Abstract
Summary: We introduce HiPub, a seamless Chrome browser plug-in that automatically recognizes, annotates and translates biomedical entities from texts into networks for knowledge discovery. Using a combination of two different named-entity recognition resources, HiPub can recognize genes, proteins, diseases, drugs, mutations and cell lines in texts, and achieve high precision and recall. HiPub extracts biomedical entity-relationships from texts to construct context-specific networks, and integrates existing network data from external databases for knowledge discovery. It allows users to add additional entities from related articles, as well as user-defined entities for discovering new and unexpected entity-relationships. HiPub provides functional enrichment analysis on the biomedical entity network, and link-outs to external resources to assist users in learning new entities and relations.
Availability and Implementation: HiPub and detailed user guide are available at http://hipub.korea.ac.kr.
Contact: kangj@korea.ac.kr, aikchoon.tan@ucdenver.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Many of the biomedical research papers are about the interactions between genes, proteins or drugs in a specific biological context. New hypotheses could be formulated by mining these papers and designing new sets of experiments to validate (or refute) hypotheses in the knowledge discovery process. However, the volume and rate of current biomedical publications are far larger and more rapid than our ability to extract and use them in this knowledge discovery cycle. For example, PubMed and PubMed Central (PMC) currently have more than 26 million abstracts and 4 million full-text articles, respectively, in their collections, and PubMed receives more than 3000 new abstracts per day. Hence, it is impossible for researchers to manually go through all the records to extract relevant information for their research.
To address these challenges, we developed HiPub, a Chrome browser plug-in that automatically highlights, annotates and translates biomedical entities (e.g. genes, proteins, drugs and diseases) from texts into networks for knowledge discovery. Compared with related tools, HiPub has the following unique features and advantages (Supplementary Table S1):
HiPub works as a seamless Chrome extension in PubMed/PMC websites for knowledge discovery and network exploration.
HiPub recognizes, highlights and annotates an array of biomedical entities such as genes, diseases, drugs, cell lines and mutations in PubMed (abstracts) and PMC (abstracts and full texts).
HiPub integrates two complementary named-entity recognition (NER) methods to achieve high precision and recall: PubTator (Wei et al., 2013) and BEST entity extractor (BEST EE) (Lee, S. et al., 2016)
HiPub visualizes texts as interactive biomedical entity networks using information from texts and external resources.
HiPub integrates biomedical entities in similar articles into the biomedical entity network.
HiPub provides an interactive User Interface for network exploration, enrichment analyses and link-outs to external databases.
2 HiPub description
The HiPub system consists of two components: the Chrome browser extension and the Biomedical NER server. HiPub is designed for PubMed (abstracts) and PMC (abstracts and full texts), and supports several journal websites. After HiPub is invoked, it recognizes the URL, and the document ID (such as PMID or PMCID) is submitted to the HiPub server for processing the document on the fly.
2.1 HiPub chrome extension
2.1.1 Biomedical entity recognition and annotation function
HiPub recognizes and highlights the following biomedical entities from texts: genes, proteins, drugs, diseases, cell lines and mutations. When a highlighted entity is selected and clicked, a pop-up window will appear with basic information about the entity and with links to various external databases for additional information. Basic information about entities was collected from NCBI Gene, MeSH and Cellosaurus (http://web.expasy.org/cellosaurus). All other detailed information about articles and functions is accessible in a HiPub window which automatically appears on the bottom right corner of the web browser.
2.1.2 Biomedical entity network
HiPub generates biomedical entity networks based on two approaches: (1) using the co-occurrence of entities in an opened article; and (2) utilizing the evidence of entity-relationships from external databases. For the co-occurrence approach, HiPub extracts the relations between entities in a text as these relations represents novel results reported in the publication. To add known relations to the publication-specific entity network, HiPub utilizes BEReX (Jeon et al., 2014) which consolidates entity-relationships from multiple data sources. By doing so, HiPub summarizes biomedical entities and their relationships within a specific context (from publication) and existing knowledge (from external databases).
2.1.3 Augmenting biomedical entity network with entities from similar articles
To explore the relationships between biomedical entities in related articles, HiPub utilizes the PubMed API to retrieve 10 articles that are the most similar to a specific paper. Using the ‘Add Similar Articles’ tab in the HiPub window, users can read and add entities to the biomedical entity network. This function allows users to expand the biomedical entity network, and use other related studies for discovering new or unexpected interactions between entities.
2.1.4 Augmenting biomedical entity network with user-defined entities
Similarly, user-defined entities can be easily added to the entity network utilizing BEReX. When a new user-defined entity is added, the shortest paths between existing entities in the network and the newly added entity will be automatically displayed, and users can also further explore the network by linking-out to the BEReX web service.
2.1.5 Detailed query for external databases
Users can send a detailed query about the entities recognized by HiPub to several external databases. HiPub provides three categories for querying a set of entities in external databases: literature (PubMed and Google Scholar), enrichment (BEST and g:Profiler) (Reimand et al., 2007) and network (KEGG, STRING and BEReX).
2.1.6 Functional enrichment analysis of entities
For genes and proteins recognized in a text, HiPub performs enrichment analysis to find the most related KEGG pathway and Gene Ontology using NCBI Biosystems (Geer et al., 2010).
2.2 Biomedical named entity recognition server
To achieve high precision and recall for named entity recognition (NER), HiPub integrates two complementary NER tools: PubTator (Wei et al., 2013) and BEST entity extractor (BEST EE) (Lee,S. et al., 2016). PubTator is a machine learning-based NER tool that achieves high recall whereas BEST EE is a dictionary-based NER tool that is fast and highly precise. Both NER tools support protein/gene, drug and disease name recognition. PubTator recognizes mutation and species information, and BEST EE recognizes cell line names. Integrating the results of both tools enables HiPub to recognize a broad range of entities. We used BEST EE for HiPub NER and combined the result with PubTator data that was obtained using PubTator’s RESTful API (Wei et al., 2016) (Supplementary Figure S1). Since PubTator provides NER data of only PubMed abstracts, it is unable to recognize entities in only non-abstract parts; however, BEST EE can recognize such entities.
To evaluate the performance of HiPub NER, we tested it on the manually curated abstracts from BRONCO (Lee,K. et al., 2016), and compared it with PubTator and TPX-plus (Joseph et al., 2015). The evaluation results of the NER methods are shown in Supplementary Table S2. HiPub achieves high precision, recall and F1-score as compared to PubTator and TPX-plus in this corpus. More details are available in Supplementary Data (Supplementary Figure S2). See Supplementary Results for details.
2.3 Implementation and installation
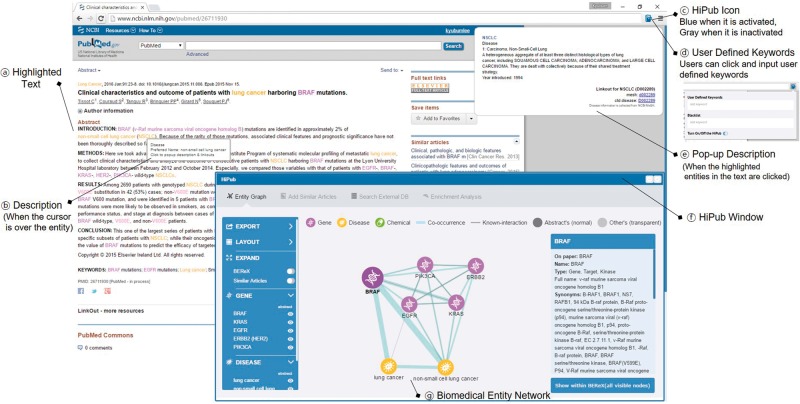
HiPub is implemented as a Chrome extension with JavaScript and is available at the Chrome Web Store. Cytoscape.js is used for network visualization and exploration. The HiPub server is implemented using Java and SPRING I/O. It stores all the PubMed abstracts in local storage for rapid and efficient text processing, with daily updates for newly published articles The HiPub project page contains the user manual, installation link and other related information (http://hipub.korea.ac.kr). Figure 1 illustrates a screen shot of HiPub with some features highlighted on an abstract. See User Manual for more details.
Fig. 1.
Overview of the HiPub application. (a) HiPub annotates and highlights biomedical entities in texts. (b) HiPub will display basic information of a highlighted entity when the cursor moves over it. (c) HiPub icon for turning on/off this application in the Chrome browser. (d) Users can click the icon to highlight and add user-defined entities to the biomedical entity network. (e) HiPub will display detailed information of a highlighted entity in a pop-up window when the entity is clicked. (f) HiPub window visualizes recognized entities in texts as a network, allows users to add entities from similar articles, provides search query links for external databases, and performs enrichment analysis for GO terms/KEGG pathways. (g) The biomedical entity network visualizes the relations between entities in a text. The network integrates context-specific entity-relationships extracted from the text and known relationships from external databases for discovering knowledge. Light blue and grey edges represent context-specific and known entity-relationships, respectively. Width of light blue edges represents the frequency of co-occurrence between two entities in the text. The darker grey edge represents the number of evidences supported by external sources
3 Use case
The research article PMID 23684607 (Andrysik et al., 2013) describes a functional genetic screen that identifies key genes and regulators in driving cells toward p53-dependent apoptosis (via BBC3 gene/PUMA protein) and cell cycle arrest (via CDKN1A gene/p21 protein). From the screen, the following two new genes that are also key were identified: (1) TCF3 which is a transcription factor that drives CDKN1A expression and suppresses BBC3 expression across multiple cancer cell types; (2) TRIAP1 which is a specific repressor of CDKN1A. Five genes (TP53, BBC3, CDKN1A, TCF3 and TRIAP1) were mentioned in the abstract, and these were correctly recognized, annotated and highlighted by HiPub (Supplementary Figure S3). However, in the result of PubTator, some of the genes were wrongly annotated (e.g. PUMA is annotated as a species, not a protein) (Supplementary Figure S4). Next, HiPub visualized the interactions of these genes as a network by integrating co-occurrence relationships (light blue edges, extracted from the abstract) and existing knowledge (grey edges, obtained from external databases). The novel interactions between TCF3 and CDKN1A, TCF3 and BBC3, TRIAP1 and CDKN1A were identified from the study. The entity network, which is the basic summary of the abstract (context-specific), faithfully captures these interactions (Supplementary Figure S5). In contrast, the existing databases (e.g. STRING) do not contain all the interactions (Supplementary Figure S6). Therefore, HiPub can be used as a tool to assist users in extracting important entity relationships from both texts and external databases, and integrating relations from related articles for knowledge discovery. See Supplementary Data for details and additional use case examples.
In summary, we have developed HiPub, a novel web browser plug-in that automatic recognizes, annotates and translates PubMed abstracts and PMC full-text articles into biomedical entity networks for knowledge discovery.
Supplementary Material
Acknowledgements
We thank the Kang lab and Tan lab members for constructive comments of this research. We thank Susan Kim for suggestions and editing of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea. NRF-2014M3C9A3063543 (to J.K.), NRF-2014R1A2A1A10051238 (to J.K.) and NRF-2015M3A9D7031070 (to J.K.), the National Institutes of Health P50CA058187 (to A.C.T.), P30CA046934 (to A.C.T.) and the David F. and Margaret T. Grohne Family Foundation (to A.C.T.).
Conflict of Interest: none declared.
References
- Andrysik Z. et al. (2013) A genetic screen identifies TCF3/E2A and TRIAP1 as pathway-specific regulators of the cellular response to p53 activation. Cell Rep., 3, 1346–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer L.Y. et al. (2010) The NCBI BioSystems database. Nucleic Acids Res., 38, D492–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon M. et al. (2014) BEReX: Biomedical Entity-Relationship eXplorer. Bioinformatics, 30, 135–136. [DOI] [PubMed] [Google Scholar]
- Joseph T. et al. (2015) Plugin for concept-assisted search and navigation on PUBMED. In: Proceedings of the 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Washington, DC, 9–12 November, pp. 1712–1714.
- Lee K. et al. (2016) BRONCO: biomedical entity Relation ONcology COrpus for extracting gene-variant-disease-drug relations. Database, 2016, baw043.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. et al. (2016) BEST: next-generation biomedical entity search tool for knowledge discovery from biomedical literature. PLoS ONE, In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J. et al. (2007) g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res., 35, W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.H. et al. (2013) PubTator: a web-based text mining tool for assisting biocuration. Nucleic Acids Res., 41, W518–W522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.H. et al. (2016) Beyond accuracy: Creating interoperable and scalable text min-ing web services. Bioinformatics, 32, 1907–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.