
| Random walk on heterogeneous network | 6948 | 3 |
| Know-GENE | 8005 | 3 |
| Know-GENE without using mutual information | 5699 | 26 |
| Know-GENE without using core gene | 7969 | 1 |
3.4 Predicting new genes associated with a given disease
In the above benchmark tests, we examine only the top 100 ranked genes for a given disease. In practice, some of the diseases are associated with fewer than 100 genes and some diseases might have associations with more than 100 genes. We thus optimize a cutoff score that will give the best binary classification (associated/not associated) measured by the Matthew’s correlation coefficient (MCC) for the training data. This results in a cutoff score value of 0.45 and a MCC of 0.878 for the training data. We then use all the 15 278 087 predicted gene–disease pair scores with unknown associations as a random score distribution and fit the distribution to an extreme value distribution with μ = 0.002 and σ = 0.0122. A cutoff of 0.45 corresponds to a P-value of 2.2 × 10−16. With a 0.45 cutoff, Know-GENE will have an average per disease recall of 53.3%, slightly worse than 65.0% within top 100 ranked genes (notice that AUC and AUPR are cutoff independent).
Now, we examine predictions of new genes for specific diseases. One example is PD (Goldman, 2014). In our test data, there are 38 genes known to be associated with PD. Using a cutoff score of 0.45, Know-GENE predicts 149 genes associated with PD. Thirty-two (84.2%) of the 38 known associated genes are among the predictions. For the top 10 predicted genes with unknown PD association, we searched the literature to support or disapprove our predictions, with the results compiled in Table 4. Eight (80%) of the top 10 genes have supporting evidence giving a prediction precision of ∼80% for this particular disease.
Table 4.
Predicted top 10 new genes for PD by Know-GENE
| Gene | Score | Evidence of support or disapprove |
|---|---|---|
| NUP62_HUMAN | 0.971 | Supported by ref. (Zatloukal et al., 2002) |
| HD_HUMAN | 0.953 | Causes Huntington’s disease that shares the same malfunctions within the motor sector of the nervous system as PD (Delcomyn, 1998); http://serendip.brynmawr.edu/bb/neuro/neuro98/202s98-paper3/Sangaramoothy3.html |
| PANK2_HUMAN | 0.953 | Disapproved by ref. (Klopstock et al., 2005) |
| TOR1A_HUMAN | 0.952 | Supported by ref. (Leung et al., 2001) |
| TOM40_HUMAN | 0.935 | Supported by ref. (Bender et al., 2013) |
| STX6_HUMAN | 0.922 | Disapprove by ref. (Trinh et al., 2013) |
| THAP1_HUMAN | 0.890 | Support: THAP1 gene is part of a family of THAP proteins that bind specific DNA sequences and regulate cell proliferation through the pRB/E2F cell cycle target genes, a pathway recently proposed to be involved in cell death in PD (Höglinger et al., 2007; Houlden et al., 2010) |
| IRF4_HUMAN | 0.875 | Supported by ref. (Soreq et al., 2008) |
| E2AK3_HUMAN | 0.875 | Supported by ref. (Dzamko et al., 2014) |
| A4_HUMAN | 0.862 | Supported by ref. (Schulte et al., 2015) |
3.5 Predicting new diseases associated with a given gene
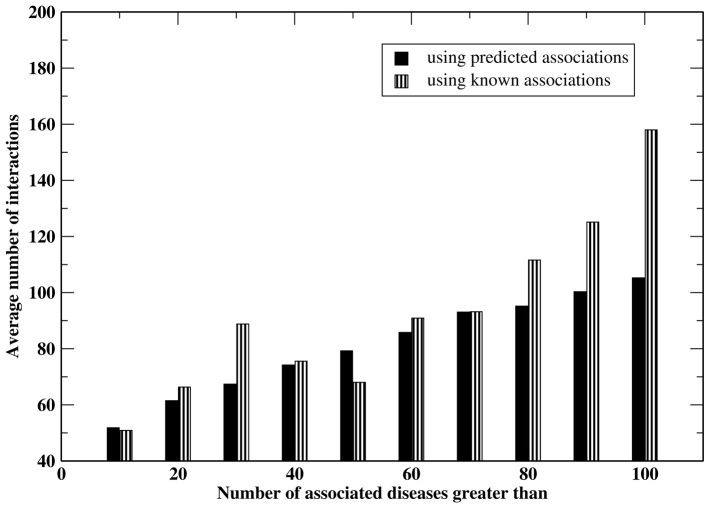
We can also examine the prediction of new diseases associated with a given gene. Diseases having a score ≥ 0.45 are used as predictions for a given gene. First, we examine the overall performance of disease prediction for a given gene by ranking diseases. The average AUC, AUPR and recall rate for 2888 genes having at least two known associated diseases are 0.969, 0.644 and 77.1%, respectively. We then examine the overall frequency of a gene to be associated with diseases and check its correlation with the number of interactions the corresponding protein has. The number of interactions of a protein can be derived from the interactome. Figure 3 shows the average number of interactions a gene has versus the number of diseases associated with a given gene ≥ given threshold. The Pearson’s correlation coefficient (C.C.) between the average number of interactions and threshold number of diseases when using known associations is 0.906, and when using predicted associations by Know-GENE with the cutoff of 0.45, it is 0.994, respectively. On average, proteins having more physical interactions tend to be associated with more diseases.
Fig. 3.
Average number of interactions a gene has versus the number of diseases associated with the gene ≥ given value. The Pearson’s C.C. between the average number of interactions and threshold number of diseases when using known gene–disease associations is 0.906; when using predicted associations by Know-GENE with a score ≥ 0.45, it is 0.994, respectively
One example of predicted new diseases for a given gene is for p53, a tumor suppressor gene. In our data, p53 is known to be associated with 100 diseases, and 84 of them have a score ≥ 0.45. Another 135 diseases also have a score ≥ 0.45 for p53 but these have unknown associations with p53. We list the top 10 new diseases predicted for the p53 gene in Table 5 along with supporting evidence. Nine diseases are supported, and one disapproved by literature.
Table 5.
Predicted top 10 new diseases for the p53 gene by Know-GENE
| Disease | Score | Evidence of support or disapprove |
|---|---|---|
| Female urogenital diseases and pregnancy complications | 0.986 | Supported by ref. (Fraga et al., 2014) |
| Intestinal diseases | 0.981 | Supported by ref. (Hussain et al., 2000) |
| Congenital abnormalities | 0.967 | Supported by ref. (Barkić et al., 2009) |
| Skin and connective tissue diseases | 0.965 | Supported by ref. (Menke et al., 2000) |
| Neoplasms, nerve tissue | 0.961 | Supported by ref. (Barbareschi et al., 1992) |
| Skin diseases | 0.960 | Supported by ref. (Batinac et al., 2004) |
| Neoplasms, germ cell and embryonal | 0.947 | Supported by ref. (Lewis et al., 1994) |
| Metabolism, inborn errors | 0.945 | Supported by ref. (Wilkins et al., 2013) |
| Neuroectodermal tumors | 0.944 | Disapprove by ref. (Raffel et al., 1993) |
| Male urogenital diseases | 0.941 | Supported by ref. (Castrén et al., 1998) |
We also find that disease recall rate is positively correlated with the number of known disease associations a given gene has. The Pearson’s C.C. is 0.173 for 2888 genes, which corresponds to a P-value of 7.7 × 10−21. For the 283 singleton genes, the average AUC, AUPR and recall rate by Know-GENE are 0.431, 0.012 and 0.0%, respectively, while the average AUC, AUPR and recall rate by Know-GENE without using mutual information are 0.841, 0.050 and 24.7%, respectively (using the cutoff = 0.32 obtained by optimizing MCC of the training set). Thus, Know-GENE is not good for predicting disease association of genes not seen in any of the diseases used for deriving the mutual information. In such cases, it would be better to apply Know-GENE without using mutual information or network propagation approach or the approach of simply walking on interactome instead of heterogeneous network.
Finally, Using Know-GENE in prediction mode, i.e. training on all data and making predictions for all genes, we provide all prediction results and a stand-alone program for academic users at http://cssb2.biology.gatech.edu/knowgene. The stand-alone program can be used to plug in new diseases with known genes for retraining the model and for predicting new genes associated with the already included diseases. Instructions as to how to use the program are also provided.
4 Discussion
We have developed a knowledge-based approach Know-GENE for prioritizing genes associated with given disease. Likewise, it can also prioritize diseases associated with a given gene. The novelty of Know-GENE is in the derivation of gene–gene mutual information from the cooccurrence frequency of pairs of genes in a large number of diseases with a known set of gene–disease associations. With this novel technique, Know-GENE performs much better than the best existing method of walking on the heterogeneous network using exactly the same input information (recall rate within top 100 ranked genes: 65.0 versus 37.9%). Both Know-GENE and walking on the heterogeneous network utilize information from the known gene–disease associations and perform much better than methods without using this information. For example, the network propagation method has a recall rate of 19.2%, which is about half that by the method of random walk on the heterogeneous network. All tested methods perform better for diseases having core genes than for those lacking core genes. When used for prioritizing diseases for a given gene, Know-GENE has an average recall rate of 77.1%.
A disadvantage of Know-GENE as well as the method of walking on the heterogeneous network is that it does not perform well for genes that are not present in any of the diseases used in deriving the mutual information (Table 3 test on singletons). In practice, for such genes, we can choose an alternative method such as Know-GENE without using mutual information or the network propagation method.
Possible further improvement of Know-GENE could come from enriching more genes with more verified disease associations and including them in deriving mutual information. These could be fulfilled by literature searches for associations with high Know-GENE prediction scores and then using the supported associations in training. This process could be iterated. For those genes without any known disease associations, other sources for deriving their mutual information are sought. For example, one possibility is the covariation of gene expression data in individuals with a given disease. Cooccurrence of gene pairs in pathways is another possible source. These and other alternatives will be explored in future work.
Acknowledgement
The authors thank Dr. Bartosz Ilkowski for managing the cluster on which this work was conducted.
Funding
This work has been supported by the National Institutes of Health [grant No GM-48835].
Conflict of Interest: none declared.
References
- Barbareschi M. et al. (1992) p53 protein expression in central nervous system neoplasms. J. Clin. Pathol., 45, 583–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkić M. et al. (2009) The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol. Cell. Biol., 29, 2489–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batinac T. et al. (2004) p53 protein expression and cell proliferation in non-neoplastic and neoplastic proliferative skin diseases. Tumori, 90, 120–127. [DOI] [PubMed] [Google Scholar]
- Becker K. et al. (2004) The genetic association database. Nat. Genet., 36, 431–432. [DOI] [PubMed] [Google Scholar]
- Bender A. et al. (2013) TOM40 mediates mitochondrial dysfunction induced by α-synuclein accumulation in Parkinson’s disease. PLoS One, 8, e62277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. (2001) Random forests . Mach. Learn., 45, 5–32. [Google Scholar]
- Castrén K. et al. (1998) Absence of p53 mutations in benign and pre-malignant male genital lesions with over-expressed p53 protein. Int. J. Cancer, 77, 674–678. [DOI] [PubMed] [Google Scholar]
- Church K.W., Hanks P. (1990) Word association norms, mutual information, and lexicography. Comput. Linguist., 16, 22–29. [Google Scholar]
- Cortes C., Vapnik V. (1995) Support-vector networks. Mach. Learn., 20, 273–297 [Google Scholar]
- Davis J., Goadrich M. (2006) The relationship between precision-recall and ROC curves. In Proceedings of the 23rd international conference on Machine learning. ACM, pp. 233–240.
- Delcomyn F. (1998) Foundations of neurobiology In: Diaz H. (ed.) Foundations of Neurobiology. W.H. Freeman and Company, New York, pp. 436-437. [Google Scholar]
- Dzamko N. et al. (2014) Parkinson’s disease-implicated kinases in the brain; insights into disease pathogenesis. Front. Mol. Neurosci., 7, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fano R. (1961) Transmission of Information: A Statistical Theory of Communications. MIT Press, Cambridge, MA. [Google Scholar]
- Fraga L. et al. (2014) p53 signaling pathway polymorphisms associated to recurrent pregnancy loss. Mol. Biol. Rep., 41, 1871–1877. [DOI] [PubMed] [Google Scholar]
- Friedman J.H., Meulman J.J. (2003) Multiple additive regression trees with application in epidemiology. Stat. Med., 22, 1365–1383. [DOI] [PubMed] [Google Scholar]
- Goh K.I. et al. (2007) The human disease network. Proc. Natl. Acad. Sci. USA, 104, 8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S.M. (2014) Environmental toxins and Parkinson’s disease. Annu. Rev. Pharmacol. Toxicol., 54, 141–164. [DOI] [PubMed] [Google Scholar]
- Hamosh A. et al. (2002) Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res., 30, 52–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höglinger G. et al. (2007) The pRb/E2F cell-cycle pathway mediates cell death in Parkinson’s disease. Proc. Natl. Acad. Sci. USA, 104, 3585–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H. et al. (2010) THAP1 mutations (DYT6) are an additional cause of early-onset dystonia. Neurology, 74, 846–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S. et al. (2000) Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer-prone chronic inflammatory disease. Cancer Res., 60, 3333–3337. [PubMed] [Google Scholar]
- Klopstock T. et al. (2005) Mutations in the pantothenate kinase gene PANK2 are not associated with Parkinson disease. Neurosci. Lett., 379, 195–198. [DOI] [PubMed] [Google Scholar]
- Köhler S. et al. (2008) Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet., 82, 949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J. et al. (2001) Novel mutation in the TOR1A (DYT1) gene in atypical early onset dystonia and polymorphisms in dystonia and early onset parkinsonism. Neurogenetics, 3, 133–143. [DOI] [PubMed] [Google Scholar]
- Lewis D. et al. (1994) Immunohistochemical expression of P53 tumor suppressor gene protein in adult germ cell testis tumors: clinical correlation in stage I disease. J. Urol., 152, 418–423. [DOI] [PubMed] [Google Scholar]
- Li Y., Patra J. (2010) Genome-wide inferring gene-phenotype relationship by walking on the heterogeneous network. Bioinformatics, 26, 1219–1224. [DOI] [PubMed] [Google Scholar]
- Lu H., Skolnick J. (2001) A distance-dependent atomic knowledge-based potential for improved protein structure selection. Proteins, 44, 223–232. [DOI] [PubMed] [Google Scholar]
- Manolio T. et al. (2009) Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menche J. et al. (2015) Uncovering disease-disease relationships through the incomplete interactome. Science, 347, 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke D. et al. (2000) Lymphomas in patients with connective tissue disease. Comparison of p53 protein expression and latent EBV infection in patients immunosuppressed and not immunosuppressed with methotrexate. Am. J. Clin. Pathol., 113, 212–218. [DOI] [PubMed] [Google Scholar]
- Morton N. (1955) Sequential tests for the detection of linkage. Am. J. Hum. Genet., 7, 277–318. [PMC free article] [PubMed] [Google Scholar]
- Mottaz A. et al. (2008) Mapping proteins to disease terminologies: from UniProt to MeSH. BMC Bioinformatics, 9 (Suppl. 5), S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan N., Dhillon I.S. (2014) Inductive matrix completion for predicting gene–disease associations. Bioinformatics, 30, i60–i68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogutu J. et al. (2011) A comparison of random forests, boosting and support vector machines for genomic selection. BMC Proc., 5 (Suppl. 3), S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro R., Di Cunto F. (2012) Computational approaches to disease-gene prediction: rationale, classification and successes. FEBS J., 279, 678–696. [DOI] [PubMed] [Google Scholar]
- Qi Z. et al. (2014) Rotenone and paraquat perturb dopamine metabolism: a computational analysis of pesticide toxicity. Toxicology, 315, 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y. et al. (2014) Identifying disease associated genes by network propagation. BMC Syst. Biol., 8 (suppl. 1), S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffel C. et al. (1993) Absence of p53 mutations in childhood central nervous system primitive neuroectodermal tumors. Neurosurgery, 33, 301–305. [DOI] [PubMed] [Google Scholar]
- Ramos E. et al. (2014) Phenotype-genotype integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur. J. Hum. Genet., 22, 144–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Lander E. (2001) On the allelic spectrum of human disease. Trends Genet., 17, 502–510. [DOI] [PubMed] [Google Scholar]
- Roe B.P. et al. (2006) Boosted decision trees, a powerful event classifier In: Lyons L., Unel M.K. (eds) Statistical Problems in Particle Physics, Astrophysics and Cosmology, Imperial College Press, London, p. 139. [Google Scholar]
- Schaefer M.H. et al. (2012) HIPPIE: integrating protein interaction networks with experiment based quality scores. PLoS One, 7, e31826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte E. et al. (2015) Rare variants in β-amyloid precursor protein (APP) and Parkinson’s disease. Eur. J. Hum. Genet., 23, 1328–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Blom U.M. et al. (2013) Prediction and validation of gene-disease associations using methods inspired by social network analyses. PLoS One, 8, e58977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq L. et al. (2008) Advanced microarray analysis highlights modified neuro-immune signaling in nucleated blood cells from Parkinson’s disease patients. J. Neuroimmunol., 201, 227–236. [DOI] [PubMed] [Google Scholar]
- Ta M. et al. (2010) Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med., 363, 166–176. [DOI] [PubMed] [Google Scholar]
- Thusberg J. et al. (2011) Performance of mutation pathogenicity prediction methods on missense variants. Hum. Mutat., 32, 356–368. [DOI] [PubMed] [Google Scholar]
- Trinh J. et al. (2013) STX6 rs1411478 is not associated with increased risk of Parkinson’s disease. Parkinsonism Relat. Disord., 19, 563–565. [DOI] [PubMed] [Google Scholar]
- van Driel M. et al. (2006) A text-mining analysis of the human phenome. Eur. J. Hum. Genet., 14, 535–542. [DOI] [PubMed] [Google Scholar]
- Vanunu O. et al. (2010) Associating genes and protein complexes with disease via network propagation. PLOS Comput. Biol., 6, e1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins B.J. et al. (2013) p53-mediated biliary defects caused by knockdown of cirh1a, the Zebrafish homolog of the gene responsible for North American Indian childhood cirrhosis. PLoS One, 8, e77670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe C. et al. (2005) Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinformatics, 6, 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zatloukal K. et al. (2002) p62 Is a common component of cytoplasmic inclusions in protein aggregation diseases. Am. J. Pathol., 160, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. (2010) Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med. Genomics, 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. (2015) Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Scientific Rep., 5, 11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Zhou Y. (2002) Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci., 11, 2714–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]