Summary
MicroRNAs (miRNAs) are non-coding RNAs that suppress translation of specific mRNAs. The miRNA machinery interacts with Fragile X Mental Retardation Protein (FMRP), which functions as translational repressor. We show that miR-125b and miR-132, as well as several other miRNAs, are associated with FMRP in mouse brain. miR-125b and miR-132 had largely opposing effects on dendritic spine morphology and synaptic physiology in hippocampal neurons. FMRP knockdown ameliorates the effect of miRNA overexpression on spine morphology. We identified NMDA receptor subunit NR2A as novel target of miR-125b and show that NR2A mRNA is specifically associated with FMRP in brain. In hippocampal neurons, NR2A expression is negatively regulated through its 3’UTR by FMRP, miR-125b and Argonaute 1. Regulation of NR2A 3’UTR by FMRP depends in part on miR-125b. Because NMDA receptor subunit composition profoundly affects synaptic plasticity, these observations have implications for the pathophysiology of Fragile X Syndrome, in which plasticity is altered.
Introduction
MicroRNAs (miRNAs) are short (~22 nucleotide) non-coding RNAs that mediate post-transcriptional gene silencing (Filipowicz et al., 2008; Rana, 2007). miRNAs are loaded into effector proteins of the Argonaute family. Once loaded the Argonaute protein is said to be “programmed” with the miRNA, which guides the Argonaute protein to specific mRNA targets. miRNAs usually bind their target mRNAs through imperfect base pairing in the 3’ untranslated region (UTR) and impact protein expression by inhibiting mRNA translation or by promoting mRNA decay.
In mammals, several hundred distinct miRNAs have been discovered, including those selectively expressed in the brain (Cao et al., 2006). miRNAs play roles in early development (Stefani and Slack, 2008) and in diseases such as cancer (Esquela-Kerscher and Slack, 2006) and neurodegeneration (Eacker et al., 2009). Yet, only a small number of miRNA targets have been validated and shown to be functionally important in vivo (Schratt, 2009). In mammals, miR-132 regulates dendrite development by targeting p250GAP (Vo et al., 2005; Wayman et al., 2008) and miR-134 and 138 have been implicated in dendritic spine development through repression of LIMK1 and APT1 (Schratt et al., 2006; Siegel et al., 2009).
Fragile X syndrome (FXS), the most common inherited cause of mental retardation (Bagni and Greenough, 2005; Bassell and Warren, 2008), is usually due to a trinucleotide expansion in the FMR1 gene that results in transcriptional silencing of FMRP (Fragile X mental retardation protein) expression. FMRP contains multiple RNA-binding domains and is widely thought to function as a translational suppressor of specific mRNAs, including MAP1b, CaMKIIα, and Arc (Bassell and Warren, 2008).
FMRP is biochemically and genetically linked to the miRNA pathway. FMRP interacts with proteins (e.g. Argonaute and Dicer) in the RNA interference silencing complex (RISC) and with miRNAs, but FMRP itself is not essential for RNAi-mediated mRNA cleavage (Bolduc et al., 2008; Caudy et al., 2002; Cheever and Ceman, 2009; Hock et al., 2007; Ishizuka et al., 2002; Jin et al., 2004b; Okamura et al., 2004; Plante et al., 2006). Heterozygous loss of AGO1 enhances the phenotype of heterozygous loss of FMRP in flies, suggesting AGO1 facilitates translational repression by FMRP (Bolduc et al., 2008; Jin et al., 2004b). One hypothesis is that specific miRNAs – as part of the FMRP translation regulatory complex – could facilitate selection and/or suppression of target mRNAs by FMRP (Jin et al., 2004a). However, no specific example of such a functional association of FMRP, mRNA and miRNA has been identified.
While ~22 nucleotide RNA has been detected in the FMRP-complex (Caudy et al., 2002; Ishizuka et al., 2002; Jin et al., 2004b), it is unknown which specific miRNAs associate with FMRP in mammals. We hypothesized that such FMRP-associated miRNAs might regulate of synaptic function and dendritic spine structure, given the well-established synaptic abnormalities found in FMR1 knockout (KO) mice that are also central to the pathogenesis of FXS in humans (Comery et al., 1997; Irwin et al., 2001). Here we report that two FMRP-associated miRNAs (miR-125b and miR-132) can affect dendritic spine morphology. For both miRNAs, we identified specific mRNA targets that are also associated with FMRP and encode key proteins involved in synaptic function.
Results
FMRP-associated miRNAs in mouse brain
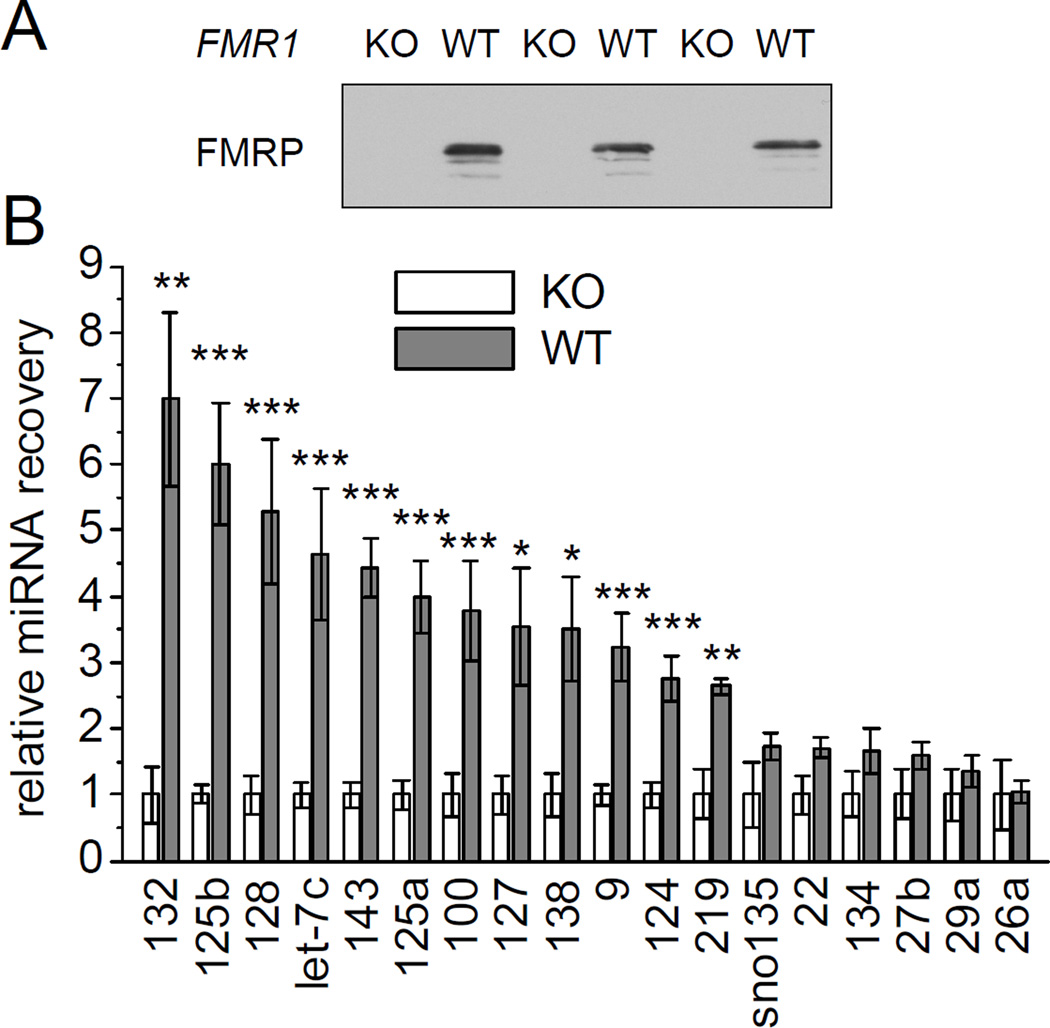
To identify FMRP-associated miRNAs, we purified FMRP from mouse brain using FMRP antibodies and isolated the associated miRNAs following a protocol commonly used for the identification of mRNA targets of FMRP (Brown et al., 2001). As a control for specificity of miRNA association, we also used FMRP antibodies to immunoprecipitate from FMR1 KO brains (Figure 1A).
Figure 1. Identification of miRNAs associated with FMRP in brain.
(A) FMRP immunoprecipitates from individual wild-type and FMR1 KO mouse brain (age 3 months) were immunoblotted with anti-FMRP antibody. (B) miRNA recovery from FMRP-immunoprecipitation of wild-type mice normalized to FMR1 KO mice. For both groups individual immunoprecipitations derived from six single mice were analyzed by TaqMan qPCR. Statistical analysis by Student’s t-test: * p < 0.05, ** p < 0.01, *** p < 0.001.
We performed quantitative PCR (qPCR) to measure the amount of mature miRNAs coimmunoprecipitated with anti-FMRP antibodies. A specific set of miRNAs were quantified: the brain-specific miRNAs 9, 124, 128; miRNAs 132 and 219, which are inducible by CREB, an important transcription factor implicated in learning and memory; and two miRNAs previously reported to regulate spine morphology, miRNAs 134 and 138 (Cao et al., 2006; Schratt et al., 2006; Siegel et al., 2009; Vo et al., 2005). Additionally, we looked for broadly expressed miRNAs of high (miRNAs 26a, 29a, 125b, 127 and let-7c), intermediate (miRNAs 22, and 125a) and low abundance (miRNAs 27b, 100 and 143) (Landgraf et al., 2007; Pena et al., 2009). Twelve of these candidate miRNAs were found to associate with FMRP (Figure 1B). That is, let-7c, miR-9, 100, 124, 125a, 125b, 127, 128, 132, 138, 143 and 219 were enriched three- to seven-fold in FMRP immunoprecipitates from age-matched wild-type brains as compared to control immunoprecipitates from FMR1 KO brains. Expression levels for all tested miRNAs were indistinguishable between brains from wild-type and FMR1 KO mice (data not shown). In addition, a small nucleolar RNA (snoRNA-135) and five other miRNAs (miR-22, 26a, 27b, 29a and 134) did not show significant co-precipitation with FMRP above the background seen in FMR1 KO brain, indicating that there is selectivity of miRNA association with FMRP (Figure 1B).
Effects of FMRP-associated miRNAs on spine morphology
We asked whether specific miRNAs associated with FMRP might be involved in the regulation of spine morphogenesis. To determine the gain-of-function phenotype of the miRNAs, we transfected dissociated rat hippocampal neurons in culture with DNA plasmids that express the precursor-miRNA hairpin under the control of a β-actin promoter (Zeng et al., 2005). Overexpression of the mature miRNA in neurons was validated using EGFP-based miRNA-sensors containing artificial miRNA target sites in their 3’UTR (Supplemental Figure S1A).
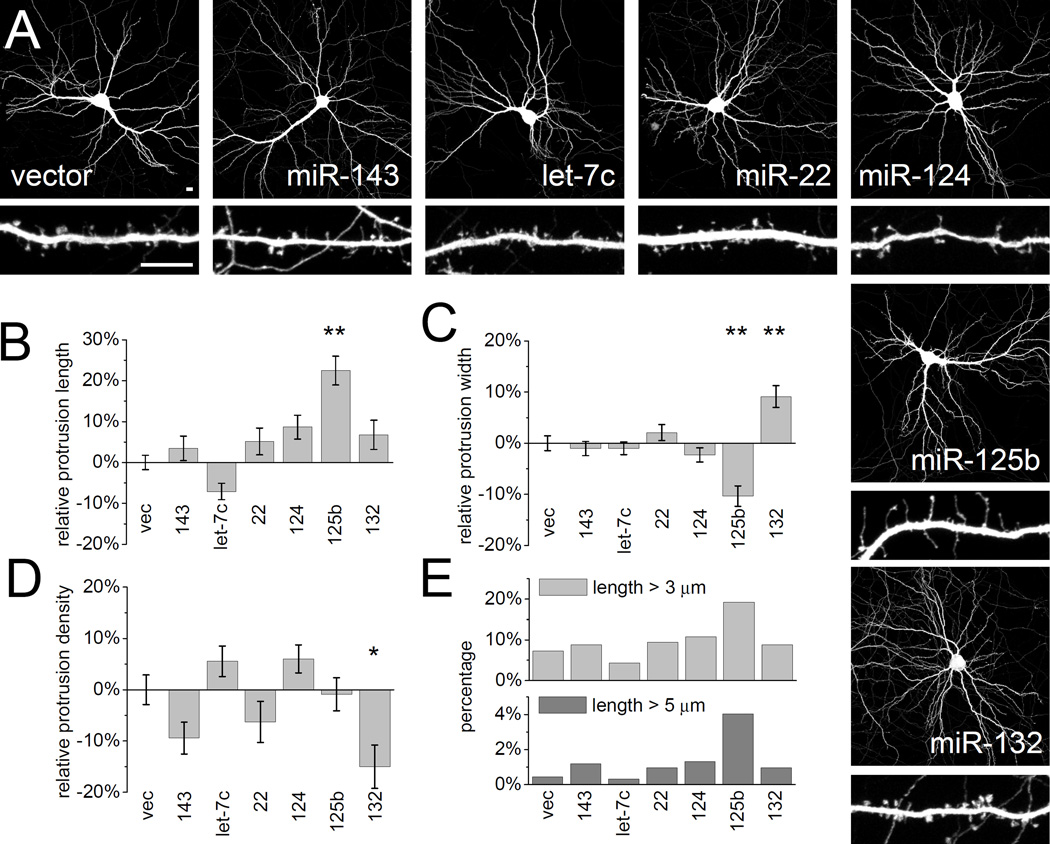
Hippocampal neurons were transfected at 14 days in vitro (DIV14) with individual miRNAs and EGFP marker for three days (DIV14+3). Dendritic protrusions were differentially affected by miR-125b and miR-132. Overexpression of miR-125b resulted in longer and thinner protrusions (Figure 2A, B, C). In neurons overexpressing miR-125b, ~20% of dendritic protrusions were very long (>3 µm), versus ~5–10% in untransfected neurons or in neurons transfected with other miRNAs (Figure 2E). This difference was even more marked for protrusions > 5 µm. Protrusion density was not affected. Neither miR-125b, let-7c, miR-22, miR-124, miR-132 nor miR-143 had a statistically significant effect on dendritic growth or arborization as measured by Sholl analysis (Figure 2A and Supplemental Figure S2).
Figure 2. Overexpression of FMRP-associated miRNAs differentially affects dendritic spine morphology.
(A) Hippocampal neurons at DIV14 were cotransfected with plasmids expressing specific miRNAs (see Supplemental Figure S1A) and EGFP to outline neuron morphology. Three days post-transfection (DIV14+3) neurons were fixed, immunostained for EGFP and imaged by confocal microscopy to visualize dendritic arborization and spine morphology. Low-magnification images show transfected whole neurons, and higher-magnification images show representative dendritic segments of these cells. Scale bars represent 10 µm (upper and lower panels). The length (B), width (C) and density (D) of dendritic protrusions were manually measured using MetaMorph software. Data was normalized to neurons transfected with empty vector (vec). Statistical analysis by one-way ANOVA with Dunnett’s post test: * p < 0.05, ** p < 0.01. n = 26 to 72 neurons each group. (E) Percentage of protrusions with length > 3 µm or length > 5 µm. See also Supplemental Figure S2.
In contrast, neurons overexpressing miR-132 were characterized by stubby and mushroom spines (Figure 2A). Overexpressed miR-132 caused an increase in average protrusion width, without affecting the average length (Figure 2B, C). In addition, average protrusion density fell significantly by ~15 % (Figure 2D). The effects of miRNAs 125b and 132 on dendritic spine width appear to be specific, because overexpression of let-7c, miR-22, miR-124, miR-9 and miR-128 (Figure 2 and data not shown) had no significant effect on spine number or shape compared with vector control. While miR-132 has been implicated in regulating neuron morphology (Siegel et al., 2009; Vo et al., 2005; Wayman et al., 2008), here we show a novel role for miR-125b in spine morphogenesis.
FMRP is required for the effect of miR-125b and miR-132 on spine morphology
To test whether regulation of spine morphology by miR-125b and 132 is dependent on FMRP, we overexpressed the miRNA constructs in neurons either with FMRP-shRNA or control luciferase shRNA constructs (Zhang and Macara, 2006). In our experiments, FMRP-knockdown (DIV14+3) by itself did not affect spine morphology (Figure 3). The lack of effect of FMRP suppression might be explained by the acute manipulation (compared with FMR1 KO mice) and by the fact that the effect of FMRP on spine morphology declines in neurons of this age (Nimchinsky et al., 2001). We validated FMRP-knockdown and specificity of two shRNA constructs (#1 and #2) in HEK293 cells and cultured hippocampal neurons (Supplemental Figure S3). shRNA FMRP #1 (and to a lesser extent #2) reduced expression of cotransfected EGFP-FMRP (Antar et al., 2004) and endogenous FMRP immunofluorescence.
Figure 3. Overexpression of FMRP-associated miRNAs differentially affects dendritic spine morphology.
(A) Hippocampal neurons were cotransfected (DIV14+3) with EGFP, miRNAs and shRNAs (compare Supplemental Figure S3). Scale bars represent 10 µm (upper and lower panels). The length (B), width (C) and density (D) of dendritic protrusions were manually quantified. Data was normalized to neurons co-transfected with miR-143 and Luc shRNA. Statistical analysis by one-way ANOVA with Bonferroni’s post test: * p < 0.05, ** p < 0.01. n = 25 to 35 neurons each group.
Control Luc shRNA expression did not affect the differential spine phenotypes induced by miR-125 or 132 overexpression (Figure 2 and 3). Compared to miR-143, a miRNA that does not affect dendritic spines (Figure 2), miR-125b overexpressing neurons had significantly longer and thinner dendritic protrusions, while miR-132 expression increased protrusion width and reduced spine density. In cells with knockdown of FMRP, however, the effects of miR-125 and 132 were abolished and spine morphology was not significantly different from miR-143 control. These data indicate that FMRP is required for miR-125b and 132 to alter spine morphology. Since FMRP is not needed for general RNAi function (Caudy et al., 2002; Hock et al., 2007), these findings support the idea that the FMRP-associated miRNAs 125b and 132 modulate dendritic spine morphology together with FMRP through common mRNA targets.
Sequestration of FMRP-associated miRNA affects neuron morphology
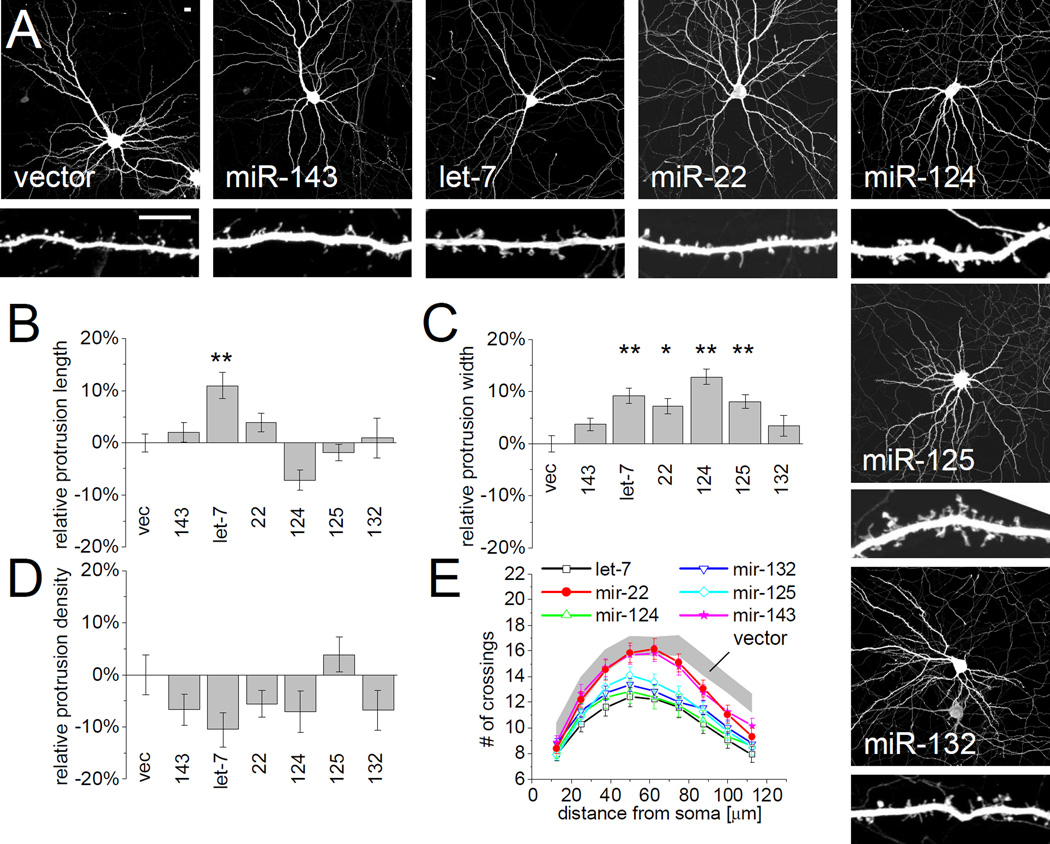
To investigate the loss-of-function phenotype of individual miRNAs on spine morphology, we applied the plasmid-based “sponge” method of Ebert et al (2007). Overexpression of mRNA constructs containing multiple (five to seven) concatenated miRNA binding sites with central mismatches (that prevent AGO2-mediated mRNA cleavage) results in the sequestration of endogenous miRNAs that bind to these sequences (Supplemental Figure S1B). Because the eight 5’-most nucleotides of a miRNA, the so-called “seed” region, thought to be critical for target sequence recognition, this approach leads to cross-capture of related miRNA isoforms with identical seed regions (e.g. let-7a through let-7i, or miR-125a and 125b). In these cases, the sponge constructs are denoted according to the miRNA family sequestered (e.g. let-7 and miR-125 sponge). Specificity and efficacy of miRNA suppression was confirmed for each sponge in hippocampal neurons, indicating sufficient sequestration and functional suppression of targeted endogenous miRNAs. For example, transfection of a sponge construct for miR-124, the most abundant miRNA in brain, upregulated expression of a sensor containing target sequences for miR-124 by ~5 fold, but had no effect on other sensors containing unrelated miRNA target sites (Supplemental Figure S1C).
The effects of individual miRNA sponges on neuron morphology were examined in hippocampal neurons three days after transfection (DIV14+3) (Figure 4). Loss-of-function of the abundant neuronal miRNAs let-7 and miR-124, and of the less abundant miR-125 and miR-132 (Landgraf et al., 2007), caused a marked pruning of the dendritic arbor, as quantified by Sholl analysis (Figure 4A, E). Primary dendrites ramified less and the dendritic arbor covered a smaller area. By contrast, sponges targeting miR-22 and miR-143 had little effect on dendrite complexity (Figure 4A, E).
Figure 4. Sponging of FMRP-associated miRNAs differentially affects dendritic spine morphology.
(A) Hippocampal neurons were cotransfected (DIV14+3) with EGFP and sponges for specific miRNAs (see Supplemental Figure S1B, C) to sequester endogenous miRNA. Scale bars represent 10 µm (upper and lower panels). The length (B), width (C) and density (D) of dendritic protrusions was manually quantified. Data was normalized to neurons expressing empty vector (vec). n = 23 to 50 cells each group. Statistical analysis by one-way ANOVA with Dunnett’s post test: * p < 0.05, ** p < 0.01. (E) Sholl analysis of sponge-transfected neurons, measuring the number of dendrites crossing concentric circles at the indicated distance around the cell body. Gray corridor represents neurons transfected with empty vector (mean +/− SEM). n = 40 to 71 neurons each group. Sponge transfected cells are significantly different from control neurons (Two-way ANOVA, p<0.05) for: miR-124 and let-7 (from 37.5 µm radius), miR-125 and miR-132 (from 63.5 µm), miR-22 and miR-143 (from 75 µm).
Conversely to miR-125b gain-of-function, which reduced spine width (Figure 2C), miR-125 loss-of-function significantly increased protrusion width (Figure 4C). Moreover, neurons transfected with sponges for let-7, miR-22 and miR-124 also showed a modest but significant increase in protrusion width. Only the let-7 sponge significantly increased protrusion length (Figure 4B) and none of the sponge constructs tested affected the density of protrusions (Figure 4D). The lack of effect of sponging of miR-132 and miR-143 could be explained by an activity-dependent role for miR-132 and low basal expression of miR-132 and miR-143 (Landgraf et al., 2007). Because overexpression and sponging of miR-125b had opposite effects on protrusion width, these data corroborate the idea that miR-125b is important for regulating spine morphology.
miR-125b and miR-132 differentially modify synaptic strength
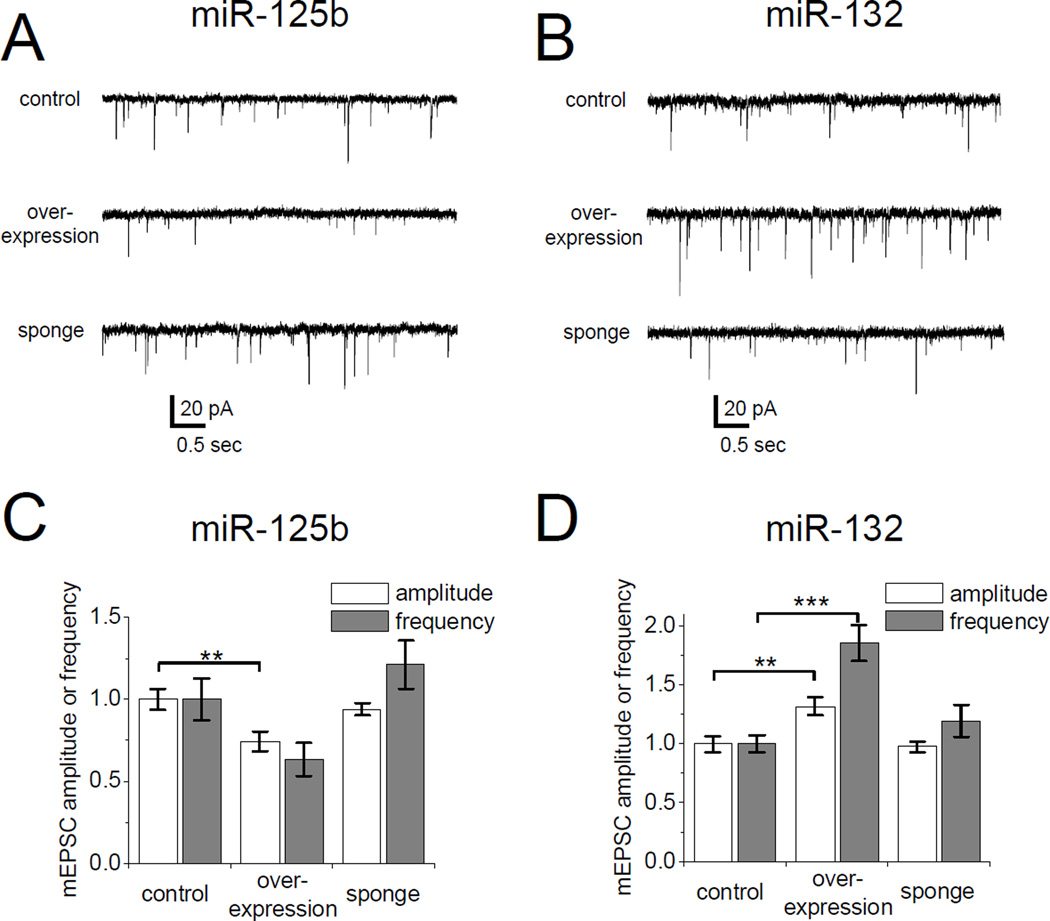
Spine size often correlates with synaptic strength. We therefore measured AMPA receptor-mediated miniature excitatory postsynaptic currents (mEPSCs) in cultured hippocampal neurons transfected with miR-125b and miR-132 constructs (Figure 5). Overexpression of miR-125b resulted in a significant 25% drop in mean mEPSC amplitude (Figure 5A, C). The weakening of synaptic transmission is consistent with the long thin protrusions and the reduced spine width induced by miR-125b (Figure 2C). In neurons overexpressing miR-125b, mEPSC frequency fell relative to control neurons transfected with empty vector, but this did not reach the level of statistical significance (Figure 5C). Suppressing endogenous miR-125b activity with the miR-125 sponge had no significant effect on mEPSC amplitude or frequency (Figure 5A, C), consistent with the modest effect of miR-125b sponging on spine morphology (Figure 4).
Figure 5. miR-125b and miR-132 modify synaptic strength.
AMPA-mediated miniature excitatory postsynaptic currents (mEPSCs) measured in cultured hippocampal neurons cotransfected (DIV14+3) with EGFP and miR-125b (A, C) or miR-132 (B, D) overexpression or sponge constructs. (A, B) Representative mEPSC traces from transfected neurons. (C, D) Mean mEPSC amplitude and frequency normalized to control neurons transfected with empty vector. Statistical analysis using Kruskal-Wallis test and Dunn’s post test: ** p < 0.01, *** p < 0.001. n = 16 to 22 neurons for each group.
In contrast to miR-125b, overexpression of miR-132 led to an increase in mean mEPSC amplitude (Figure 5B, D), consistent with the wider spines induced by miR-132 (Figure 2C). mEPSC frequency was also considerably elevated by miR-132 overexpression (Figure 5D), even though spine density was decreased by this miRNA (Figure 2D and 3D). This could reflect a greater number of miniature events detected with the increased amplitude of EPSC. The miR-132 sponge did not affect either amplitude or frequency of mEPSCs. Overall, these electrophysiological findings are consistent with and corroborate the effects of miR-125b and miR-132 on the structure of dendritic spines.
mRNAs encoding for synaptic proteins are targeted by FMRP-associated miRNAs
We asked which target genes might mediate the bidirectional effects of miR-125b and miR-132 on synaptic physiology. RNAi-mediated knockdown of the known miR-132 target p250GAP was recently found to increase dendritic spine size (Nakazawa et al., 2008; Vo et al., 2005; Wayman et al., 2008). This is in accord with the spine enlargement upon miR-132 expression in our experiments (Figure 2).
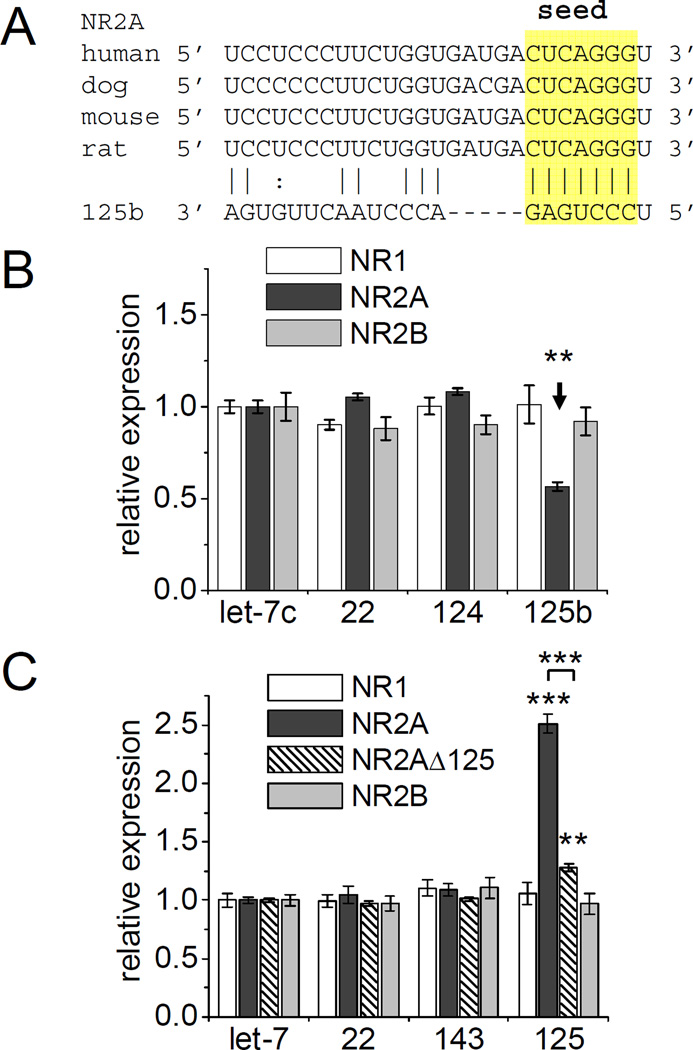
We focused on miR-125b target mRNAs predicted by TargetScan (Lewis et al., 2005) that might contribute to the induction of long thin protrusions and altered synaptic function. Target prediction mainly relies on the presence of a seed match to a miRNA in the 3’UTR of an mRNA that is conserved between species. For example, the 3’UTR of the NR2A subunit of NMDA receptors contains such a conserved sequence complementary to the seed region of miR-125b (Figure 6A).
Figure 6. NR2A 3’UTR is regulated by miR-125b.
FF-luc 3’UTR reporters for NMDA receptor subunits NR1, NR2A or N2B were cotransfected with miRNA overexpressing or sponging constructs, as well as RR-luc. Relative expression was determined by normalizing the ratio of FF-luc and RR-luc activity to the effect of each miRNA on a control FF-luc reporter (and let-7c). (A) Alignment of the 3’UTR sequence of NR2A in four mammalian species with miR-125b. (B) Relative expression of NMDA receptor FF-luc reporters constructs cotransfected with miRNA-expressing constructs in HEK293 cells. n =18 to 24. One-way ANOVA with Dunnett’s post test: * p < 0.05, ** p < 0.01. n = 12 to 54. See Supplemental Figure S4 and S5. (C) Relative expression of FF-luc reporters cotransfected with miRNA-sponges in hippocampal neurons (DIV4+3). NR2A Δ125 completely lacks the miRNA-target site shown above. Two-way ANOVA with Bonferroni’s post test: ** p < 0.01, *** p < 0.001. n = 14 to 56.
To validate candidate miRNA targets, we fused the 3’UTRs of the specific mRNAs to the coding sequence of firefly luciferase (FF-luc). The FF-luc reporter construct was then cotransfected with plasmids expressing individual miRNAs in HEK293 cells, along with a plasmid encoding Renilla reniformis luciferase (RR-luc) for normalization. miRNA expression was validated using FF-luc reporters containing perfect-match miRNA target-sites (Supplemental Figure S4).
Most of the tested FF-luc 3’UTR reporters of predicted target genes were unaffected by miRNA coexpression (Supplemental Figure S5). We found slight suppression of GluR2 (by miR-124a) and Eph receptor A4 (EphA4, by let-7c, miR-22 and possibly miR-125b). Only the NR2A 3’UTR reporter was strongly inhibited by coexpression of miR-125b (~45%), consistent with the conserved seed match (Figure 6A, B). By contrast, co-expression of let-7c, miR-22 and miR-124 had no effect on NR2A reporter expression, and no seed matches to these miRNAs exist in the NR2A 3’UTR. FF-luc reporter constructs containing the 3’UTR of the other NMDA receptor subunits NR1 and NR2B (containing no such seed region) were not affected by the tested miRNAs (Figure 6B).
To corroborate the regulation of NR2A by miR-125b in a loss-of-function experiment in neurons, we co-expressed FF-luc reporter constructs together with miRNA sponges in hippocampal neurons. The NR2A 3’UTR FF-luc reporter was strongly and specifically upregulated (~2.5 fold) by sponging of endogenous miR-125 (Figure 6C). Deleting the putative miR-125b target site in the 3’ UTR (NR2A Δ125) removed more than 80% of the induction of NR2A reporter expression by miR-125 sponge, indicating that this site mediates the majority of the inhibitory effect of miR-125. A second, non-canonical potential miR-125b target site was observed in the NR2A 3’UTR (UCUCAGG), which might account for the minor residual effect of miR-125 sponging. Reporters containing NR1 or NR2B 3’UTR were unaffected by miR-125 inhibition. Together these data indicate that neuronal miR-125 can specifically regulate NR2A reporter expression.
NR2A is a target of miR-125 in neurons
To validate NR2A as a bona fide miR-125b target in neurons, we transduced hippocampal neurons in culture (DIV4+4) with lentiviral vectors expressing either miRNA precursor hairpins or concatenated miRNA binding sites (Supplemental Figure S1B) and analyzed NMDAR subunit expression by immunoblotting (Figure 7A). We used miR-143 as a negative control, because it did not affect spine morphology and NR2A reporter expression (Figures 2 to 6).
Figure 7. NR2A is a miR-125b target in neurons.
(A) Cultured hippocampal neurons (DIV4+4) were infected with miR-143 (−) or 125b (+) overexpressing or sponging viral vectors. Immunoblots show NMDA receptor subunits expression. Immunoblot signals were quantified by densitometry and normalized to total protein amounts and miR-143 expressing controls. Student’s t-test: ** p < 0.01. *** p < 0.001. n = 6. (B) NMDA-receptor EPSC half-width measured in CA1 pyramidal cells in organotypic hippocampal slice culture transfected with miR-143 (ctrl), miR-125b alone (with empty vector), miR-125b together with NR2A or miR-125b sponge (DIV11+4). Paired recording of transfected and untransfected neighboring neurons in the same slice, labeled + and − respectively. Paired Student’s t-test: * p < 0.05. n = 12 to 14 pairs.
Overexpression of miR-125b reduced NR2A expression by ~60% compared to control neurons infected with miR-143 expressing virus, but had no significant effect on the expression levels of NR1 and NR2B, PSD-95 or β3-tubulin. Conversely, sequestering endogenous miR-125 by sponging specifically enhanced endogenous NR2A levels by ~60% compared to miR-143 sponging (Figure 7A). These data extend the findings using FF-luc reporters (Figure 6) and demonstrate that miR-125b specifically regulates endogenous NR2A in neurons.
During postnatal brain development, NR2A expression rises while NR2B falls (Monyer et al., 1994; Sheng et al., 1994). In hippocampal cultures, NR2A mRNA levels increased ~4-fold within the first 14 days in vitro, while NR2B mRNA tapered to a plateau after an initial peak (Supplemental Figure S6A). During this period miR-125b levels dropped ~50% (Supplemental Figure S6B), which may contribute to the induction of NR2A protein expression at the post-transcriptional level (Yashiro and Philpot, 2008).
NR2A-containing NMDA receptors (NMDARs) have faster deactivation kinetics than NR2B-containing NMDA receptors. Could miR-125b affect the subunit composition of NMDA receptors and result in a functional change in NMDAR channel kinetics? Evoked NMDAR-EPSC kinetics were measured in pairs of transfected and untransfected neighboring CA1 neurons of organotypic hippocampal slice culture transfected with miRNA-expressing constructs. miR-125b transfected cells showed prolonged EPSC half-width relative to untransfected neighboring cells, consistent with relative loss of synaptic NR2A-containing NMDARs (Figure 7B). Control miR-143 overexpression had no significant effect on NMDAR-EPSC kinetics (Figure 7B). In contrast, sequestering endogenous miR-125b using sponge transfection accelerated the NMDAR-EPSC kinetics consistent with increased NR2A expression. Furthermore, co-transfecting an NR2A construct lacking most of the 3’UTR including the miR-125b target region reversed the effect of miR-125b overexpression on the NMDAR-EPSC half-width. The overall NMDAR EPSC amplitude was not significantly changed in miRNA-overexpressing neurons (data not shown). Together these data indicate that endogenous miR-125 directly and specifically regulates NR2A expression and affects NMDA receptor function accordingly.
Regulation of NR2A by FMRP
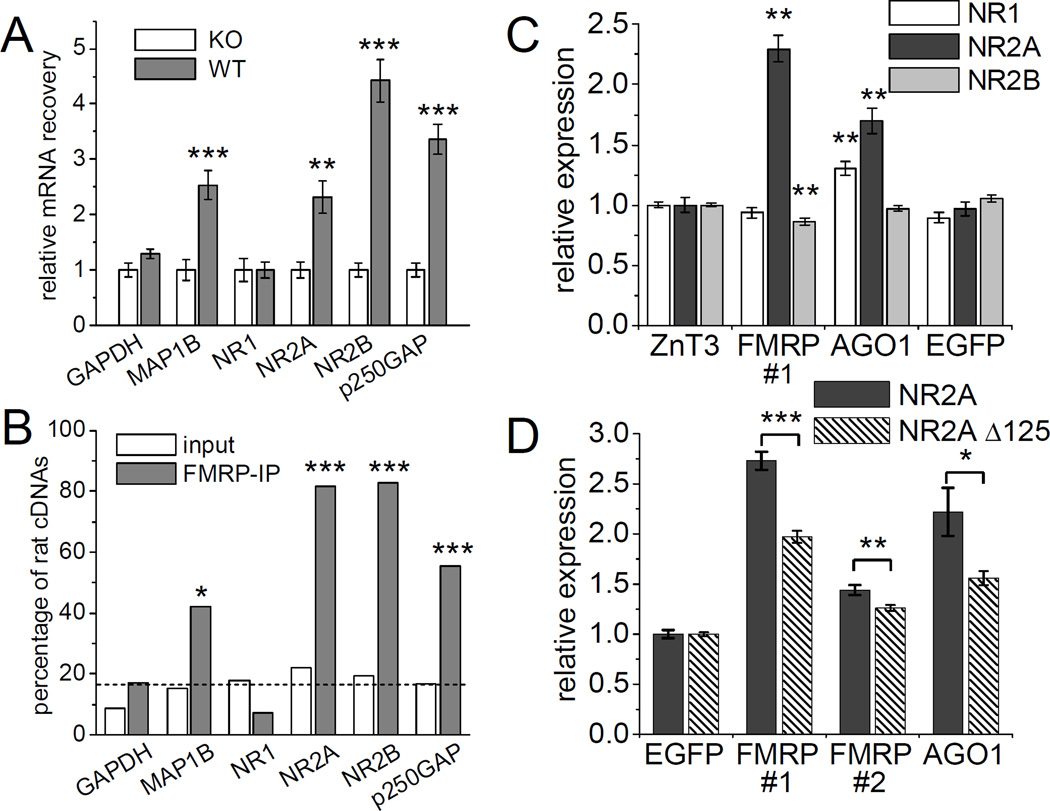
In the experiments above, we found that miR-125b is associated with FMRP, and that miR-125b regulates the expression of NR2A. So is the NR2A transcript an mRNA regulated by FMRP? To address this question, we performed quantitative RT-PCR to measure the amount of NR2A mRNA that was coimmunoprecipitated with FMRP. NR2A mRNA was 2.3-fold more abundant in anti-FMRP immunoprecipitates from wild-type mice compared to the background level immunoprecipitated from FMR1 KO mice, indicating specific association for NR2A mRNA with FMRP (Figure 8A). NR2A recovery with FMRP immunoprecipitation was similar to that measured here for MAP1B mRNA (2.5-fold), a reported FMRP target mRNA. Interestingly, NR2B mRNA was also enriched in FMRP immunoprecipitates from wild-type mouse brains (~4 fold over the background level from FMR1 KOs), even though NR2B is not a predicted target of any of the miRNAs we tested in this study. NR2B may be regulated by other miRNAs or associate with FMRP independent of miRNAs.
Figure 8. miRNA target genes associate with FMRP in mouse brain.
(A) Enrichment of specific mRNAs in FMRP-immunoprecipitates from wild-type mouse brain extracts relative to those derived from FMR1 KO mice (age 3 months) measured by RT-qPCR. RNA-samples are identical to those used in Figure 1. Statistical analysis by Student’s t-test: ** p < 0.01, ** p < 0.001. n = 6 immunoprecipitations from individual mice each group. (B) Enrichment of rat mRNAs in FMRP-immunoprecipitates from wild-type rat brain mixed with FMR1 KO mouse brain during homogenization.indicates in vivo interaction of FMRP with specific mRNAs (see text). Statistical analysis using Fisher’s exact test ** p < 0.01, *** p < 0.001. n = 29 to 45. Dashed line indicates the frequency (~17%) of rat cDNAs in the input material averaged among all tested genes. See also Supplemental Figure S7. (C, D) Hippocampal neurons (DIV4+3) were cotransfected with FF-luc 3’UTR reporters for the indicated NMDA receptor subunits as well as RR-luc and shRNA constructs targeting ZnT3, FMRP, AGO1 or EGFP (see Supplemental Figure S3). Graphs indicate expression of the reporter constructs normalized to the effect of each shRNA on a control FF-luc construct. Statistical analysis: (C) One-way ANOVA with Dunnett’s post test: * p < 0.05, ** p < 0.01. n = 24 to 72. (D) Student’s test comparing the response of both NR2A reporter variants: * p < 0.05, ** p < 0.01, *** p < 0.001. n = 18. See also Supplemental Figures S6, S7 and S8.
In contrast to NR2A and NR2B, the NR1 mRNA was not associated with FMRP by the coimmunoprecipitation assay (Figure 8A), nor was there coprecipitation of GAPDH mRNAs, a commonly used negative control that is believed not to be regulated by FMRP (Brown et al., 2001). However, we found p250GAP (which is reported to be regulated by miR-132) to be significantly associated with FMRP (Figure 8A). p250GAP had not been implicated in FXS previously.
To rule out artificial association between FMRP and mRNAs during immunoprecipitation, we utilized a novel co-mixing assay that confirms interaction in the intact brain. We homogenized wild-type rat brain together with ~5-fold excess FMR1 KO mouse brain and determined the species distribution of mRNAs in FMRP-immunoprecipitates using diagnostic restriction analysis of cloned cDNA fragments (Figure 8B and Supplemental Figure S7). Thus, only rat FMRP protein, but both rat and mouse mRNAs are present during immunoprecipitation. For mRNAs interacting with FMRP in vivo mainly rat mRNA should be found in the immunoprecipitation of FMRP. For GAPDH and NR1, which were not enriched in FMRP-immunoprecipitates (Figure 8A), the ratio of rat versus mouse cDNA clones was not significantly different in FMRP-immunoprecipitates and input material, indicating non-specific contamination with these mRNAs (Figure 8B). In contrast, for the positive control MAP1B and the putative FMRP-target genes NR2A, NR2B and p250GAP, the fraction of rat cDNA clones was much higher in the FMRP-immunoprecipitates than in the mixed rat/mouse brain extract. Thus, “species-tagging” confirmed that the interaction of MAP1B, NR2A, NR2B and p250GAP mRNA with FMRP occurred prior to homogenization in vivo.
Among the tested genes only NR2B showed a small change in mRNA level in the FMR1 KO (133+/−10% compared to 100+/−5% in the wild-type, p < 0.05), which is in keeping with the belief that FMRP mainly regulates the translation of its target genes rather than mRNA abundance. We therefore measured the protein levels of NMDA receptor subunits in hippocampus of FMR1 KO mice at 7 and 14 days after birth. Consistent with previous reports (Giuffrida et al., 2005), we found no significant difference in NR2A, NR2B or p250GAP total protein levels between wild-type and KO (Supplemental Figure S8).
Inappropriate development or compensatory mechanisms (e.g. on the level of transcription or degradation) might obscure dysregulation of NR2A in FMR1 KO mice (Yashiro and Philpot, 2008). Thus we used acute knockdown of FMRP in hippocampal neurons and FF-luc 3’UTR reporters to circumvent these confounding factors. Acute knockdown of FMRP using shRNA #1 strongly stimulated the expression of the NR2A 3’UTR reporter (by ~2.5 fold) compared to control shRNAs targeting zinc transporter 3 (ZnT3) or EGFP (Figure 8C). In contrast, FMRP knockdown did not affect the NR1 reporter and even slightly reduced NR2B reporter expression. These data suggest that FMRP has a suppressive action on the 3’ UTR of NR2A in neurons.
Because Argonaute proteins interacting with FMRP may contribute to miRNA-dependent translational repression of FMRP target genes, we asked whether knockdown of Argonaute 1 (AGO1) also affects the NR2A 3’UTR reporter. AGO1, which is highly expressed in the central nervous system, lacks endonuclease activity but supports translational repression of mismatch-containing miRNA targets (Filipowicz et al., 2008; Lu et al., 2005). Therefore we took advantage of an shRNA target sequence with validated specificity for AGO1 (Meister et al., 2004b). AGO1 knockdown increased expression of the NR2A 3’UTR reporter by ~70%, but had no effect on the NR2B reporter (Figure 8C). There was also a slight increase in NR1 3’UTR reporter expression with AGO1 suppression.
To test whether the NR2A 3’UTR is regulated by FMRP in a miR-125b-dependent manner, we compared regulation of NR2A reporter constructs containing or lacking the miR-125b target site. Basal translation of the NR2A Δ125 reporter was ~4-fold higher than wildtype, consistent with derepression due to loss of miR-125 binding. In fold terms, the NR2A Δ125 reporter was significantly less induced by FMRP knockdown than wild-type NR2A (Figure 8D), implying that miR-125 contributes in part to FMRP-dependent NR2A regulation. We attribute the differential effect of FMRP knockdown on NR2A wild-type versus NR2A Δ125 to a functional interaction between FMRP and miR-125 on the control of NR2A 3’UTR. Other mechanisms independent of miRNAs or dependent on other miRNAs likely exist to regulate NR2A expression. A second shRNA (FMRP #2) that is less effective in knocking down FMRP also induced the NR2A 3’UTR reporter dependent on the miR-125b target site, but to a weaker extent than shRNA FMRP #1 (Supplemental Figure S3 and Figure 8D). Together these data indicate that the 3’UTR of NR2A (but not NR1 or NR2B) is normally suppressed by FMRP and AGO1 as well as by miR-125, and these mechanisms show partial functional interaction.
Discussion
FMRP-associated miRNAs in brain
The interaction of FMRP with Argonaute proteins led to the idea that miRNAs could facilitate the selection and/or silencing of mRNA targets by FMRP (Caudy et al., 2002; Ishizuka et al., 2002; Jin et al., 2004b). We found that many brain miRNAs (12 out of 17 tested) are associated with FMRP. So far, enrichment of specific miRNAs (including miR-125) had only been reported in FMRP-immunoprecipitates from fly ovaries (Yang et al., 2009).
How do miRNAs interact with FMRP? The lack of a canonical miRNA/siRNA binding domain in FMRP suggests that miRNAs are indirectly associated with FMRP, possibly through Argonaute proteins which contain a miRNA binding domain and interact with FMRP in an RNA-independent manner (Caudy et al., 2002; Ishizuka et al., 2002). We found that AGO1 knockdown enhanced NR2A reporter expression similar to FMRP knockdown, which is consistent with the genetic interaction between FMRP and Argonaute found in flies (Bolduc et al., 2008; Jin et al., 2004b). Direct interaction with miRNA has been reported for purified FMRP in vitro (Plante et al., 2006). It is possible that miRNA and mRNA together could act as the “kissing complex” RNA structure proposed to bind the KH2 domain (Bassell and Warren, 2008; Darnell et al., 2005a). Our data cannot tell whether miRNAs associate with FMRP directly or indirectly (e.g. through independent binding of miRNA and FMRP to the same mRNA complex). Nor do they reveal the molecular mechanism by which miR-125b and FMRP collaborate to suppress NR2A 3’UTR-regulated translation.
The data that suggest a functional interaction of FMRP and specific miRNAs in neurons can be summarized as follows. First, knockdown of FMRP prevented the very different effects of overexpression of FMRP-associated miRNAs 125b and 132 on spine morphology. Second, removing the miR-125b target site from the NR2A 3’UTR dampens regulation of the latter by FMRP, implying that miR-125b assists FMRP in regulating NR2A. We emphasize that miRNAs 125b and 132 could also have multiple effects in neurons that are independent of FMRP.
Regulation of synapse structure and function by miR-125b and miR-132
Remarkably, miR-125b and miR-132 showed largely opposing effects on dendritic spines. Overexpression of miR-125b induced long narrow spines, which correlated with a reduction in mEPSC amplitude. Conversely, sponging of endogenous miR-125 increased the average width of dendritic protrusions. These results provide strong evidence that miR-125b regulates synapse structure and function. Interestingly, endogenous miR-125b levels are highest in young neurons (DIV3), when filopodia-like protrusions are predominant. Recent studies using gene ontology analysis of putative miR-125b targets suggests a role for this miRNA in neuronal differentiation and cytoskeletal organization (Chi et al., 2009; Le et al., 2009).
We identified NR2A as a novel target gene of miR-125b, but unlike NR1 and NR2B, this NMDAR subunit has so far not been linked to spine morphogenesis (Alvarez et al., 2007; Kim et al., 2005; Ultanir et al., 2007). NR2A overexpression did not rescue the effect of miR-125b overexpression on spine morphology (data not shown), suggesting NR2A downregulation is not a major cause of altered spine morphology. In reporter assays the EphA4 3’UTR was modestly repressed by miR-125b (Supplementary Figure S5). Because loss of EphA4 promotes the formation of filopodia-like protrusions in hippocampal neurons (Murai et al., 2003), it is possible that the suppression of EphA4 (together with other unidentified targets) contributes to the miR-125b overexpression phenotype.
In contrast to miR-125b, overexpressing miR-132 hippocampal neurons increased dendritic protrusion width and increased mEPSC amplitude. Recently, Siegel et al. (2009) reported that miR-132 inhibition caused a slight reduction in spine volume. In our hands sponging of miR-132 had little effect on dendritic spines, but caused a significant reduction in dendritic complexity. This is consistent with the positive effect of miR-132 on dendrite branching through repression of p250GAP, which has been described in younger neurons (Vo et al., 2005; Wayman et al., 2008). Knockdown of p250GAP has recently been shown to enlarge dendritic spines in mouse neurons (Nakazawa et al., 2008), which could help explain the increased spine width we observed upon miR-132 overexpression.
The fact that miR-125b and miR-132 associate with FMRP and affect dendritic spine morphology does not necessarily mean that these miRNAs play a key role in the spine phenotype of FXS. The complexity of this issue is highlighted by the opposing effects of miR-125b and miR-132 on spine size and shape. Indeed it is unlikely that a single or a few FMRP-associated miRNAs (or mRNAs) can fully explain the morphological and functional features of FXS
Regulation of NR2A by miR-125b and FMRP
The 3’ UTR of NR2A contains an evolutionarily conserved site that matches the miR-125b target sequence and that largely mediates suppression by endogenous miR-125b in neurons. We confirmed regulation of NR2A upon miR-125b overexpression and sponging by use of luciferase reporters, immunoblotting of endogenous protein levels and NMDAR-EPSC recordings. In addition to being controlled by miR-125, the NR2A 3’UTR reporter was also upregulated by knockdown of FMRP or AGO1. Importantly, deleting the major miR-125b target site within the NR2A 3’UTR impairs upregulation by FMRP knockdown, suggesting that miR-125b participates in FMRP regulation of NR2A. Consistent with NR2A mRNA being a target of FMRP, we found NR2A mRNA to be associated with FMRP in rodent brain. Together, these data suggest that NR2A translation may be regulated by miR-125b and FMRP in vivo.
We could not detect significant changes in total protein levels of NR1, NR2A, or NR2B in the hippocampus of FMR1 KO mice (Supplementary Figure S8). This is perhaps not surprising, given that FMR1 KO usually has subtle or no overt effects even on expression of “established” FMRP target genes (Bassell and Warren, 2008; Darnell et al., 2005b; Muddashetty et al., 2007). Moreover, FMRP may regulate its target mRNAs only at a specific subcellular location without altering total protein levels. Given that the NR2A/NR2B ratio appears to be an important determinant of NMDA receptor signaling, even subtle effects on NR2A expression by miRNAs might influence synaptic plasticity (Barria and Malinow, 2005; Kim et al., 2005; Philpot et al., 2007). It should be pointed out, however, that miR-125b and FMRP impact multiple targets and pathways, and altered expression of NR2A per se is unlikely to be the key mediator of miR-125b and FMRP effects on synapse structure and function.
What is known about NMDA receptor function in FMR1 KO mice? Loss of FMRP enhances NMDAR function in young (Pfeiffer and Huber, 2007; Pilpel et al., 2008), but not in adult mice (Pilpel et al., 2008; Zhao et al., 2005). Moreover, several forms of NMDA-receptor-dependent plasticity are impaired by loss of FMRP. LTP is abolished in cortex and amygdala (but not hippocampus), the threshold for spike-timing dependent plasticity is increased, and ocular dominance plasticity is altered (Desai et al., 2006; Dolen et al., 2007; Li et al., 2002; Meredith et al., 2007; Zhao et al., 2005). These are changes that could conceivably be due, in part, to altered NMDA receptor function or signaling at the synapse. Investigating NMDA receptor function in greater detail in FMR1 KO mice should be informative and may lead to novel intervention strategies for FXS treatment.
Experimental Procedures
Antibodies
NR1 (mouse, BD Bioscience), NR2A (rabbit, Covance), NR2B (mouse, Neuromab), PSD-95 (mouse, Neuromab), FMRP (rabbit, ab17722, Abcam), β-galactosidase (rabbit, ICN; mouse, Promega), EGFP (rabbit, MBL), β3-tubulin (mouse, Sigma), MAP1B (mouse, Santa Cruz), EphA4 (rabbit, Santa Cruz). p250GAP antibody (Nakazawa et al., 2003).
DNA Constructs
For miRNA expression in neurons, we cloned the complete miRNA precursor hairpin with ten nucleotide genomic context extending to the 5’ and 3’ side of the stem in each direction (to allow proper processing) using synthetic oligonucleotides (Supplemental Table S1) into a vector containing the chicken β-actin promoter (Zeng et al., 2005). These constructs were used in Supplemental Figure S1 and S2; Figure 2, 3, 5, 7B. A second generation of miRNA expression constructs based on the FhSynW vector (Nakagawa et al., 2006) allowed robust expression in HEK293 cells and lentiviral packaging. Genomic miRNA precursor sequences (165 to 376 nucleotide fragments) were amplified by PCR and cloned into the 3’UTR of mCherry (Shaner et al., 2004) driven by the human synapsin 1 promoter. These constructs were used for transfection (Supplemental Figures S4 and S5; Figure 6B) and lentiviral expression (Figure 7A). Viral particles were packaged using a third-generation system (Tiscornia et al., 2006).
For single cell analysis, EGFP-based miRNA sensors were constructed by cloning several perfect match miRNA target sites into the 3’UTR of a TK promoter-driven EGFP. mOrange (Shaner et al., 2004) was expressed from the same plasmid to identify transfected cells regardless of EGFP expression level. For quantification using luciferase assays, the miRNA target sites were subcloned into the 3’UTR of FF-luc (driven by human synapsin 1 promoter).
Specific miRNA sponges (Ebert et al., 2007) were constructed by concatemerizing annealed oligonucleotides containing a bulged-match miRNA binding site inside of two linkers, each containing restriction sites for cloning (see Supplemental Table 1). After ligation, the appropriate size concatemers were PCR-amplified and cloned (NotI-SalI) into the 3’UTR of mCherry cDNA driven by β-actin promoter.
3’UTRs fragments of putative miRNA targets (Supplemental Table S1) were cloned from rat brain cDNA into a modified pGL3-control vector (Promega). In neurons, NR1, NR2A and NR2B 3’UTR FF-luc reporter were expressed under the control of human synapsin 1 promoter to boost FF-luc activity.
shRNA targeting FMRP (rat and mouse, #1 GTGATGAAGTTGAGGTTTA and #2 CCACCAAATCGTACAGATA), AGO1 (rat and human, GAGAAGAGGTGCTCAAGAA) (Meister et al., 2004a), Luc (CGTACGCGGAATACTTCGA) (Zhang and Macara, 2006), ZnT3 (rat and mouse, GCCTCATCCCGGCTCTATT, gift from J. Jaworski) and EGFP (GCAAAGACCCCAACGAGAA, gift from J. Jaworski) at the indicated sites were cloned into pSUPER (Oligoengine). All constructs were verified by DNA-sequencing.
FMRP-IP, RNA extraction, reverse transcription and quantitative PCR
We followed the standard procedure for FMRP immunoprecipitation used to identify mRNA targets (Brown et al., 2001). Mouse brains were homogenized in 2 ml ice cold lysis buffer (10 mM Hepes, pH 7.4, 200 mM NaCl, 30 mM EDTA, 0.5% Triton X-100, 1 U/µl SUPERase inhibitor, protease and phosphatase inhibitor cocktail 1:100). After removing the nuclear fraction (3000 g, 10 min), the NaCl concentration was raised to 400 mM to dissociate FMRP from ribosomes and insoluble material was removed (70,000 g, 20 min). After adding 100 µg/ml yeast tRNA (Sigma) to block non-specific RNA binding, the extracts were precleared with Protein A-Sepharose (30 min at 4°C). One part of the extract was saved and ten parts were used for immunoprecipitation with FMRP antibodies (15 µg with 40 µl Protein A-Sepharose slurry) for two hours at 4°C. Beads were washed five times with lysis buffer containing 0.1 U/ml SUPERase inhibitor and 10 µg/ml yeast tRNA (for the first 4 washes only). Bound RNA was extracted with mirVana miRNA isolation kit (Applied Biosystems) and subjected to real-time RT-PCR using TaqMan assays to quantify mature miRNAs (Applied Biosystems) and SYBR green PCR (Applied Biosystems) for mRNA quantification (see Supplemental Table 1 for oligonucleotides). For the latter, RNA was reverse transcribed using random hexamer primers with the Message Sensor kit (Applied Biosystems). miRNA recovery was calculated as the ratio of miRNA found in FMRP-immunoprecipitation compared to brain extracts and normalized to spliceosomal RNA U6 (also quantified using a TaqMan assay) to compensate for unspecific RNA pulldown. mRNA recovery was calculated accordingly and normalized to PGK1, all quantified by SYBR green PCR.
Analysis of “species-tagged” mRNA in FMRP-immunoprecipitates
Half a rat brain (containing FMRP-bound mRNA) was homogenized together with four FMR1 KO mouse brains (lacking FMRP). An aliquot of the resulting lysate and the FMRP immunoprecipitate (prepared as above) was subjected to RNA extraction and reverse transcription. Specific gene fragment were PCR-amplified until saturation using bi-specific primers matching rat and mouse mRNAs (50 cycles using taq, Sigma). PCR-products were TOPO-cloned into pCR4 vector (Invitrogen). After transformation the inserts from 48 individual colonies were PCR-amplified (with backbone-specific primers CACACAGGAAACAGCTATGACCATG and GACGTTGTAAAACGACGGCCAGTG). 5 µl of the PCR reaction was analyzed using species-specific restriction enzymes for GAPDH (rat MboI, mouse HindIII), MAP1b (rat BanII, mouse MboII), NR1 (rat BanII, mouse PflFI), NR2A (rat MboI, mouse BsrBI), NR2B (rat MspI, mouse BsaHI) or p250GAP (rat MboII, mouse HinP1I).
Neuron culture, transfection and immunostaining
Hippocampal neurons were cultured from embryonic day 19 rat embryos as described previously (Tada et al., 2007). After transfection (Lipofectamine 2000, Invitrogen) neuron morphology was visualized by cotransfected EGFP or β-galactosidase. Confocal microscopy and analysis of neuron morphology were as described (Tada et al., 2007). Image acquisition and analysis were performed blind to the experimental conditions.
Electrophysiology
Miniature synaptic events from sister cultures of hippocampal neurons cotransfected with EGFP and miRNA overexpression or sponge constructs were recorded and analyzed as described (Seeburg et al., 2008). Organotypic hippocampal slice cultures from postnatal day 8 rats were prepared, cultured and biolistically transfected (DIV11+4) as described (Futai et al., 2007). Evoked NMDA receptor currents from CA1 pyramidal neurons were recorded at room temperature (Futai et al., 2007).
Luciferase Assays
HEK293 cells were cotransfected with FF-luc 3’UTR reporters and constructs expressing RR-luc, miRNA and EGFP at a DNA ratio of 40:40:80:10 using Lipofectamine 2000. All experiments were performed in 96-well plates with six replicates for each condition. Two days after transfection luciferase activity was quantified using Dual-Glo Luciferase Assay (Promega). Relative expression of reporter constructs was determined by normalizing the ratio of FF-luc and RR-luc activity to a control miRNA (let-7c or miR-124) and the effect of each miRNA on a control FF-luc reporter (not containing the heterologous 3’UTR). Similarly, hippocampal neurons (DIV4) were cotransfected with plasmids expressing shRNA or sponges, FF-luc reporters, RR-luc and EGFP at a DNA ratio of 60:40:40:10 using Lipofectamine 2000 and analyzed three days later as above.
FMR1 KO mice and quantitative immunoblotting
FMR1 KO mice (Dutch–Belgian Fragile X Consortium, 1994) in C57/B6 background were obtained from S. Tonegawa. Mouse housing and euthanasia were performed in compliance with federal and institutional guidelines. For each experiment age-matched male mice, if possible from the same litter, were used. Hippocampi were homogenized in 0.32 M sucrose, 4 mM HEPES (pH 7.4) together with protease and phosphatase inhibitor mix (Sigma). Immunoblots were quantified by densitometry and normalized to loaded protein (SYPRO Ruby, Invitrogen).
Supplementary Material
Acknowledgments
We are grateful for brain tissue from FMR1 KO mice provided by S. Tonegawa. We thank G. Bassell, J. Jaworski, T. Nakazawa, R. Tsien and T. Yamamoto for reagents. We thank B. Bingol, Y. Chen, Y. Hayashi, H. Hsin, C. Nelson and L.-H. Tsai for critical reading of the manuscript. D.E. received a postdoctoral fellowship from Deutsche Forschungsgemeinschaft (ED157/1). J.R.N. and P.A.S. were supported by NCI PO1-CA42063 to P.A.S. and partially by NCI P30-CA14051 Cancer Center Support (Core) Grant. J.R.N. was additionally supported by NCI K99-CA131474. M.S. was Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alvarez VA, Ridenour DA, Sabatini BL. Distinct structural and ionotropic roles of NMDA receptors in controlling spine and synapse stability. J Neurosci. 2007;27:7365–7376. doi: 10.1523/JNEUROSCI.0956-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24:2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. doi: 10.1016/j.neuron.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc FV, Bell K, Cox H, Broadie KS, Tully T. Excess protein synthesis in Drosophila fragile X mutants impairs long-term memory. Nat Neurosci. 2008;11:1143–1145. doi: 10.1038/nn.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever A, Ceman S. Phosphorylation of FMRP inhibits association with Dicer. Rna. 2009;15:362–366. doi: 10.1261/rna.1500809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci U S A. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005a;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Mostovetsky O, Darnell RB. FMRP RNA targets: identification and validation. Genes Brain Behav. 2005b;4:341–349. doi: 10.1111/j.1601-183X.2005.00144.x. [DOI] [PubMed] [Google Scholar]
- Desai NS, Casimiro TM, Gruber SM, Vanderklish PW. Early postnatal plasticity in neocortex of Fmr1 knockout mice. J Neurophysiol. 2006;96:1734–1745. doi: 10.1152/jn.00221.2006. [DOI] [PubMed] [Google Scholar]
- Dolen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56:955–962. doi: 10.1016/j.neuron.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch-Belgian Fragile X Consortium. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell . 1994;78:23–33. [PubMed] [Google Scholar]
- Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10:837–841. doi: 10.1038/nrn2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Futai K, Kim MJ, Hashikawa T, Scheiffele P, Sheng M, Hayashi Y. Retrograde modulation of presynaptic release probability through signaling mediated by PSD-95-neuroligin. Nat Neurosci. 2007;10:186–195. doi: 10.1038/nn1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffrida R, Musumeci S, D’Antoni S, Bonaccorso CM, Giuffrida-Stella AM, Oostra BA, Catania MV. A reduced number of metabotropic glutamate subtype 5 receptors are associated with constitutive homer proteins in a mouse model of fragile X syndrome. J Neurosci. 2005;25:8908–8916. doi: 10.1523/JNEUROSCI.0932-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004a;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004b;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A-and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46:745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. microRNA-125b Promotes Neuronal Differentiation in Human Cells by Repressing Multiple Targets. Mol Cell Biol. 2009 doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li J, Pelletier MR, Perez Velazquez JL, Carlen PL. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Mol Cell Neurosci. 2002;19:138–151. doi: 10.1006/mcne.2001.1085. [DOI] [PubMed] [Google Scholar]
- Lu J, Qian J, Chen F, Tang X, Li C, Cardoso WV. Differential expression of components of the microRNA machinery during mouse organogenesis. Biochem Biophys Res Commun. 2005;334:319–323. doi: 10.1016/j.bbrc.2005.05.206. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. Rna. 2004a;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004b;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Meredith RM, Holmgren CD, Weidum M, Burnashev N, Mansvelder HD. Increased threshold for spike-timing-dependent plasticity is caused by unreliable calcium signaling in mice lacking fragile X gene FMR1. Neuron. 2007;54:627–638. doi: 10.1016/j.neuron.2007.04.028. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ. Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci. 2007;27:5338–5348. doi: 10.1523/JNEUROSCI.0937-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nat Neurosci. 2003;6:153–160. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Feliu-Mojer MI, Wulf P, Lois C, Sheng M, Hoogenraad CC. Generation of lentiviral transgenic rats expressing glutamate receptor interacting protein 1 (GRIP1) in brain, spinal cord and testis. J Neurosci Methods. 2006;152:1–9. doi: 10.1016/j.jneumeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Kuriu T, Tezuka T, Umemori H, Okabe S, Yamamoto T. Regulation of dendritic spine morphology by an NMDA receptor-associated Rho GTPase-activating protein, p250GAP. J Neurochem. 2008;105:1384–1393. doi: 10.1111/j.1471-4159.2008.05335.x. [DOI] [PubMed] [Google Scholar]
- Nakazawa T, Watabe AM, Tezuka T, Yoshida Y, Yokoyama K, Umemori H, Inoue A, Okabe S, Manabe T, Yamamoto T. p250GAP, a novel brain-enriched GTPase-activating protein for Rho family GTPases, is involved in the N-methyl-d-aspartate receptor signaling. Mol Biol Cell. 2003;14:2921–2934. doi: 10.1091/mbc.E02-09-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J Neurosci. 2007;27:3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpot BD, Cho KK, Bear MF. Obligatory role of NR2A for metaplasticity in visual cortex. Neuron. 2007;53:495–502. doi: 10.1016/j.neuron.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Kolleker A, Berberich S, Ginger M, Frick A, Mientjes E, Oostra BA, Seeburg PH. Synaptic ionotropic glutamate receptors and plasticity are developmentally altered in the CA1 field of FMR1 KO mice. J Physiol. 2008 doi: 10.1113/jphysiol.2008.160929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante I, Davidovic L, Ouellet DL, Gobeil LA, Tremblay S, Khandjian EW, Provost P. Dicer-Derived MicroRNAs Are Utilized by the Fragile X Mental Retardation Protein for Assembly on Target RNAs. J Biomed Biotechnol. 2006;2006:64347. doi: 10.1155/JBB/2006/64347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, Sheng M. Role of Septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- Ultanir SK, Kim JE, Hall BJ, Deerinck T, Ellisman M, Ghosh A. Regulation of spine morphology and spine density by NMDA receptor signaling in vivo. Proc Natl Acad Sci U S A. 2007;104:19553–19558. doi: 10.1073/pnas.0704031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Xu S, Xia L, Wang J, Wen S, Jin P, Chen D. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009;5:e1000444. doi: 10.1371/journal.pgen.1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cai X, Cullen BR. Use of RNA polymerase II to transcribe artificial microRNAs. Methods Enzymol. 2005;392:371–380. doi: 10.1016/S0076-6879(04)92022-8. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- Zhao MG, Toyoda H, Ko SW, Ding HK, Wu LJ, Zhuo M. Deficits in trace fear memory and long-term potentiation in a mouse model for fragile X syndrome. J Neurosci. 2005;25:7385–7392. doi: 10.1523/JNEUROSCI.1520-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.