Abstract
The major histocompatibility complex (MHC) has genetic associations with many diseases, often due to differences in presentation of antigenic peptides by polymorphic MHC molecules to T lymphocytes of the immune system. In chickens, only a single classical class I molecule in each MHC haplotype is expressed well due to co-evolution with the polymorphic transporters associated with antigen presentation (TAPs), which means that resistance and susceptibility to infectious pathogens are particularly easy to observe. Previously, structures of chicken MHC class I molecule BF2*2101 from B21 haplotype showed an unusually large peptide-binding groove that accommodates a broad spectrum of peptides to present as epitopes to cytotoxic T lymphocytes (CTL), explaining the MHC-determined resistance of B21 chickens to Marek's disease. Here, we report the crystal structure of BF2*0401 from the B4 (also known as B13) haplotype, showing a highly positively-charged surface hitherto unobserved in other MHC molecules, as well as a remarkably narrow groove due to the allele-specific residues with bulky side chains. Together, these properties limit the number of epitope peptides that can bind this class I molecule. However, peptide-binding assays show that in vitro BF2*0401 can bind a wider variety of peptides than are found on the surface of B4 cells. Thus, a combination of the specificities of the polymorphic TAP transporter and the MHC results in a very limited set of BF2*0401 peptides with negatively charged anchors to be presented to T lymphocytes.
Keywords: disease, susceptibility/chicken, MHC, BF2*0401/crystal, structure/peptide-binding, groove/TAP
Introduction
The major histocompatibility complex (MHC) is the genetic region with the most associations with disease in humans. Much of this preponderance of genetic association is due to the classical MHC molecules, which bind proteolytic fragments of proteins and present these peptides (called epitopes) to T lymphocytes of the immune system(1, 2). The classical MHC molecules have high levels of allelic polymorphism and sequence diversity, thought to be driven mostly by a molecular arms race with pathogens(3). Most of the sequence diversity is located in and around the peptide binding site, which allows each allele to present different peptides(4–7).
Nearly all of the detailed work leading to this description of the MHC, MHC molecules and antigen presentation has been developed from experiments in humans and mice. However, the enormous global poultry industry has keen interests in the genetics and mechanisms of responses to pathogens and vaccines, and much evidence for MHC determination of resistance and susceptibility to pathogens is from work with chickens. Over decades of experimentation, it has been found that some MHC haplotypes (including B21) generally confer resistance to a variety of infectious viral diseases, while other haplotypes (such as B4, also known as B13) often confer susceptibility(8–11).
Compared to mammals, the region containing the classical MHC molecules in chickens (variously called “BF-BL region”, “classical MHC” or “core MHC”) is simple and compact, with only a single classical class I molecule and a single class II molecule expressed at high levels(12–18). The chicken MHC is also organised differently than the MHC of typical mammals. In particular, the genes for the transporter associated with antigen presentation (TAP), a peptide transporter that pumps peptides from the cytoplasm to the lumen of the endoplasmic reticulum for loading onto class I molecules, are polymorphic and very closely linked with the class I genes, with co-evolution resulting in a single dominantly-expressed class I molecule(19). The “minimal essential MHC” hypothesis proposes that the properties of the single dominantly-expressed class I and class II molecules are responsible for the strong genetic associations with pathogen and vaccine responses, in comparison to typical mammals with a multigene family of MHC molecules(13, 14). Moreover, evidence suggests that many if not most non-mammalian vertebrates have the salient properties discovered in chickens(20).
Peptide motifs for the classical class I molecules from chicken blood cells of a few chicken MHC haplotypes have been determined, and can explain the resistance and susceptibility of chickens to economically-important viral diseases such as Rous sarcoma virus (RSV) and Marek’s disease virus (MDV)(14, 15, 18). These chicken peptide motifs look very different from those reported for humans, mice and rats, since most mammalian motifs are nonamer peptides with two so-called “anchor residues” at which only a few related amino acids are used to bind into deep pockets of the class I molecule(4–7).
The peptides from chicken BF2*2101 on blood cells (BF2 is the gene name; 2101 is the allele) are mostly 10mers and 11mers, with co-variation of anchor (and other) residues such that an astonishing variety of peptides will bind. The structure of BF2*2101(15), the only one for a classical class I molecule outside of mammals, shows that it is the interplay of arginine at MHC position 9 (R9, with R being the single letter amino acid code for arginine) and aspartic acid at MHC position 24 (D24) within a very wide binding groove that allows the BF2*2101 molecule to remodel its binding site to accommodate peptides with completely different sequences.
The peptides from all other chicken MHC molecules on blood cells(14, 18) are almost exclusively octamers, and most have only one or two closely-related amino acids specified in each of three anchor positions. BF2*0401 is one such molecule, with a peptide motif of aspartic acid (D) or glutamic acid (E) at peptide position 2, D or E at position 5 and E (with a low level of hydrophobic amino acids) at position 8 (that is, DE2-DE5-E8). This motif is quite fastidious, and in fact the B4 (also called B13) haplotype in the BF-BL region is often associated with susceptibility to particular pathogens, presumably because no protective peptide from the pathogen can be presented(14, 18). In this paper, the crystal structure of BF2*0401 is reported, and shown to explain much of the specificity of the molecule. However, peptide binding studies show that BF2*0401 can bind a wider variety of peptides than is found on the surface of cells. This observation, in conjunction with previous work on TAP specificities(19), shows that chicken TAP molecules can restrict the peptides actually presented to the immune system.
Materials and Methods
Peptide synthesis
The octamer IE8 from FUS proto-onc gene(18) and twenty-two peptides with single, double or triple substitutions (Table II) were synthesised using an Apex 396 peptide synthesizer and purified by HPLC reverse phase chromatography (SciLight Biotechnology).
Table II. Binding assays of BF2*0401 with IE8 and its mutants.
| parent peptide | IE8: DDE +++ | ||
| single substitution peptides | P2E: EDE +++ | P2A: ADE + | |
| P5E: DEE +++ | P5A: DAE +++ | ||
| P8D: DDD +++ | P8A: DDA ++ | ||
| double substitution peptides | P2E5E: EEE ++ | P2A5E: AEE - | P2E5A: EAE - |
| P2E8D: EDD + | P2A8D: ADD - | P2E8A: EDA - | |
| P5E8D: DED + | P5A8D: DAD - | P5E8A: DEA - | |
| triple sub peptide | P2E5E8D: EED - | P2A5E8D: AED - | |
| P2E5A8D: EAD - | P2E5E8A: EEA - | ||
| other sub peptide | P5F: DFE - | P5Y: DYE - | P5R: DRE - |
In the in vitro refolding assay, if an elution peak representing the peptide-BF2*0401 complex appears in gel filtration, the peptide is defined as being able to bind to BF2*0401, with “+++” indicating that the refolding efficiency was comparable to that of wild type IE8 and the complex was stably eluted under anion-exchange conditions, “++” indicating that the refolding efficiency was about 50%-60% of IE8 and the complex partially dissociated under anion-exchange conditions, “+” indicating that the refolding efficiency was much lower than IE8 and the complex was not stable under anion-exchange conditions, and “–” indicating no refolded product.
Protein preparation
DNA fragments encoding extracellular domains of BF2*0401 (Genbank: AM282693, residues 1-270 of the mature protein with restriction sites EcoR I and Hind III) and chicken β2m (Genbank: AB178590, residues 1-98 of the mature protein) were cloned into pET21a(+) vectors and expressed in E. coli BL21 (DE3) pLysS (Novagen). The BF2*0401 and β2m inclusion bodies were prepared essentially as previously described by Garboczi et al. with minor modifications(21–23). After refolding with IE8 (and substituted peptides), the monomers were purified using a Superdex 200 16/60 column, followed by Resource Q anion-exchange chromatography (GE Healthcare). Purified proteins were buffer-exchanged with 10 mM Tris-HCl, 10 mM NaCl pH 8.0.
Crystallization and data collection
Mixed with reservoir buffer at a 1:1 ratio, the purified proteins were screened for crystallization trials using the hanging-drop vapour-diffusion method with Crystal Screens I and II (Hampton Research) at 291 K. Crystals (BF2*0401-IE8, BF2*0401-P8D and BF2*0401-P5E) from protein concentration 10 mg/mL were observed in 0.1 M HEPES buffer (pH 7.0, 5% MPD and 20% PEG6000). Data collection was performed on a Rigaku MicroMax007 rotating-anode X-ray generator operated at 40kV and 20 mA (Cu kα; λ = 1.5418 Å) equipped with an R-AXIS VII++ image-plate detector. Diffraction data collected to 2.26 Å was indexed and scaled using HKL2000(24).
Structure determination and refinement
The crystal of BF2*0401-IE8 belongs to the C2 space group with unit cell constants: a = 166.20 Å, b = 40.29 Å, C = 131.59 Å, α = γ = 90.00°, β = 119.71° (Table I). The structure was determined by molecular replacement using Molrep and Phaser in the CCP4 package, with BF2*2101-11mer structure (PDB code: 3BEV, with the peptide excluded) as the search model(25–27). Residues that differ between BF2*0401 and the search model were manually rebuilt in the program Coot under the guidance of 2Fo-Fc and annealed omit maps, varying with the reciprocal space refinement in Refmac5(27–29). After adding peptide, subsequent refinements were conducted for energy minimisation, individual B factor restriction, and water molecule addition, with non-crystallographic symmetry (NCS) restraints applied to the two copies of BF2*0401-IE8/P5E/P8D in the asymmetric unit. Ramachandran plot and secondary structure assignments were generated by SFCHECK(30). The final structure composes of two BF2*0401-IE8 complexes, with Rfactor = 19.7% and Rfree = 25.5%, and the two copies both contained residues 1-270 of BF2*0401 and 1-97 of β2m. The structures of mutant peptides P8D and P5E were determined using BF2*0401-IE8 as the model (Table I). The atomic coordinates of the crystal structures of BF2*0401-IE8/P8D/P5E have been deposited in the Protein Data Bank (http://www.pdb.org/pdb/home/home.do) with accession no. 4E0R (http://www.rcsb.org/pdb/search/structidSearch.do?structureId=4E0R), 4G42 (http://www.pdb.org/pdb/search/structidSearch.do?structureId=4G42) and 4G43 (http://www.pdb.org/pdb/search/structidSearch.do?structureId=4G43), respectively.
Table I. Data collection and refinement statistics.
| BF2*0401-IE8 | BF2*0401-P8D | BF2*0401-P5E | |
|---|---|---|---|
| Data collection | |||
| Space group | C2 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 166.20, 40.29, 131.59 | 165.83, 40.11, 130.95 | 166.92, 40.24, 132.62 |
| α, β, γ (°) | 90.00, 119.71, 90.00 | 90.00, 119.56, 90.00 | 90.00, 120.35, 90.00 |
| Resolution (Å) | 50.00 - 2.26 (2.34 - 2.26) | 50.00-2.30 (2.38-2.30) | 50.00-1.80 (1.86-1.80) |
| Rmerge | 0.055 (0.196) | 0.118 (0.485) | 0.008 (0.563) |
| I / σI | 19.6 (6.8) | 13.3 (3.0) | 19.9 (4.8) |
| Completeness (%) | 96.6 (94.0) | 98.8 (97.6) | 99.6 (98.7) |
| Redundancy | 3.1 (3.1) | 3.6 (3.7) | 4.3 (4.2) |
| Refinement | |||
| Resolution (Å) | 43.83 - 2.26 | 40.63-2.30 | 41.31-1.80 |
| No. reflections | 34927 | 33771 | 69202 |
| Rwork / Rfree | 0.197/0.255 | 0.196/0.254 | 0.183/0.218 |
| No. atoms | |||
| Protein | 6118 | 6108 | 6107 |
| Ligand/ion | 0 | 0 | 8 |
| Water | 517 | 541 | 606 |
| B-factors | |||
| Protein | 27.8 | 28.0 | 22.4 |
| Ligand/ion | 0 | 0 | 16.4 |
| Water | 31.9 | 31.7 | 29.6 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.003 | 0.005 | 0.005 |
| Bond angles (°) | 0.771 | 0.942 | 0.975 |
Values in parentheses are for highest-resolution shell.
Structural analysis and figure generation
Peptide-contacting residues were identified using the program CONTACT and were defined as residues containing an atom within 3.3 Å of the target partner(25). Structural figures and electrostatic potential surfaces were generated using the PyMOL molecular graphics system (DeLano Scientific, http://www.pymol.org).
Determination of complex thermostability using CD spectroscopy
The thermostabilities of BF2*0401 with seven mutant peptides were tested by circular dichroism (CD) spectroscopy. CD spectra were measured at 20°C on a Chirascan spectropolarimeter (Applied Photophysics) equipped with a water-circulating cell holder. The protein concentration was 7 µM in pH 8.0 Tris buffer (20 mM Tris and 50 mM NaCl). Thermal denaturation curves were determined by monitoring the CD value at 218 nm by using a 1-mm-optical-path-length cell as the temperature was raised from 25°C to 94°C at a rate of 1°C/min. The temperature of the sample solution was directly measured with a thermistor. The fraction of unfolded protein was calculated from the mean residue ellipticity (θ) by the standard method. The unfolded fraction (%) is expressed as (θ-θN)/(θU-θN), where θN and θU are the mean residue ellipticity values in the fully folded and fully unfolded states. The midpoint transition temperature (Tm) was determined by fitting data to the denaturation curves using the Origin 8.0 program (OriginLab).
Results
Overall structure of chicken BF2*0401 and its comparison with other MHC molecules
The crystal structure of BF2*0401 bound to peptide IE8 (IDWFDGKE from ref. 18) was solved by molecular replacement using the BF2*2101 structure (PDB code: 3BEV) as a search model, with data collection and refinement statistics of the crystal structure listed in Table I. There are two BF2*0401-IE8 molecules per asymmetric unit. The overall structure is similar to that of the previously determined BF2*2101. Briefly, the α1 and α2 domains together form the binding groove, with two α-helices atop an eight-stranded β-sheet. The α3 domain and the non-covalently associated light chain β2-microglobulin (β2m) beneath the α1/α2 platform have immunoglobulin-like (Ig-like) folds and consist primarily of anti-parallel β-sheets. The IE8 peptide binds to the groove at the top of the BF2*0401 molecule in a position to interact with the T cell receptor (Fig. 1A).
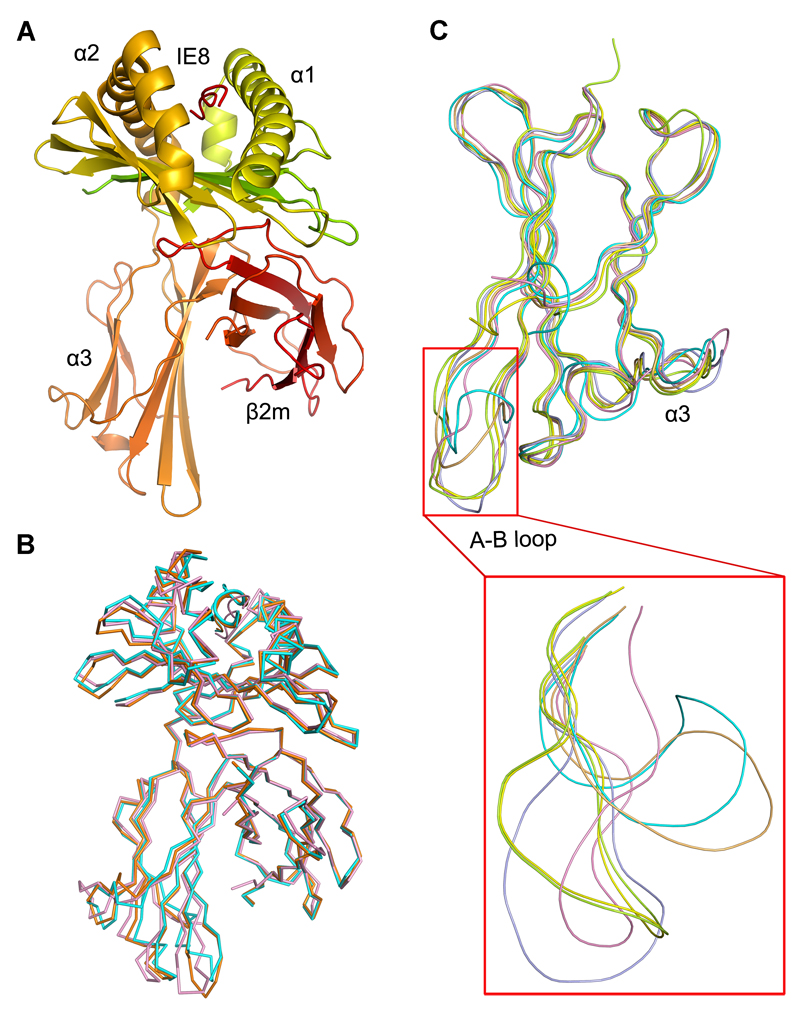
Figure 1. The structure of the chicken MHC class I molecule BF2*0401.
(A) The overall structure of chicken BF2*0401, a typical class I MHC structure. (B) Superimposed Cα-traces of chicken BF2*0401-IE8 (pink), BF2*2101-10mer (PDB code: 3BEW, orange) and BF2*2101-11mer (PDB code: 3BEV, cyan) with the peptides. It shows that the overall structures of the two chicken class I MHC are very similar. (C) The Cα-trace comparison of the α3 domains of BF2*0401-IE8 (pink), BF2*2101-10mer (orange), BF2*2101-11mer (cyan), HLA-A*0201 (PDB code: 1HHK, limon), H-2Kb (PDB code: 1G7Q, blue) and Mamu-A*01 (PDB code: 1ZVS, yellow), with the A-B loop zoomed in the red box. BF2*0401-IE8 structure does not show a different orientation of the A-B loop as observed in that of BF2*2101, rather similar to other known mammalian structures.
Superposition demonstrates that the BF2*0401 structure closely agrees with the BF2*2101 (3BEV with one structure, 3BEW with two structures), with root-mean-square deviation (r.m.s.d.) between Cα positions of α1/α2 domains of 0.727 Å, and an identical orientation of the β2m domains (Fig. 1B). However, the A-B loop of the α3 domain in BF2*0401 is much like the structure of mammalian class I molecules, unlike the large rotation seen for all three structures of BF2*2101 (Fig. 1C). In any case, the two residues Q222(226) and D223(227) [amino acid positions are with and without parentheses for human and chicken classical class I heavy chains, respectively] predicted to be most involved in binding the co-receptor CD8 are conserved(31–34).
Compared to the chicken BF2*2101 molecule, BF2*0401 has a very narrow groove
Similar to most mammalian classical class I molecules but unlike BF2*2101, BF2*0401 has distinct pockets named A-F (Fig. 2A-2C). At the N-terminus of IE8, the main chain atoms of P1 Ile make strong hydrogen bonds with the side chains of Y7, Y156 and Y168 in A pocket. This network of interactions is highly conserved in BF2*2101 and most mammalian MHC class I molecules. Tyr(84) invariant among mammalian classical class I molecules is replaced with an R83 in pocket F of BF2*0401 (as in all other non-mammalian vertebrates) (Fig. 2F).
Figure 2. The peptide-binding groove of BF2*0401.
Comparison of the peptide-binding grooves between BF2*0401 and BF2*2101, illuminating an extremely narrow groove of BF2*0401 and an unusually large binding groove in BF2*2101. (A-C) Molecular surfaces (grey) of the peptide-binding grooves of BF2*2101-11mer (A), BF2*0401-IE8 (B) and BF2*2101-10mer (C) with the peptides (11mer, IE8 and 10mer coloured in cyan, pink and orange, respectively). The N- (P1) and C- (P11, P8, and P10) termini of the peptides are marked. Pockets in each groove are sequentially labelled A to F. (D and E) The surface of BF2*0401-IE8 binding groove is superimposed onto that of BF2*2101-11mer (D) or BF2*2101-10mer (E). BF2*0401 is shown in pink, while BF2*2101-11mer and BF2*2101-10mer are coloured in cyan and orange, respectively. (F) Structure-based amino acid sequence alignment of the α1-α2 domains of BF2*0401 and BF2*2101, with the secondary structure elements indicated above. The cysteine residues involved in the first disulfide bond are marked by green 1. Conserved residues are high-lightened in red. Residues forming the binding groove are marked by orange stars for BF2*0401 and blue stars for BF2*2101.
Pocket B in many (although not all) mammalian class I molecules is very deep(35–38), reaching under the α1 helix so that the peptide P2 residue touches residue 45, one of the most polymorphic residues of mammalian class I molecules. In contrast (Fig. 2 and Fig. 3C), pocket B in BF2*0401 is relatively shallow with accessibility to the equivalent position Y43(45) mostly blocked off by the T24(24). In BF2*0401, pocket B is formed by T24(24), Y43(45), Q62(63) and I65(66), along with R9(9) which is shared with pocket C. This relatively shallow pocket is similar in BF2*2101, but as discussed below, the boundary of pockets B and C is more rigid in BF2*0401.
Figure 3. IE8 bound to BF2*0401 and the positively charged pockets in the BF2*0401 groove.
(A) Interactions between IE8 and BF2*0401. Side chains of residues involved are labelled and coloured in pink for the BF2*0401 and green for the IE8. Salt bridges and hydrogen bonds are illustrated as dashed lines in teal. (B) The BF2*0401 peptide-binding groove demonstrates an entirely positive charged surface. The electrostatic surface potential was generated with PyMOL. Red is electronegative and blue is electropositive. Yellow arrows represent the pockets holding the anchor residues. (C) The P2 Asp is shown as a stick in green. Residues forming the B pocket in BF2*0401 are shown in pink on the surface. (D) The P5 Asp and residues forming the C pocket in BF2*0401 are shown. (E) The P8 Glu and residues forming the F pocket in BF2*0401 are shown.
The C, D and E pockets in the centre of the groove are far better defined in BF2*0401 than in BF2*2101 (Fig. 2 and Fig. 3D). Pocket C includes N69(70), N73(74), W95(97) and R111(114), along with R9(9). In addition, L68(69), N69(70), F97(99) and R111(114) pointing up from the β-sheet of the α2 domain divide pockets C, D and E into small separated sections. Finally (Fig. 2 and Fig. 3E), pocket F is narrower and shallower in BF2*0401 than either BF2*2101 or other mammalian class I molecules, with the sides lined with N76(77), I79(80), R83(84) and T140(143), and the very bottom occupied mainly by R80(81) with some contribution of F120(123) and I121(124).
Compared to BF2*2101, BF2*0401 has a much narrower groove, particularly after pocket B, toward the carboxyl end of the peptide (Fig. 2A-2E and Fig. 4A, 4B). The nearest and farthest distances between the cavity walls are 7.00 Å and 10.23 Å for BF2*0401-IE8 compared to 12.12 Å and 14.42 Å for BF2*2101-11mer. Indeed, the groove of BF2*0401 seems narrower even in comparison with mammalian class I molecules.
Figure 4. Detailed comparison of the particular amino-acid residues in the peptide-binding grooves between BF2*0401 and BF2*2101.
(A and B) Pockets C, D and E of the peptide-binding grooves in BF2*0401-IE8 (A) and BF2*2101-11mer (B), respectively, are highlighted, showing the size differences, with the amino-acid residues with different side chains labelled on the surfaces. (C) Superposition of the Cα-traces of the α1 and α2 domains in BF2*0401 (red) and BF2*2101 (green), with the α1 helix fixed. (D) Side chains of the amino-acid residues contributing to the binding groove in BF2*0401 (pink) are relatively larger than their counterparts in BF2*2101 (cyan). (E) Interactions of Arg9 with the peptides in BF2*0401-IE8 (pink), BF2*2101-10mer (orange) and BF2*2101-11mer (cyan), showing Arg9 in BF2*0401-IE8 adopts a fixed conformation, unlike in BF2*2101 where the unusually-large cavity makes Arg9 mobile. The peptides are viewed with the α1 helices fixed. Salt bridges formed between Arg9 and peptide are shown as dashed lines in pink for IE8, orange for 10mer and cyan for 11mer. (F) Conformation of Arg9 with constrained space in BF2*0401-IE8 (pink), compared to those in BF2*2101-10mer (orange) and BF2*2101-11mer (cyan). Side chains of residues around each Arg9 are labelled and shown as sticks; clearly shown are bulky side chains in BF2*0401.
The mainchain atoms of the α1/α2 platform of BF2*0401 and BF2*2101 are nearly superimposable (Fig. 4C), so that the narrow groove of BF2*0401 and the wide groove of BF2*2101 are due entirely to the different side chains of amino acids pointing into the groove. In particular, the large residues L68(69) and N69(70) from the α1 helix and R152(155) and W153(156) from the α2 helix of BF2*0401 are replaced by the much smaller G68(69), S69(70), G152(155) and L153(156) in BF2*2101 (Fig. 4A, 4B, 4D). These large overhanging residues along with bulky side chains of F97(99) and R111(114) constrain the position of R9(9) to stand up straight, high and relatively rigid in BF2*0401. In contrast, R9(9) is free to adopt various conformations in wider groove of BF2*2101, allowing the remodelling of the groove to accept peptides of completely different sequences (Fig. 4E, 4F).
A unique positively-charged groove of BF2*0401 binds several negatively-charged anchor residues of the peptide
In typical mammalian classical class I molecules, the peptides are nonamers tightly bound at the N- and C-termini into pockets A and F, respectively, and with extensive interactions of their anchor residues at position 2 and the final position into deep pockets B and F, respectively. The peptide rears up out of the groove after position 2, so that pockets C, D and E are generally shallower and bind so-called secondary anchor residues (4–7).
Some features of the IE8 peptide are similar to the typical pattern in mammals, but some are strikingly different. The N-terminal amino group and C-terminal carboxy group bind to the extensive H-bond networks in pockets A and F. However, the IE8 peptide lies flatter than most peptides in mammalian class I molecules as well as in BF2*2101 (Fig. 4E). Compared to the octamer peptides presented by mammalian MHC molecules, including Kb-SIINFEKL, IE8 is still flatter than these peptides. The IE8 octamer does not rear up out of the groove in BF2*0401 as much as the nonamers typically found in mammals, and far less than the 10mer and 11mer in BF2*2101. The IE8 peptide also does not sink down so far back into the groove at the C-terminal end as in BF2*2101 due to a bulky side chain of R80(81) at the bottom of F pocket. Pockets B, C, D and E are shallow, contributing to the more extended conformation of the bound peptide.
As predicted from the sequence and model(18), the structure of BF2*0401 shows that there are several arginines located in the peptide-binding site (Fig. 3A). Indeed, the whole groove is strongly positively-charged (Fig. 3B), unlike any mammalian class I molecule for which the structure is known (Fig. 5). Three arginines point upwards from the β-sheet of the groove, all of which interact with the peptide anchor residues. R9(9) in pocket B makes two salt bridges with peptide position D2, along with a hydrogen bond from Y43(45) and two hydrogen bonds with the mainchain from Q62(63) (Fig. 3A, 3C). R9(9) and R111(114) in pocket C both make salt bridges with peptide position D5, along with a hydrogen bond with the mainchain from N69(70) (Fig. 3A, 3D). R80(81) in pocket F makes two salt bridges with peptide position E8, along with a hydrogen bond from N76(77) (Fig. 3A, 3E). These interactions are consistent with the negative charges found at positions 2, 5 and 8 in the motif determined for B4 cells. Some B4 peptides (but not IE8) also have a negatively-charged residue at P4, which would interact with R152(155) on the α2 helix. Also, a very few peptides found on B4 cells have a hydrophobic amino acid at the C-terminus, which is compatible with the lining of pocket F by hydrophobic amino acids I79(80), F120(123) and I121(124). Thus the structure of BF2*0401 bound to IE8 would appear to explain the peptide motif found for B4 cells.
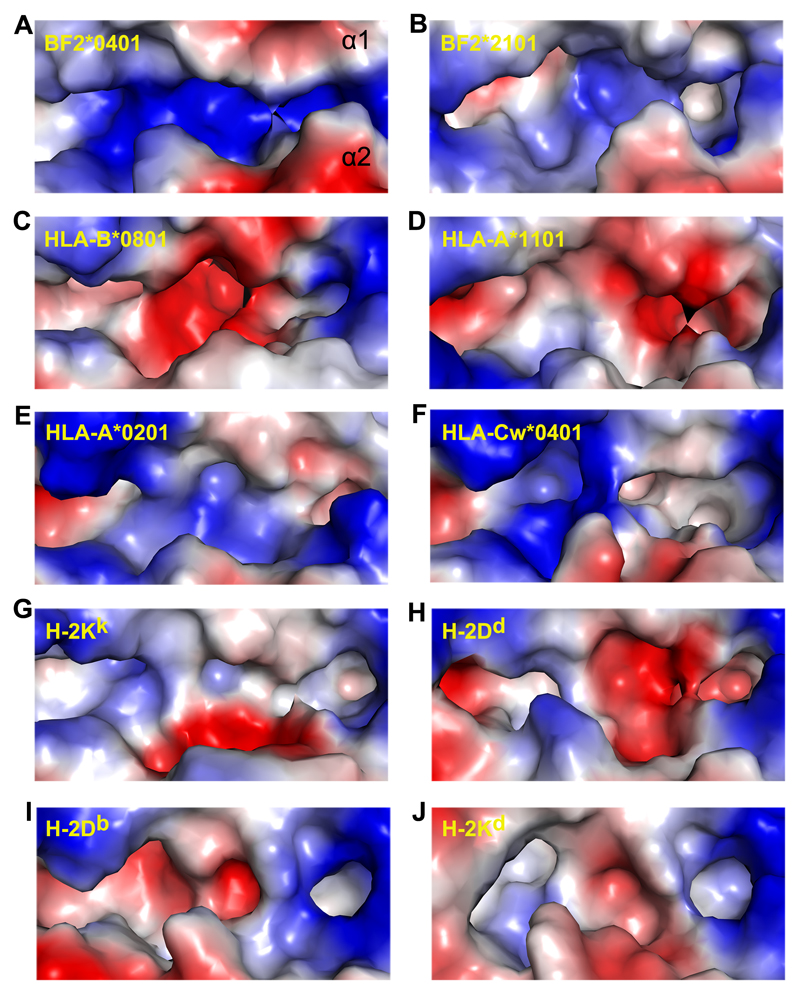
Figure 5. Electrostatic potential surface of the BF2*0401 groove compared to those of other MHC class I structures.
Comparison of the electropotential surface of BF2*0401 (A) with those of BF2*2101 (B), HLA-B*0801 (C; PDB code: 1M05), HLA-A*1101 (D; PDB code: 1QVO), HLA-A*0201 (E; PDB code: 1S9X), HLA-Cw*0401 (F; PDB code: 1IM9), H-2KK (G; PDB code: 1ZT1), H-2Dd (H; PDB code: 1BII), H-2Db (I; PDB code: 1JPG) and H-2Kd (J; PDB code: 1VGK), demonstrating the unique positively charged peptide-binding groove in BF2*0401, which has not been seen in any other known class I MHC structures. The figures were generated with PyMOL. Red is electronegative and blue is electropositive. The grooves are illustrated as α1 helix on the top.
Compared to the peptides found on B4 cells, BF2*0401 binds a wider variety of peptides in vitro
The sequence motif for peptides presented by B4 cells was determined by eluting peptides from BF2*0401 isolated from erythrocytes and spleen cells, and sequencing both individual peptides and pools of peptides(14, 15, 18). All experiments gave the same answer: the peptides were octamers with mostly aspartic acid (and some glutamic acid) at positions 2 and 5, and glutamic acid (and a very low level of some hydrophobic amino acids) at position 8.
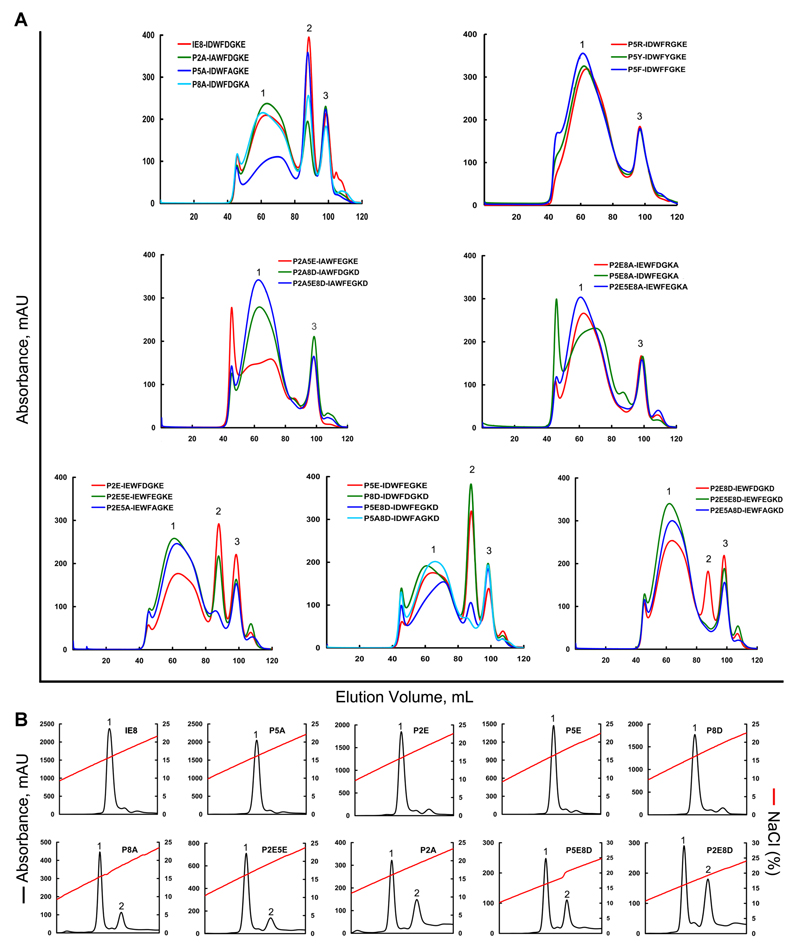
In order to verify that the peptide motif is determined by the binding specificity of the BF2*0401 molecule, renaturation of the heavy chain, β2m and synthetic peptides followed by size exclusion chromatography (SEC, gel filtration) was performed (Table II, Fig. 6). The original peptide IE8 (D2-D5-E8) gave a high yield of refolded material at the size expected for a monomer (that is, heavy chain, β2m and peptide). Single substitutions of negatively-charged residues at the anchor residues (E2-D5-E8, D2-E5-E8, D2-D5-D8) also gave high yields, but double substitutions led to lower yields and the triple substitution (E2-E5-D8) did not yield a monomer peak. Single substitutions of alanine at the anchor residues of IE8 gave high yields for one peptide (D2-A5-E8), but lower yields for the other two peptides (A2-D5-E8, D2-D5-A8) and no monomer peak for any alanine substitutions of the charge substituted peptides. Finally, substitution at peptide position 5 with large residues (D2-R5-E8, D2-Y5-E8 and D2-F5-E8) resulted in no binding.
Figure 6. Refolding of BF2*0401 and β2m with IE8 and its mutants.
Peptide-induced assembly and stabilization assay of BF2*0401 molecules by in vitro refolding. (A) Gel filtration chromatograms of the refolded products. Peak 1, peak 2 and peak 3 represent the aggregated heavy chain, the correctly refolded BF2*0401 complex (45 kDa) and the extra β2m, respectively. The refolding efficiencies are represented by the relevant concentration ratio and height of the peak 2 for IE8 and each mutant, the more the better. Otherwise, if little or none peak 2 will be observed, the peptide can not be considered to stabilize the complex, thereof treated a non-presenting peptide. The IE8 mutants were listed in Table II. (B) Results of further stabilization assays of the refolded complexes by anion exchange. Under the anion-exchange conditions, complexes of IE8, P5A, P2E, P5E and P8D can be eluted normally at NaCl concentration of 15%-17% (peak 1). With the peptides of P8A, P2E5E, P2A, P5E8D and P2E8D, the refolded complex proteins can be partially dissociated at NaCl concentration of 18%-20% (peak 2), implying less stable.
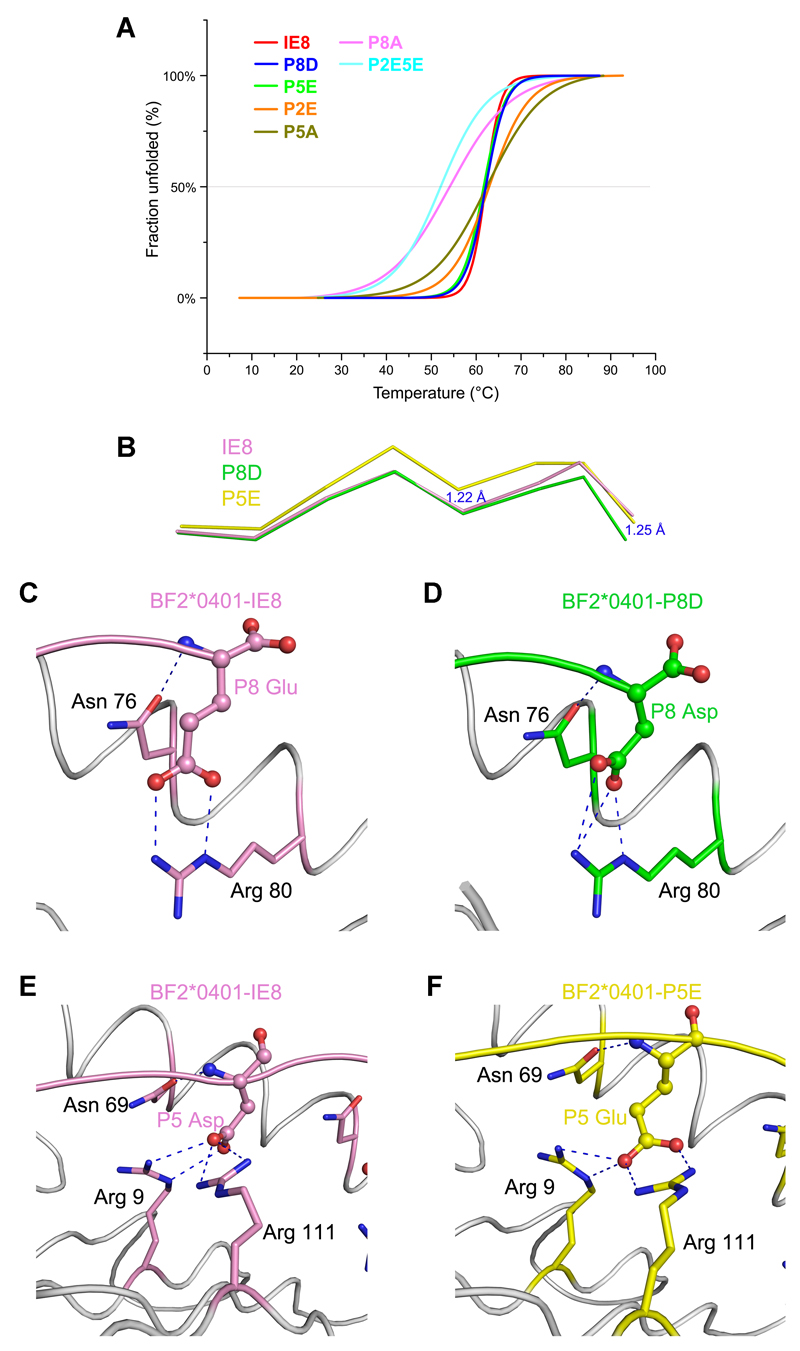
The binding stabilities of peptide IE8 and its substitutions (P2E, P5E, P8D, P5A, P8A and P2E5E) were further analyzed by using CD spectra (Fig. 7A). Tms were determined from melting curves. The complexes with well refolded peptides (IE8, P2E, P5E, P8D and P5A) have similar thermostabilities, with Tms approximate 62°C, meaning these BF2*0401 complexes are very stable. However, the curves of IE8, P5E and P8D are almost overlapping and are steeper than the curves of P2E and P5A (Fig. 7A). The starting unfolded temperatures of these three peptides are nearly 50°C, which are higher than P2E and P5A (40°C and 35°C). The Tms of BF2*0401 complexed with low yield peptides (P8A and P2EP5E) are approximate 54°C and 52°C, respectively. The thermostabilities of BF2*0401 complexes with low yield peptides are lower than those of well refolded peptides. The CD results are in accord with renaturation results, indicating the peptide motif determined by the binding specificity of BF2*0401 is correct.
Figure 7. Thermostabilities of BF2*0401 complexes and their structural bases.
(A) The thermostabilities of BF2*0401 with IE8 peptide and its substitutions (P2E, P5E, P8D, P5A, P8A, P2EP5E) were tested by CD spectroscopy. The temperature was increased by 1°C/min. The curves for the unfolded fractions were determined by monitoring the CD value at 218 nm. The midpoint transition temperatures (Tm) of different peptides are indicated by the grey line at 50% Fraction Unfolded. (B) The structural alignment of the peptides of P8D, P5E and the cognate peptide IE8. The C terminal portion of the peptide P8D descends to the F pocket 1.25Å and the middle part of the peptide P5E rises up 1.22 Å. (C) and (D) Residue P8 Asp in peptide P8D (D, green) forms the similar salt bridge and hydrogen bonds as P8 Glu in the structure of peptide IE8 (C, pink). (E) and (F) P5 Glu in peptide P5E (E, yellow) forms the similar salt bridge and hydrogen bonds as P5 Asp in the structure of peptide IE8 (F, pink).
Thus, the peptide-binding motif of the BF2*0401 molecule is wider than the peptide motif found from B4 cells. Based on the motif which is derived from the in vivo acid eluted peptides, the predicted BF2*0401 binding peptides from MDV are about 3. However, we can predict 11 BF2*0401 binding peptides from MDV based on the results of Table II in this structural study. The fact that large amino acids in position 5 did not support renaturation is consistent with the narrow groove at that position. However, the finding that aspartic acid at position 8 would support renaturation (when one of the other two positions was an aspartic acid) was unexpected. Furthermore, the finding that alanine in any of the three anchor positions would support renaturation was also unexpected. These results argue for some but not total flexibility in the binding site, and suggest that the peptides supplied to the class I molecule are restricted in this MHC haplotype.
To elucidate the different roles of the major anchor residues of IE8 in its binding to BF2*0401, we determined the crystal structures of BF2*0401-P8D and BF2*0401-P5E at 2.3 Å and 1.8 Å, respectively (Table I). The superimposing of the two newly defined structures with BF2*0401-IE8 through main-chain carbon atoms indicated that the similar overall structures. The r.m.s.d is 0.153 Å and 0.264 Å when BF2*0401-P8D and BF2*0401-P5E were aligned with BF2*0401-IE8, respectively. The major distinctions of the three structures locate in the main-chain conformation of the peptide IE8 and its mutants (Fig. 7B), while the residues comprising the C and F pockets of BF2*0401 retain similar conformations. Conformation comparison of the peptides IE8 and P8D shows that the main-chain carbon atom of residue P8 Asp drops 1.25 Å at the C-terminus of the peptide P8D, and the depth of the side chain of P8 Asp in pocket F is almost the same as the IE8. Thus, the Glu8Asp substitution does not change the peptide binding strength (including salt bridge and hydrogen bonds) (Fig. 7C, 7D). Especially, the residue R80 of BF2*0401 molecule in pocket F adopts the same conformation in both the BF2*0401-peptide structures (Fig. 7C, 7D). As the P5 Asp in the IE8 of the BF2*0401-IE8 structure, P5 Glu in the BF2*0401-P5E forms the similar salt bridge and hydrogen bonds with the residues R9, N76 and R111 in the C pockets. The medium portion of the main-chain of peptide P5E is raised about 1.22 Å (Fig. 7E, 7F).
The structural analyses of the two mutant peptides P8D and P5E complexed BF2*0401 indicated the structural basis of binding affinity profiles of the peptide motif proposed in Table II. The narrow peptide binding groove including the major pockets of BF2*0401 retains a rigid conformation. Peptides with different mutant anchor residues need to modify their conformation to adapt the pockets. As in BF2*0401-P8D, the C terminal portion of the peptide descends to the F pocket and in BF2*0401-P5E, the middle part of the peptide rises up about the length of one covalent bond. Based on these observations, we can indicate that other peptide mutants which comprise two mutated residues between D and E, may also remain some binding affinity to BF2*0401 by the minor modification of conformation of the peptides. The mutants including single D/E to A replacement would also support renaturation to BF2*0401 due to the retainment of the main-chain conformation of the peptide. The concordance between peptide binding capabilities and the conclusions of our structural analyses complement our findings on the peptide presentation of BF2*0401.
Discussion
The structure of the classical class I molecule BF2*0401 in complex with the self peptide IE8, along with the binding assays of substituted peptides, goes a long way towards explaining the peptides presented by the chicken B4 haplotype. Four points illuminate principles of MHC-peptide interaction either unusual or not found at all in humans and other mammals, each of which leads to questions requiring further study.
First, in contrast to many classical class I molecules in humans and mice, cells of the B4 haplotype present octamer peptides with the motif DE2-DE5-E8 (with small amounts of hydrophobic amino acids at peptide position 8). The structure of BF2*0401 shows that the peptide-binding groove is flatter and shallower than those of typical mammals, with no deep pocket B leading to the equivalent of human residue 45. Based on peptide motifs determined for certain other chicken class I molecules (such as B12, B15 and B19) and simple wire models (and consistent with the structure of BF2*2101 for pocket B) (14, 15, 18), this is likely to be a general feature of chicken classical class I molecules.
Second, the centre of the groove is narrower compared to BF2*2101, being filled and even overhung by the large residues L68(69), N69(70), R152(155) and W153(156). Similarly large residues are found in the sequences of the dominantly-expressed class I molecules from the B6, B12, B15 and B19 haplotypes(16, 18), so this is also likely to be a feature of many chicken classical class I molecules.
Third, three positively-charged residues [R9(9), R80(81), and R111(114)] point up from the β-strands that make up the floor of the binding groove, along with another [R152(155)] pointing in from the α2 helix. The resulting groove is extremely positively-charged, which should explain the negatively-charged peptide motif determined from B4 cells. Although there are mammalian class I molecules with negatively-charged anchor residues (notably H-2Kk, HLA-A1, B37, B38, B40 and B44), BF2*0401 is the only class I molecule reported with more than one negatively-charged anchor residue. Moreover, it is the only class I molecule reported with a negatively-charged anchor residue in the C-terminal position. Indeed, the constitutive proteasome specificity in mammals is altered to the immunoproteasome by replacing constitutive components which include a peptidyl-glutamyl peptide-hydrolyzing activity with the inducible proteasome components which cleave poorly after acidic residues(39, 40). Inducible proteasome components have not yet been identified in chickens, but if they do exist it is likely that they cleave after acidic residues, at least in the B4 haplotype.
Fourth, perhaps the most important point to come out of this analysis is the fact that the peptide-binding specificity found by renaturation assays is wider than the peptide motif from B4 cells. Since the specificity of the TAP molecule from the B4 haplotype matches the peptide motif(18, 19), it is clear that the B4 TAP is restricting the peptides which reach the endoplasmic reticulum to load onto class I molecules. In contrast, TAP genes in typical mammals have no functional polymorphism, instead pumping a wide range of peptides from which the particular class I molecules present select the peptides to bind(41, 42). In rats, there are two allelic lineages of the TAP2 gene which affect the translocation specificity of peptides depending on the C-terminal amino acid. In this species, it is reported that the peptide-binding specificity of the class I molecule(s) converge with the translocation specificity of the TAPs(41, 43). Thus, the restriction of peptides by the translocation specificity of TAP alleles in chickens has never been described in mammals. The other components in the antigen processing/presentation pathway may also contribute to the restricted B4 haplotype peptide repertoire, which need further exploration.
The fact that at least some alleles of the polymorphic chicken TAP molecules restrict the peptides loaded onto the dominantly-expressed class I molecule has some profound implications. It may be that the grooves of many chicken class I molecules are generally shallower than those in typical mammals, with the peptide-binding specificities correspondingly less fastidious. Moreover, more and different peptides might be presented by a dominantly-expressed class I molecule in an MHC heterozygote compared to a homozygote, adding a new dimension and power to the notion of MHC heterozygote advantage compared to typical mammals.
Indeed, this may explain the peptide motif reported for the BF2*1301 molecule (identical in sequence to BF2*0401) transfected into RP9 cells which are derived from a B2/B15 heterozygote chicken(44). This motif looks like the BF2*0401 motif, but with a penultimate acidic residue often followed by a C-terminal hydrophobic residue. These results are easily understood if, in the absence of the appropriate TAP molecule, peptides with a C-terminal hydrophobic residue were translocated into the endoplasmic reticulum, but only peptides with the requisite acidic residues bound to the BF2*1301 molecule, with the C-terminal hydrophobic residue hanging out the end of the groove, as has been seen occasionally in structures of human class I molecules(45). Further experiments are needed to understand the general principles of this co-evolutionary interaction.
Finally, the structure of BF2*0401 (along with the peptide-translocation motif of TAP in B4 cells) leads to a truly fastidious motif at the cell surface for presentation of peptides to T cells. Thus, there are generally few peptides presented by the class I molecule of B4 cells, leading to susceptibility to pathogens whose sequences contain no protective peptide with the right motif. Indeed, B4 (and B13) haplotypes are generally reported to confer susceptibility to a variety of infectious pathogens. However, these haplotypes are not rare in the chicken populations, so there must be some selective pressure leading to their relatively high gene frequency. Whether this is due to selective breeding, to resistance conferred to some important (strains of) pathogen(s), or to some other selective pressure is the subject of ongoing investigations.
Acknowledgements
We thank Hao Cheng, Tianyao Yang, Weihong Chen and Mingwei Sun for assistance in experimentation, Yi Shi, Guangwen Lu, Nianzhi Zhang and Fuliang Chu for thoughtful discussion, Di Liu for bioinformatics analysis, and Christopher J. Vavricka for help with English.
Grant support
This work was supported by grants from the Ministry of Science and Technology of China (MOST) (Project 973, Grant Nos. 2011CB504703 and 2007CB815805), the National Natural Science Foundation of China (Grant No. 30671566), as well as the Wellcome Trust (Programme grant 089305 to JK). GFG is a leading principal investigator of the Innovative Research Group of the National Natural Science Foundation of China (NSFC, Grant No. 81021003).
Footnotes
Author contributions
G.F.G., C.X. and J.Z. conceived and designed the research, J.Z. carried out the protein expression, purification and crystallization, J. Z. and Y. L. measured the thermostabilities of MHC complexes, Y.C., J.Q. and F.G. solved the crystal structure, G.F.G., C.X., J.Z., X. Z., J.K., Y.C. and J. L. performed the data analysis. G.F.G., C.X., J.K. and J.Z. wrote the manuscript.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Ghodke Y, Joshi K, Chopra A, Patwardhan B. HLA and disease. Eur J Epidemiol. 2005;20:475–488. doi: 10.1007/s10654-005-5081-x. [DOI] [PubMed] [Google Scholar]
- 2.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nature reviews Immunology. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 3.Meyer D, Thomson G. How selection shapes variation of the human major histocompatibility complex: a review. Annals of human genetics. 2001;65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x. [DOI] [PubMed] [Google Scholar]
- 4.Jones EY. MHC class I and class II structures. Current opinion in immunology. 1997;9:75–79. doi: 10.1016/s0952-7915(97)80162-8. [DOI] [PubMed] [Google Scholar]
- 5.Madden DR. The three-dimensional structure of peptide-MHC complexes. Annual review of immunology. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 6.Stern LJ, Wiley DC. Antigenic peptide binding by class I and class II histocompatibility proteins. Structure. 1994;2:245–251. doi: 10.1016/s0969-2126(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 7.Wilson IA, Fremont DH. Structural analysis of MHC class I molecules with bound peptide antigens. Seminars in immunology. 1993;5:75–80. doi: 10.1006/smim.1993.1011. [DOI] [PubMed] [Google Scholar]
- 8.Bacon LD, Witter RL. Influence of B-haplotype on the relative efficacy of Marek's disease vaccines of different serotypes. Avian diseases. 1993;37:53–59. [PubMed] [Google Scholar]
- 9.Bacon LD, Witter RL, Crittenden LB, Fadly A, Motta J. B-haplotype influence on Marek's disease, Rous sarcoma, and lymphoid leukosis virus-induced tumors in chickens. Poultry science. 1981;60:1132–1139. doi: 10.3382/ps.0601132. [DOI] [PubMed] [Google Scholar]
- 10.Lamont SJ. Immunogenetics and the major histocompatibility complex. Veterinary immunology and immunopathology. 1991;30:121–127. doi: 10.1016/0165-2427(91)90013-3. [DOI] [PubMed] [Google Scholar]
- 11.Plachy J, Pink JR, Hala K. Biology of the chicken MHC (B complex) Critical reviews in immunology. 1992;12:47–79. [PubMed] [Google Scholar]
- 12.Jacob JP, Milne S, Beck S, Kaufman J. The major and a minor class II beta-chain (B-LB) gene flank the Tapasin gene in the B-F /B-L region of the chicken major histocompatibility complex. Immunogenetics. 2000;51:138–147. doi: 10.1007/s002510050022. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman J, Milne S, Gobel TW, Walker BA, Jacob JP, Auffray C, Zoorob R, Beck S. The chicken B locus is a minimal essential major histocompatibility complex. Nature. 1999;401:923–925. doi: 10.1038/44856. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman J, Volk H, Wallny HJ. A “minimal essential Mhc” and an “unrecognized Mhc”: two extremes in selection for polymorphism. Immunol Rev. 1995;143:63–88. doi: 10.1111/j.1600-065x.1995.tb00670.x. [DOI] [PubMed] [Google Scholar]
- 15.Koch M, Camp S, Collen T, Avila D, Salomonsen J, Wallny HJ, van Hateren A, Hunt L, Jacob JP, Johnston F, Marston DA, et al. Structures of an MHC class I molecule from B21 chickens illustrate promiscuous peptide binding. Immunity. 2007;27:885–899. doi: 10.1016/j.immuni.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Shaw I, Powell TJ, Marston DA, Baker K, van Hateren A, Riegert P, Wiles MV, Milne S, Beck S, Kaufman J. Different evolutionary histories of the two classical class I genes BF1 and BF2 illustrate drift and selection within the stable MHC haplotypes of chickens. J Immunol. 2007;178:5744–5752. doi: 10.4049/jimmunol.178.9.5744. [DOI] [PubMed] [Google Scholar]
- 17.Shiina T, Briles WE, Goto RM, Hosomichi K, Yanagiya K, Shimizu S, Inoko H, Miller MM. Extended gene map reveals tripartite motif, C-type lectin, and Ig superfamily type genes within a subregion of the chicken MHC-B affecting infectious disease. J Immunol. 2007;178:7162–7172. doi: 10.4049/jimmunol.178.11.7162. [DOI] [PubMed] [Google Scholar]
- 18.Wallny HJ, Avila D, Hunt LG, Powell TJ, Riegert P, Salomonsen J, Skjodt K, Vainio O, Vilbois F, Wiles MV, Kaufman J. Peptide motifs of the single dominantly expressed class I molecule explain the striking MHC-determined response to Rous sarcoma virus in chickens. Proc Natl Acad Sci U S A. 2006;103:1434–1439. doi: 10.1073/pnas.0507386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker BA, Hunt LG, Sowa AK, Skjodt K, Gobel TW, Lehner PJ, Kaufman J. The dominantly expressed class I molecule of the chicken MHC is explained by coevolution with the polymorphic peptide transporter (TAP) genes. Proc Natl Acad Sci U S A. 2011;108:8396–8401. doi: 10.1073/pnas.1019496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaufman J. Co-evolving genes in MHC haplotypes: the “rule” for nonmammalian vertebrates? Immunogenetics. 1999;50:228–236. doi: 10.1007/s002510050597. [DOI] [PubMed] [Google Scholar]
- 21.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhou M, Xu Y, Lou Z, Cole DK, Li X, Liu Y, Tien P, Rao Z, Gao GF. Complex assembly, crystallization and preliminary X-ray crystallographic studies of MHC H-2Kd complexed with an HBV-core nonapeptide. Acta Crystallogr D Biol Crystallogr. 2004;60:1473–1475. doi: 10.1107/S0907444904013587. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Gao F, Peng H, Xia C, Gao GF. Preparation of selenomethionine-labelled chicken beta2m and its x-ray crystallographic studies. Sci Tech Rev (in Chinese) 2009;27:38–41. [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Lebedev AA, Vagin AA, Murshudov GN. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta Crystallogr D Biol Crystallogr. 2008;64:33–39. doi: 10.1107/S0907444907049839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 31.Cole DK, Rizkallah PJ, Boulter JM, Sami M, Vuidepot AL, Glick M, Gao F, Bell JI, Jakobsen BK, Gao GF. Computational design and crystal structure of an enhanced affinity mutant human CD8 alphaalpha coreceptor. Proteins. 2007;67:65–74. doi: 10.1002/prot.21176. [DOI] [PubMed] [Google Scholar]
- 32.Gao GF, Tormo J, Gerth UC, Wyer JR, McMichael AJ, Stuart DI, Bell JI, Jones EY, Jakobsen BK. Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2. Nature. 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 33.Kaufman J, Andersen R, Avila D, Engberg J, Lambris J, Salomonsen J, Welinder K, Skjodt K. Different features of the MHC class I heterodimer have evolved at different rates. Chicken B-F and beta 2-microglobulin sequences reveal invariant surface residues. J Immunol. 1992;148:1532–1546. [PubMed] [Google Scholar]
- 34.Kaufman J, Salomonsen J, Flajnik M. Evolutionary conservation of MHC class I and class II molecules--different yet the same. Seminars in immunology. 1994;6:411–424. doi: 10.1006/smim.1994.1050. [DOI] [PubMed] [Google Scholar]
- 35.Madden DR, Garboczi DN, Wiley DC. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993;75:693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- 36.Madden DR, Gorga JC, Strominger JL, Wiley DC. The three-dimensional structure of HLA-B27 at 2.1 A resolution suggests a general mechanism for tight peptide binding to MHC. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- 37.Matsumura M, Fremont DH, Peterson PA, Wilson IA. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992;257:927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- 38.Smith KJ, Reid SW, Harlos K, McMichael AJ, Stuart DI, Bell JI, Jones EY. Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53. Immunity. 1996;4:215–228. doi: 10.1016/s1074-7613(00)80430-6. [DOI] [PubMed] [Google Scholar]
- 39.Driscoll J, Brown MG, Finley D, Monaco JJ. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365:262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 40.Gaczynska M, Rock KL, Goldberg AL. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365:264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 41.Momburg F, Roelse J, Howard JC, Butcher GW, Hammerling GJ, Neefjes JJ. Selectivity of MHC-encoded peptide transporters from human, mouse and rat. Nature. 1994;367:648–651. doi: 10.1038/367648a0. [DOI] [PubMed] [Google Scholar]
- 42.Obst R, Armandola EA, Nijenhuis M, Momburg F, Hammerling GJ. TAP polymorphism does not influence transport of peptide variants in mice and humans. European journal of immunology. 1995;25:2170–2176. doi: 10.1002/eji.1830250808. [DOI] [PubMed] [Google Scholar]
- 43.Joly E, Le Rolle AF, Gonzalez AL, Mehling B, Stevens J, Coadwell WJ, Hunig T, Howard JC, Butcher GW. Co-evolution of rat TAP transporters and MHC class I RT1-A molecules. Current biology: CB. 1998;8:169–172. doi: 10.1016/s0960-9822(98)70065-x. [DOI] [PubMed] [Google Scholar]
- 44.Sherman MA, Goto RM, Moore RE, Hunt HD, Lee TD, Miller MM. Mass spectral data for 64 eluted peptides and structural modeling define peptide binding preferences for class I alleles in two chicken MHC-B haplotypes associated with opposite responses to Marek's disease. Immunogenetics. 2008;60:527–541. doi: 10.1007/s00251-008-0302-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins EJ, Garboczi DN, Wiley DC. Three-dimensional structure of a peptide extending from one end of a class I MHC binding site. Nature. 1994;371:626–629. doi: 10.1038/371626a0. [DOI] [PubMed] [Google Scholar]