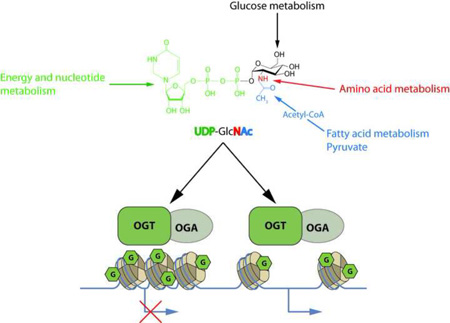
Abstract
O-GlcNAcylation is a dynamic post-translational modification that is responsive to nutrient availably via the hexosamine biosynthetic pathway and its endproduct UDP-GlcNAc. O-GlcNAcylation serves as a nutrient sensor to regulate the activities of many proteins involved in nearly all biological processes. Within the last decade, OGT, OGA and O-GlcNAcylation have been shown to be at the nexus of epigenetic marks controlling gene expression during embryonic development, cell differentiation, in the maintenance of epigenetic states and in the etiology of epigenetic related diseases. OGT O-GlcNAcylates histones and epigenetic writers/erasers, and regulates gene activation, as well as gene repression. Here, we highlight recent work documenting the important roles O-GlcNAcylation and its cycling enzymes play in the nutrient regulation of epigenetic partners controlling gene expression.
Graphical Abstract
Introduction
Can what we eat change our genetics? Beside toxic compounds that have the ability to induce mutations, our eating habits don’t change the nucleotide sequence of our genes. However, organisms with a predefined set of genes have to maintain proper homeostasis and adapt to their environment, especially to adapt to nutrient availability. Phenotype adaptation is a long-term adjustment to the environment and can be done by heritable encoded information on DNA without changes in the gene sequence [1]. This second layer of information is called epigenetics and includes DNA methylation, post-translational modifications (PTMs) of histones and chromatin remodeling. Epigenetics is also an important feature of embryogenesis and cell fate, controlling and defining transcriptional pattern crucial for cellular lineage.
The first evidence that link O-GlcNAcylation to chromatin and transcription was found in Drosophila [2]. O-GlcNAcylation is a versatile PTM controlled by two non-redundant enzymes: the O-GlcNAc transferase (OGT) transfers the GlcNAc moiety from UDP-GlcNAc to a serine or a threonine residue, while the O-GlcNAcase (OGA) removes the modification. UDP-GlcNAc is a main cellular nutrient sensor since its synthesis through the hexosamine biosynthetic pathway (HBP) depends on flux through every major metabolic pathway (graphical abstract). Since OGT’s enzymatic activity and substrate specificity varies according UDP-GlcNAc concentration, variation in metabolism that feed the HBP have profound effects on protein O-GlcNAcylation [3]. Within the last decade, studies have defined O-GlcNAcylation as an epigenetic mark and linked its cycle to the regulation of chromatin modifications.
Multiple roles of histone O-GlcNAcylation
The histone code is written by molecular complexes that add or remove part of the code in response to various cellular stimuli or metabolism. Although a recent paper called into question histone O-GlcNAcylation [4], the presence of the sugar on each subunit of the nucleosome has been reported independently by many laboratories and some sites have been mapped (reviewed in [2]). Some of the site-specific functions have been documented (Figure 1).
Figure 1. Nucleosome O-GlcNAcylation.
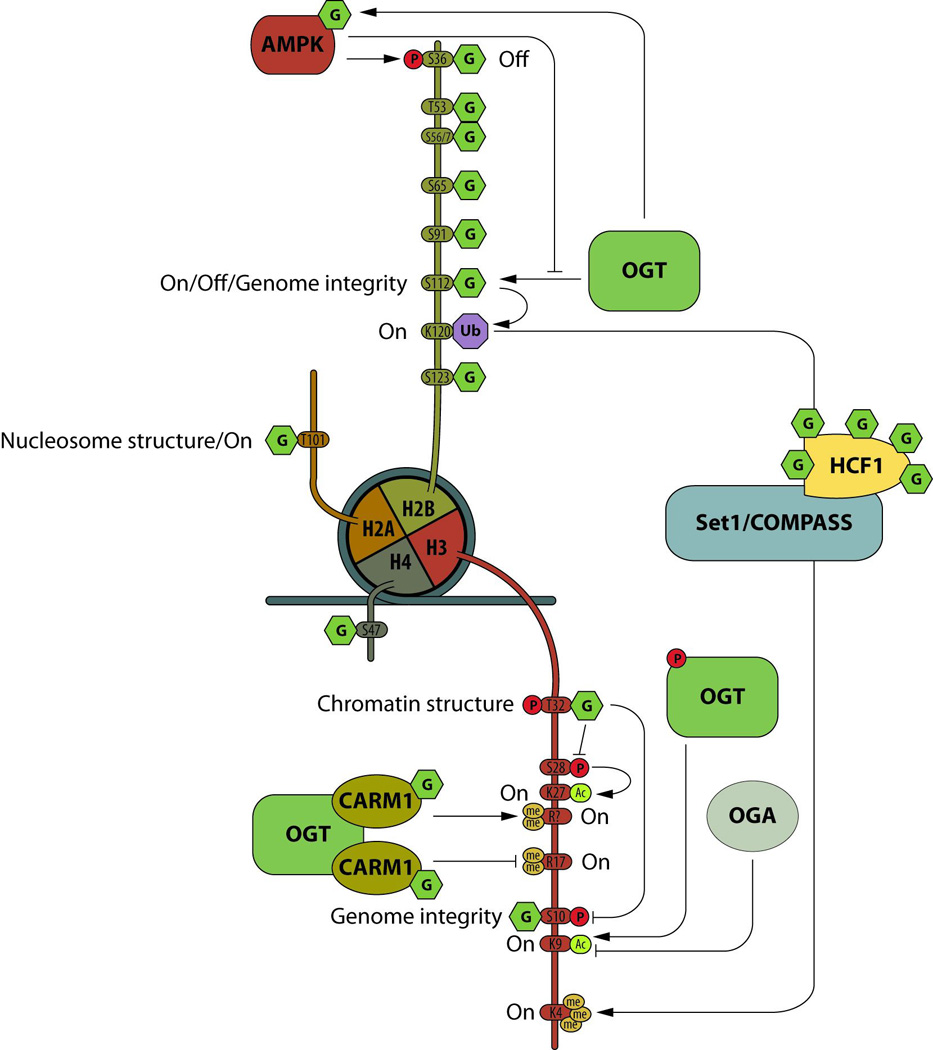
The O-GlcNAcylation (G) of the histone core is extensive and interplays with other PTMs, such as phosphorylation (P), methylation (me), acetylation (Ac), or ubiquitination (Ub). All four subunits of the nucleosome are modified by the sugar, and site specific O-GlcNAcylation is involved in gene transcription activation (on) or repression (off), chromatin structure or genomic stability.
The O-GlcNAc/phosphorylation interplay on histone H3 is essential for mitosis. Overexpression of OGT reduces phosphorylation of H3S10 and leads to errors in chromosomal segregation, while OGA inhibition impairs G2-M transition [5,6]. The H2BS112O-GlcNAc mark is associated with DNA damage response and genomic stability [7]. O-GlcNAcylation at H2BS112 is increased at DNA double strand breaks. Down-regulation of OGT or H2BS112A mutant over-expression impairs homologous repair (HR) and non-homologous end joining. Since H2BS112O-GlcNAc stimulates H2BK120 ubiquitination that activates the ring finger protein 20 [8], OGT and O-GlcNAcylation could be key initiators for the recruitment of the HR complex in response to DNA damage.
Histone O-GlcNAcylation is linked to gene transcription. The sugar at T101 of H2A destabilized H2A/H2B dimmers in the nucleosome, promoting an open chromatin state [9]. This suggests that O-GlcNAcylation at H2AT101 would lower the barrier for RNA polymerase passage and hence increase transcription. H2B O-GlcNAcylation at S112 has been reported to have multiple roles. In HepG2 cells, activated AMPK phosphorylates OGT, which lowers H2BS112 O-GlcNAcylation and inhibits expression of genes regulated by H2BS112O-GlcNAc [10]. In HeLa cells, H2BS112 O-GlcNAcylation co-localizes with H2BK120Ub mark. The H2BK120Ub mark acts as a platform for the SET1/COMPASS complex that stimulates H3K4 trimethylation and gene transcription. Conversely, H2B O-GlcNAcylation is a stable chromatin landmark during adipocyte differentiation [11]. Ronningen et al. identified long H2BS112O-GlcNAc enriched domains, called GADs, ranging from 60kb to about 10Mb. At the early stage of adipogenesis, lamin-associated domains rearrange following GADs pattern, releasing the repression of genes mainly related to metabolic processes, but repressing genes within GADs [11], suggesting a repressive role for H2BS112O-GlcNAc in cell fate.
While yeast apparently lack O-GlcNAcylation and O-GlcNAcylation enzymes, it was recently reported that O-Man glycosylation of nuclear and cytoplasmic proteins mirror mammals O-GlcNAcylation [12]. A peptide covering the K123 of yH2B (ubiquitination of yH2BK123 is homologue of the mammalian H2BK120Ub) is O-Man glycosylated. Considering that glucose metabolism increases both mammalian H2BK120ub and yeast H2BK123Ub [13,14], O-Man glycosylation of yH2B could mimic the molecular mechanism of H2BK120 ubiquitination mediated by H2BS112O-GlcNAc observed in mammals.
As mentioned above, discrepancies and lack of reproducibility regarding Histone O-GlcNAcylation have been observed recently [4]. These issues could arise because investigations have used different models and different techniques to assess histone O-GlcNAcylation. As different studies have suggested, some of if not all histone O-GlcNAcylation could be cell type specific, highly dynamic and cell cycle stage specific and/or restricted to a specific developmental stage [5,6,7,11]. Although the H2BS112O-GlcNAc site-specific antibody is commercially available, some concerns have been raised regarding its specificity [2,4]. Re-analysis of five randomly picked loci in the top 20 hits of the H2BS112-O-GlcNAc ChIP-seq data from Fujiki et al., using this antibody by Gambetta et al., has pointed out that, except for the locus of GSK3B, the signal over background of theses specific loci is low and might not be of sufficient quality to justify the authors’ conclusions. In general, the conditions to work with O-GlcNAc site-specific antibodies, which often have low affinities, are very precise and specific due to the nature of the antigen and the lack of counter selection (i.e. antibody must recognize both peptide backbone and GlcNAc moiety but none of them separately). It is essential when developing and using O-GlcNAc site-specific antibodies, that the specificity be well documented and as many details as possible should be provided in the methods section when publishing with these antibodies in order to ensure reproducibility.
OGA, a histone acetyltranferase?
The histone acetyltransferase (HAT) activity of OGA has been controvertial. Its putative HAT domain is located in the C-terminal domain, while the O-GlcNAcase activity resides in the N-terminal domain of the molecule. Although our lab was not able to observe this activity in vitro [15], others have reported it in different publications. Toleman et al. have shown that mouse OGA acetylates histones in vitro and mutations within the C-terminal domain lead to substantial loss of its HAT activity [16]. Hayakawa et al. over-expressed OGA in emybryonic stem cells (ESC) and observed a decreased HAT activity toward H3K14 and H4K8 upon Thiamet-G treatment or with the expression of OGAD175A, which also showed a significant decrease in O-GlcNAcase activity [17], suggesting that O-GlcNAcase activity is required for HAT activity in vivo. Conversely, a structural study of the HAT domain of human OGA has shown that it shouldn’t be able to bind Acetyl-CoA [18]. Since recombinant OGA purified from bacteria acetylated H3K14 and H4K8 but only when pre-incubated with mammalian cell lysates [16], it is then possible that the observed HAT activity could be due to interaction and activation with a third HAT. Nevertheless the HAT domain appears to be important and could act as a scaffold for HAT interactions and OGA function.
OGA function is fundamental during embryogenesis since its genetic disruption leads to nearly complete neonatal lethality with developmental delay [19,20]. At the cellular level, OGA KO induces increased O-GlcNAcylation and leads to mitotic defects associated with cytokinesis failure and binucleation, accompanied by increased lagging chromosome and micronuclei formation [19]. The antagonistic relationship between OGT and OGA toward the histone code is likely important for the regulation and the maintenance of genomic stability, especially considering that H2BS112 O-GlcNAcylation is associated with and facilitates DNA homologous recombination, as well as the essential de-GlcNAcylation of H3 for proper mitosis [5,6].
OGT and O-GlcNAcylation regulate epigenetic marks
Much evidence strongly suggest that nutrients impact epigenetic modifications of chromatin [21] and recent publications have highlighted the important role of the nutrient sensors OGT and O-GlcNAcylation modulating chromatin marks.
Nutrient and energy sensor interplay
The energy sensor 5’-AMP-Activated Protein Kinase (AMPK), phosphorylates OGT at T444 and targets it to the nucleus, which is correlated with increased H3K9Ac in C2C12 myoblast cells. H3K9Ac levels are significantly decreased with nutrient/growth factor deprivation and increased with an AMPK activator [22]. Phosphorylation of OGT inhibits OGT:chromatin association, histone O-GlcNAcylation and gene transcription. Using a KO/KI strategy, OGT phosphorylation by AMPK was shown to impair H2BS112 O-GlcNAcylation [10]. Phosphorylation of H2B at S36 by AMPK is essential for transcription and survival in response to metabolic stress [23]. Since H2BS36 is also a target of OGT [24], O-GlcNAc/phosphorylation interplay may occur at H2BS36 in a responsive to nutrient/energy availability manner. Also, since phosphorylation of OGT at T444 by AMPK affects OGT’s substrate selectivity [22] and O-GlcNAcylation of AMPK positively regulates its activity [10], the crosstalk between both enzymes is not limited to site-specific competition but also results from a regulatory feedback loop (figure 1).
Coactivator-associated arginine methyltransferase 1 (CARM1) is regulated by O-GlcNAcylation (Figure 1). CARM1 catalyzes the methylation of the histone H3 at R2, R17 and R36. In diabetes, CARM1 activates expression of key genes involved in gluconeogenesis and glycogen metabolism. Over-expression of OGT decreases H3R17me2 [5], suggesting that O-GlcNAcylation interplays with histone marks via CARM1. Although O-GlcNAcylation does not appear to regulate the methyltransferase activity of CARM1, it does appear to control its substrate specificity since enriched O-GlcNAcylated CARM1 isoform methylates a different set of targets compared to the non-O-GlcNAcylated enzyme [25].
OGT, Tet proteins and 5hmC
Ten-eleven translocation (Tet) family proteins are enzymes that catalyze oxidation of 5-methylcytosine (5mC) in DNA to produce 5-hydroxymethylcytosine (5hmC). 5hmC is predominant at transcription start sites and on promoters of genes with bivalent chromatin, harboring activating and repressive marks, or throughout the bodies of transcriptionally active genes. Simultaneous active and repressive marks, i.e. H3K4me3 and H3K27me3 respectively, are an important feature that controls gene expression supervising cell fate and differentiation. Tet1, Tet2 and Tet3 interact with OGT [3] and are extensively O-GlcNAcylated. Tet1 could have at-least 6 O-GlcNAcylation sites, while Tet2 and Tet3 could have at-least 20 O-GlcNAcylated serine or threonine moieties, some of which are in competition with phosphorylation [26].
Knockdown of OGT reduces Tet1 protein levels [27] and 5hmC on its targets [27,28], while a Tet1 mutant at the T535 putative O-GlcNAcylation site is no longer stabilized by OGT over-expression [27]. However, O-GlcNAcylation of Tet3 is responsive to increased glucose concentration, which was correlated with the nuclear export of the proteins and therefore associated with a decrease of 5hmC catalyzed by Tet3 [29], suggesting that the increased glucose effect on DNA demethylation via O-GlcNAcylation could be restricted to Tet1. It was also reported that, although Tet1 and Tet2 are O-GlcNAcylated, neither their localization nor their activity was affected by the sugar [29]. Since the authors of these studies have used different models, the regulation of Tet proteins’ activity by O-GlcNAcylation could be cell/tissue specific.
O-GlcNAcylation associated with gene activation (figure 2)
Figure 2. OGT and O-GlcNAcylation are key regulators of transcription activation.
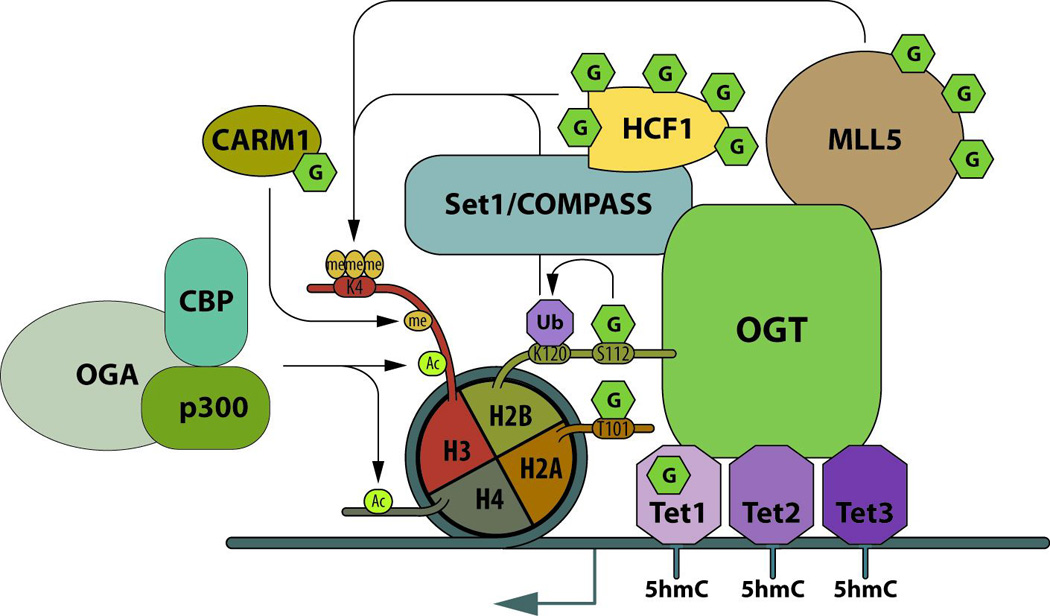
OGT is targeted to chromatin by Tet proteins, where OGT and O-GlcNAcylation regulates the conversion of 5mC to 5hmC. At promoters, OGT is a scaffold at the center of epigenetic regulators and O-GlcNAcylation of some of these proteins regulates their activity leading to gene activation. At gene specific promoters, OGA is recruited to the chromatin, where it forms a complex with the histone methyltransferases: p300 and CBP. Acetylation of H3 and H4 are mainly associated with gene activation. O-GlcNAcylation: G; methylation: me; ubiquitination: Ub; and acetylation: Ac.
Tet proteins target OGT to chromatin and O-GlcNAcylation of chromatin proteins is decreased upon Tet protein knockdown [30–32]. At gene specific loci, Tet3 targets OGT to GlcNAc transferase 9 (GnT-XI) promoters and the complex acts as a dock for the recruitment of NeuroD1, required for the expression of GnT-XI in a brain-specific manner [33]. Chip-seq analyses show that Tet proteins interact with OGT at the transcription start site (TSS) near CpG-rich regions and co-localized with the H3k4me3 active mark [28,30,31]. Tet2 and Tet3 target OGT to TSS in association with Host Cell Factor C1 (HCF-1) (a member of the SETI/COMPASS complex). Both Tets and OGT activity favor integrity of SET1/COMPASS and recruitment of SET Domain containing 1A to chromatin, which favors tri-methylation of H3K4 at target promoters [30]. Tet2 drives OGT to chromatin and enhances H2BS112 O-GlcNAcylation [31]. O-GlcNAcylation of H2B increases its ubiquitination at K120 that stimulates H3K4 tri-methylation through the SETI/COMPASS complex. OGT:HCF-1 also form a complex with Lysine-(K)-Specific Methyltransferase 2E (MLL5), the enzyme that catalyses the mono- and di-methylation of H3K4. The complex OGT:HCF-1:MLL5 is recruited to E2F transcription factor 1 (E2F1)-responsive promoters and stimulates H3K4 trimethylation at the promoters, and promotes activation of E2F1 target genes during G1/S transition [34]. OGT stabilizes and heavily O-GlcNAcylates MLL5 [35]. Since deep sequencing data show that MLL5 preferentially binds at TSS and CpG rich regions, suggesting a link with DNA methylation, OGT could be a scaffold protein for Tet proteins, SET1/COMPASS and MLL5. In Drosophila, dmOGT is encoded by the polycomb group gene: scx, and, like in mammals, its knockout is lethal at the pharate stage. Over-expression of dmOGTWT, as well as dmOGTD955A (a catalytic dead mutant), in scx1/scx6 genetic background rescue the lethal phenotype [36], which strongly suggests that not only O-GlcNAc transferase activity is important for embryonic development but the protein itself is as well. All together, these studies indicate crucial roles of Tet proteins for targeting OGT to chromatin in order to catalyze chromatin modification favorable to transcription activation.
O-GlcNAcylation associated with gene repression (figure 3)
Figure 3. Gene transcription repression by OGT and O-GlcNAcylation.
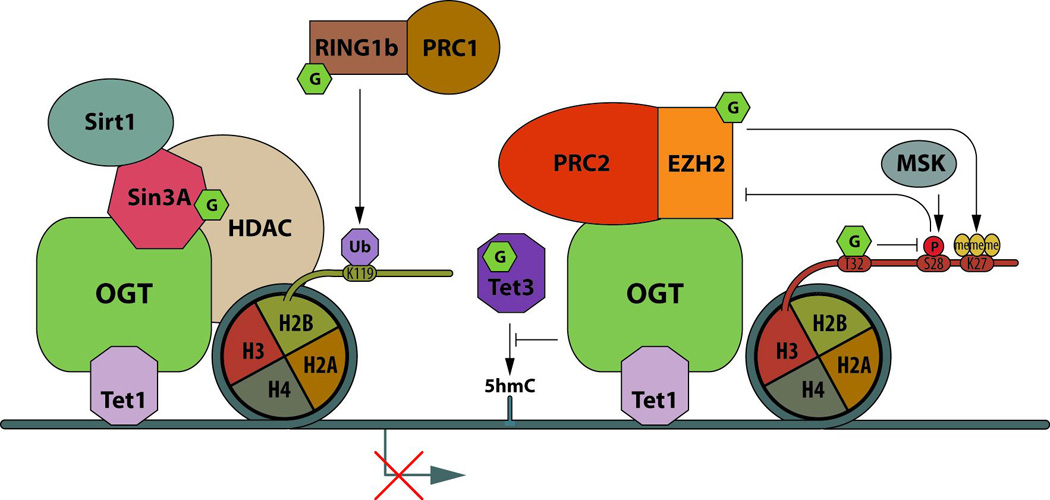
OGT is targeted to gene promoters by Tet1. Upstream of the TSS (around −39bp, near the area where the TFIID complex is recruited to DNA), OGT:Tet complex recruits the transcription repressor sin3A, as well as the histone deacetylases, HDAC1 and Sirt1. Downstream of the TSS (around +455bp), OGT:Tet1 are associated with the PRC2 complex. O-GlcNAcylation of the methyltransferase subunit of PRC2, EZH2, enhances its activity by stabilization of the protein. O-GlcNAcylation of RING1b (the catalytic subunit of the PRC1 complex) also increases its activity leading to H2BK119 ubiquitination. All of these PMTs crosstalk and converge to result in gene transcription repression.
Catalytic subunits of polycomb repressive complex 1 and 2 (PRC1 and PRC2), which are required for maintenance of repression of homeotic genes during embryonic development, cell proliferation and differentiation, are regulated by O-GlcNAcylation. In hESC, the protein Ring Finger Protein 2 (RING1b; a subunit of the PRC1 complex that catalyzes H2BK119 ubiquitination, a histone mark associated with transcription repression) is O-GlcNAcylated at T250 and/or T251 and S278. Non-O-GlcNAcylated RING1b complex is enriched at the promoter of genes mainly related to metabolism and cell cycle, while O-GlcNAcylated RING1b complex is enriched at promoters of genes mainly related to neural differentiation process [37], suggesting that O-GlcNAcylation targets the PRC1 complex to specific sets of genes.
During neuronal differentiation, an epigenetic switch at the locus of the orexin gene (Hcrt), involving DNA methylation and histone acetylation, leads to the generation of orexin producing neurons. At the locus Hcrt, in its inactive state, O-GlcNAcylation and OGT co-localize with Sirtuin 1 (Sirt1), with SIN3 Transcription Regulator Family Member A (Sin3A) and with Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit (EZH2), a subunit of PRC2 complex that carries a H3K27 methyltransferase activity. In contrast, OGA, E1A Binding Protein p300 (p300) and CREB Binding Protein (CBP) are present at the active Hcrt locus [17] (Figure 2). Interestingly, Sin3A and Histone Deacetylase 1 (HDAC1), thought to functionally repress transcription in parallel with histone deacetylation, are part of the complex formed by OGT, Tet1 and Tet3 [27,28,30]. At bivalent promoters that are considered to be poised for expression of developmental genes, Tet1 is enriched with Sin3A or with PRC2 about 39bp upstream and 455bp downstream of the TSS, respectively [38]. Considering that OGT stabilizes EZH2 by O-GlcNAcylation at S75, which promotes H3K27me3 [39], Tet1:OGT would lead to repress transcription via sin3A:HDAC1:Sirt1 and PRC2:EZH2 at both upstream and downstream TSS. Remarkably, the kinase MSK phophorylates H3S28 in the presence of H3K27me3, which leads to the dissociation of the PRC2 complex and gene transcription without removal of the tri-methyl mark on residue 27 of H3 [40]. Owing to the fact that the O-GlcNAcylation of H3S32 impairs phosphorylation at S28, OGT activity could act at multiple levels to lock transcription in an “off” state at specific loci.
Conclusions and future outlook
It is clear that O-GlcNAcylation and its cycling enzymes are important regulators of epigenetics. The sugar itself is a chromatin mark and its interplay with other PTMs to histones establishes a specific pattern recognized by protein complexes to activate or repress gene expression. Deregulation of O-GlcNAcylation, observed in diabetes, cancer and neurodegenerative diseases [41], could, by altering chromatin marks and gene expression, underlie the etiology of these diseases.
As a nutrient sensor, O-GlcNAcylation is at the nexus between food intake and epigenetics. In fact, western-diet or exercise modulates OGT’s association with epigenetic writers in mice [42,43]. Since epigenetic changes can be inherited during cell division and maintained as an acquired phenotype, our eating habits and even life style have the potential to be imprinted in our genes. Many studies have highlighted the impact of nutrients or diabetes during pregnancy affecting offspring. Some of these effects are still detectable at the F3 and therefore transgenerational [44–48]. In the light that O-GlcNAcylation is deregulated under diabetes and obesity and considering its crucial role as a nutrient sensor and modulator of epigenetics, O-GlcNAcylation, as an epigenetic mark, could be one of the molecular mechanisms of nutrient- and stress-dependent non-DNA sequenced encoded inheritance phenotypes.
Highlights.
Nearly all transcription proteins are dynamically O-GlcNAcylated
Histone O-GlcNAcylation interplays with other histone marks
Epigenetic writers/erasers are regulated by the sugar
OGT and OGA are required for the transcription cycle
Acknowledgments
The authors are supported by R01DK61671 and P01HL107153. G.W.H. receives a share of royalties received by Johns Hopkins University on sales of the CTD110.6 antibody, which is managed by JHU.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gambetta MC, Muller J. A critical perspective of the diverse roles of O-GlcNAc transferase in chromatin. Chromosoma. 2015;124:429–442. doi: 10.1007/s00412-015-0513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardiville S, Hart GW. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014;20:208–213. doi: 10.1016/j.cmet.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagnon J, Daou S, Zamorano N, Iannantuono NV, Hammond-Martel I, Mashtalir N, Bonneil E, Wurtele H, Thibault P, Affarel B. Undetectable histone O-GlcNAcylation in mammalian cells. Epigenetics. 2015;10:677–691. doi: 10.1080/15592294.2015.1060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakabe K, Hart GW. O-GlcNAc transferase regulates mitotic chromatin dynamics. J Biol Chem. 2010;285:34460–34468. doi: 10.1074/jbc.M110.158170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, Sifers RN. beta-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem. 2012;287:12195–12203. doi: 10.1074/jbc.M111.315804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang P, Peng C, Liu X, Liu H, Chen Y, Zheng L, Han B, Pei H. OGT mediated histone H2B S112 GlcNAcylation regulates DNA damage response. J Genet Genomics. 2015;42:467–475. doi: 10.1016/j.jgg.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 9. Lercher L, Raj R, Patel NA, Price J, Mohammed S, Robinson CV, Schofield CJ, Davis BG. Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat Commun. 2015;6:7978. doi: 10.1038/ncomms8978. Using a synthetic O-GlcNAcylated histone H2A, the authors have shown for the first that the sugar can regule the stabilite of the nucleosome, suggesting that O-GlcNAcylation has an impact on chromatin structure.
- 10.Xu Q, Yang C, Du Y, Chen Y, Liu H, Deng M, Zhang H, Zhang L, Liu T, Liu Q, et al. AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res. 2014;42:5594–5604. doi: 10.1093/nar/gku236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ronningen T, Shah A, Oldenburg AR, Vekterud K, Delbarre E, Moskaug JO, Collas P. Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 2015;25:1825–1835. doi: 10.1101/gr.193748.115. The authors have shown with high throughput methods that O-GlcNAcylation of histone H2B is a stable chromatin mark important for chromatin structure and gene expression during adipogenesis.
- 12. Halim A, Larsen IS, Neubert P, Joshi HJ, Petersen BL, Vakhrushev SY, Strahl S, Clausen H. Discovery of a nucleocytoplasmic O-mannose glycoproteome in yeast. Proc Natl Acad Sci U S A. 2015;112:15648–15653. doi: 10.1073/pnas.1511743112. The absence of O-GlcNAcylation in yeast has been difficult to understand within the field for many years. The discovery that the O-man glycosylation would mimic O-GlcNAcylation in yeast, if confirmed, would be a powerful model to help understand the epigenetic role of histone O-GlcNAcylation in mammals and plants.
- 13.Dong L, Xu CW. Carbohydrates induce mono-ubiquitination of H2B in yeast. J Biol Chem. 2004;279:1577–1580. doi: 10.1074/jbc.C300505200. [DOI] [PubMed] [Google Scholar]
- 14.Gao Z, Xu CW. Glucose metabolism induces mono-ubiquitination of histone H2B in mammalian cells. Biochem Biophys Res Commun. 2011;404:428–433. doi: 10.1016/j.bbrc.2010.11.138. [DOI] [PubMed] [Google Scholar]
- 15.Butkinaree C, Cheung WD, Park S, Park K, Barber M, Hart GW. Characterization of beta-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J Biol Chem. 2008;283:23557–23566. doi: 10.1074/jbc.M804116200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toleman C, Paterson AJ, Whisenhunt TR, Kudlow JE. Characterization of the histone acetyltransferase (HAT) domain of a bifunctional protein with activable O-GlcNAcase and HAT activities. J Biol Chem. 2004;279:53665–53673. doi: 10.1074/jbc.M410406200. [DOI] [PubMed] [Google Scholar]
- 17.Hayakawa K, Hirosawa M, Tabei Y, Arai D, Tanaka S, Murakami N, Yagi S, Shiota K. Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. J Biol Chem. 2013;288:17099–17110. doi: 10.1074/jbc.M113.455899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao FV, Schuttelkopf AW, Dorfmueller HC, Ferenbach AT, Navratilova I, van Aalten DM. Structure of a bacterial putative acetyltransferase defines the fold of the human O-GlcNAcase C-terminal domain. Open Biol. 2013;3:130021. doi: 10.1098/rsob.130021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang YR, Song M, Lee H, Jeon Y, Choi EJ, Jang HJ, Moon HY, Byun HY, Kim EK, Kim DH, et al. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell. 2012;11:439–448. doi: 10.1111/j.1474-9726.2012.00801.x. [DOI] [PubMed] [Google Scholar]
- 20.Keembiyehetty C, Love DC, Harwood KR, Gavrilova O, Comly ME, Hanover JA. Conditional knock-out reveals a requirement for O-linked N-Acetylglucosaminase (O-GlcNAcase) in metabolic homeostasis. J Biol Chem. 2015;290:7097–7113. doi: 10.1074/jbc.M114.617779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi SW, Friso S. Epigenetics: A New Bridge between Nutrition and Health. Adv Nutr. 2010;1:8–16. doi: 10.3945/an.110.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bullen JW, Balsbaugh JL, Chanda D, Shabanowitz J, Hunt DF, Neumann D, Hart GW. Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK) J Biol Chem. 2014;289:10592–10606. doi: 10.1074/jbc.M113.523068. This paper demonstrates for the first time that the nutrient sensor OGT interplays with the energy sensor AMPK. AMPK regulates OGT localization and substrate specificity, which can have a profound impact on cell physiology.
- 23.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci U S A. 2010;107:19915–19920. doi: 10.1073/pnas.1009023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Charoensuksai P, Kuhn P, Wang L, Sherer N, Xu W. O-GlcNAcylation of coactivator-associated arginine methyltransferase 1 regulates its protein substrate specificity. Biochem J. 2015;466:587–599. doi: 10.1042/BJ20141072. The authors show that CARM1 is O-GlcNAcylation by top-down and middle-down mass spectrometry. They also bring evidences showing that O-GlcNAcylation controls the specificity of CARM1.
- 26.Bauer C, Gobel K, Nagaraj N, Colantuoni C, Wang M, Muller U, Kremmer E, Rottach A, Leonhardt H. Phosphorylation of TET proteins is regulated via O-GlcNAcylation by the O-linked N-acetylglucosamine transferase (OGT) J Biol Chem. 2015;290:4801–4812. doi: 10.1074/jbc.M114.605881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi FT, Kim H, Lu W, He Q, Liu D, Goodell MA, Wan M, Songyang Z. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J Biol Chem. 2013;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vella P, Scelfo A, Jammula S, Chiacchiera F, Williams K, Cuomo A, Roberto A, Christensen J, Bonaldi T, Helin K, et al. Tet proteins connect the O-linked N-acetylglucosamine transferase Ogt to chromatin in embryonic stem cells. Mol Cell. 2013;49:645–656. doi: 10.1016/j.molcel.2012.12.019. The tow studies above as well as [30] and [31] have described simultaneously for the first time the mechanism that target OGT to specific chromatin loci. These studies also point out that OGT can act as a scaffold protein for epigenetic writer/erasers.
- 29.Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, Wong J. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked beta-N-acetylglucosamine transferase (OGT) J Biol Chem. 2014;289:5986–5996. doi: 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–655. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito R, Katsura S, Shimada H, Tsuchiya H, Hada M, Okumura T, Sugawara A, Yokoyama A. TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells. 2014;19:52–65. doi: 10.1111/gtc.12107. [DOI] [PubMed] [Google Scholar]
- 33.Kizuka Y, Kitazume S, Okahara K, Villagra A, Sotomayor EM, Taniguchi N. Epigenetic regulation of a brain-specific glycosyltransferase N-acetylglucosaminyltransferase-IX (GnT-IX) by specific chromatin modifiers. J Biol Chem. 2014;289:11253–11261. doi: 10.1074/jbc.M114.554311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou P, Wang Z, Yuan X, Zhou C, Liu L, Wan X, Zhang F, Ding X, Wang C, Xiong S, et al. Mixed lineage leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with host cell factor-1 (HCF-1) J Biol Chem. 2013;288:17532–17543. doi: 10.1074/jbc.M112.439729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding X, Jiang W, Zhou P, Liu L, Wan X, Yuan X, Wang X, Chen M, Chen J, Yang J, et al. Mixed Lineage Leukemia 5 (MLL5) Protein Stability Is Cooperatively Regulated by O-GlcNac Transferase (OGT) and Ubiquitin Specific Protease 7 (USP7) PLoS One. 2015;10:e0145023. doi: 10.1371/journal.pone.0145023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mariappa D, Zheng X, Schimpl M, Raimi O, Ferenbach AT, Muller HA, van Aalten DM. Dual functionality of O-GlcNAc transferase is required for Drosophila development. Open Biol. 2015:5. doi: 10.1098/rsob.150234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maury JJ, El Farran CA, Ng D, Loh YH, Bi X, Bardor M, Choo AB. RING1B O-GlcNAcylation regulates gene targeting of polycomb repressive complex 1 in human embryonic stem cells. Stem Cell Res. 2015;15:182–189. doi: 10.1016/j.scr.2015.06.007. This study describes the role of the O-GlcNAcylation of RING1B (subunit of PRC1). The authors bring evidences that RING1B O-GlcNAcylation decreases with cell differentiation and is responsible for targeting the PRC1 complex, a major epigenetic repressor complex, to specific loci.
- 38.Neri F, Incarnato D, Krepelova A, Rapelli S, Pagnani A, Zecchina R, Parlato C, Oliviero S. Genome-wide analysis identifies a functional association of Tet1 and Polycomb repressive complex 2 in mouse embryonic stem cells. Genome Biol. 2013;14:R91. doi: 10.1186/gb-2013-14-8-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu CS, Lo PW, Yeh YH, Hsu PH, Peng SH, Teng YC, Kang ML, Wong CH, Juan LJ. O-GlcNAcylation regulates EZH2 protein stability and function. Proc Natl Acad Sci U S A. 2014;111:1355–1360. doi: 10.1073/pnas.1323226111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gehani SS, Agrawal-Singh S, Dietrich N, Christophersen NS, Helin K, Hansen K. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol Cell. 2010;39:886–900. doi: 10.1016/j.molcel.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 41.Bond MR, Hanover JA. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medford HM, Cox EJ, Miller LE, Marsh SA. Consuming a Western diet for two weeks suppresses fetal genes in mouse hearts. Am J Physiol Regul Integr Comp Physiol. 2014;306:R519–R526. doi: 10.1152/ajpregu.00253.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cox EJ, Marsh SA. Exercise and diabetes have opposite effects on the assembly and O-GlcNAc modification of the mSin3A/HDAC1/2 complex in the heart. Cardiovasc Diabetol. 2013;12:101. doi: 10.1186/1475-2840-12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith CJ, Ryckman KK. Epigenetic and developmental influences on the risk of obesity, diabetes, and metabolic syndrome. Diabetes Metab Syndr Obes. 2015;8:295–302. doi: 10.2147/DMSO.S61296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jimenez-Chillaron JC, Ramon-Krauel M, Ribo S, Diaz R. Transgenerational epigenetic inheritance of diabetes risk as a consequence of early nutritional imbalances. Proc Nutr Soc. 2016;75:78–89. doi: 10.1017/S0029665115004231. [DOI] [PubMed] [Google Scholar]
- 46.Somer RA, Thummel CS. Epigenetic inheritance of metabolic state. Curr Opin Genet Dev. 2014;27:43–47. doi: 10.1016/j.gde.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci U S A. 2014;111:1873–1878. doi: 10.1073/pnas.1321195111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benyshek DC, Johnston CS, Martin JF. Glucose metabolism is altered in the adequately-nourished grand-offspring (F3 generation) of rats malnourished during gestation and perinatal life. Diabetologia. 2006;49:1117–1119. doi: 10.1007/s00125-006-0196-5. [DOI] [PubMed] [Google Scholar]