Abstract
After more than a decade of instrument and method development, broadband coherent anti-Stokes Raman scattering (CARS) micro-spectroscopy is beginning to live up to its potential as a label-free imaging modality that can rapidly generate high resolution images with full vibrational spectra at each image pixel. Presently these instruments are able to obtain quantitative, spatially resolved information on lipids from the CH stretch region of the Raman spectrum, and some instrument designs facilitate acquisition of high quality fingerprint spectra, containing information on a host of molecular species including structural proteins, nucleotides, and metabolites. While most of the existing instruments are research projects themselves, it appears that the relevant technologies are maturing so that commercially available instruments may not be too far in the future, making this remarkable imaging modality widely available.
Body
The Raman effect occurs when light scatters inelastically from matter so that light energy is converted to or taken from molecular vibrational energy. This type of interaction relies only on changes in polarizability of molecular bonds during vibration, and occurs to some extent for almost all chemical bonds. Thus, not only does this light scattering occur ubiquitously, the inelastically scattered light contains detailed information about chemical composition of materials it has interacted with, providing a label-free chemical contrast mechanism. Perhaps the only reason this contrast mechanism is not widely used for biological microscopy is that the scattering cross section is typically very small. Typically, no more than one in every 107 photons that interact with matter will spontaneously scatter in this way.
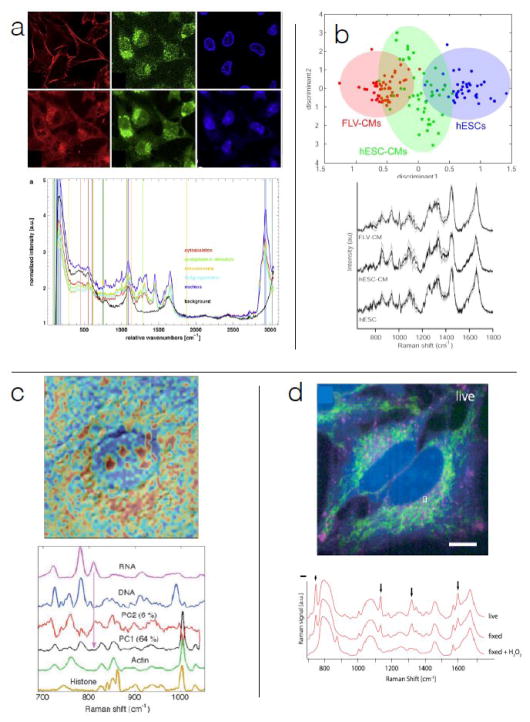
In spite of its small scattering cross section, significant effort has been expended to develop applications of spontaneous Raman (SR) scattering and micro-spectroscopy in biology. These efforts are motivated by the wealth of label-free chemical information available. Figure 1 shows examples of structural and functional information that can be extracted from SR scattering. Unless stated otherwise, contrast used to construct all images in this review is derived from combination of Raman spectral peaks. In Figure 1a, we see that the SR spectroscopic signature can be used to substitute for image contrast generated by fluorophores targeting myosin, DNA, Golgi apparatus 1 and mitochondria 2. As exemplified in Figure 1b, vibrational spectra from SR also provide information about cell phenotype 3–5; SR has also been used to objectively discriminate between clinically distinct tumor types and grades in many tissues such as breast 6, lung 7, lymph node 8,9, and skin 10. SR spectroscopy has also been used to characterize engineered tissues with respect to proper bone formation 11,12. We note that each of these applications has used spectral information that comes exclusively from sets of multiple peaks in the fingerprint region (500 to 1800 cm−1).
Figure 1.
Spontaneous Raman spectroscopy and microscopy: (a) SR spectroscopy-derived image contrast essentially recapitulates fluorescence staining for some structural motifs (top row are fluorescence images of cytoskeleton, Golgi apparatus, and nuclei, bottom row are Raman-derived images with similar contrast); 75 ms spectral acquisition time, 10 mW, 532 nm excitation 1. (b) SR spectra contain functional information required to determine cell lineage commitment; 60 s spectral acquisition time, 70 mW, 785 nm excitation 5. (c) SR provides spatial distributions of chemical species in organelles; 80 s spectral acquisition time, 80 mW, 785 nm extitation 15. (d) SR captures spatial distribution of metabolism-related species; 30 ms effective spectral acquisition time for parallelized acquisition, 532 nm, 500 mW 57.
Applications of SR spectroscopy to biological samples are hindered primarily by two factors: one is that intrinsic fluorescence presents a significant background signal, and the other is that the samples are quite susceptible to photodamage, which limits the amount of excitation light and thus the Raman signal level. Several approaches have been developed to deal with the former problem, including reducing fluorescence by optical bleaching of samples, and careful selection of substrate materials 13, using near-infrared excitation wavelengths, subtracting collected fluorescence by comparing SR spectra acquired with two closely spaced excitation laser frequencies, or by rejecting fluorescence through time-gated signal detection 14. The problem of sensitivity to photodamage is managed by using low excitation power, leading to spectral acquisition times on the order of seconds or minutes as shown in Figure 1. (Variation in time required for signal acquisition among the examples in Figure 1 is due in part to λ−4 scaling of Raman scattering intensity with excitation wavelength.) Due to the weak SR signal, high resolution Raman images from point scanning can require 10s of hours for acquisition 15. The most effective approach to ameliorate this problem for SR has been parallel spectral acquisition. This allows greater amounts of laser power on the sample by illuminating larger areas, and can lead to imaging speed improvements of up to 100-fold 16–19, so high quality Raman spectra can be acquired at an equivalent rate of 30 ms / spectrum. This increase in imaging speed comes at the cost of slightly increased instrument complexity.
Rapid Raman-based image generation is a primary driver for development of coherent Raman imaging (CRI) techniques. Narrow-band CRI methods have demonstrated image acquisition at video rate 20,21 but these derive contrast from single Raman bands, typically from the CH stretch region (2800 to 3200 cm−1). These narrow-band coherent Raman methods are limited in their utility for obtaining the types of functional and structural information referred to above, which comes from multiple peaks in the fingerprint region.
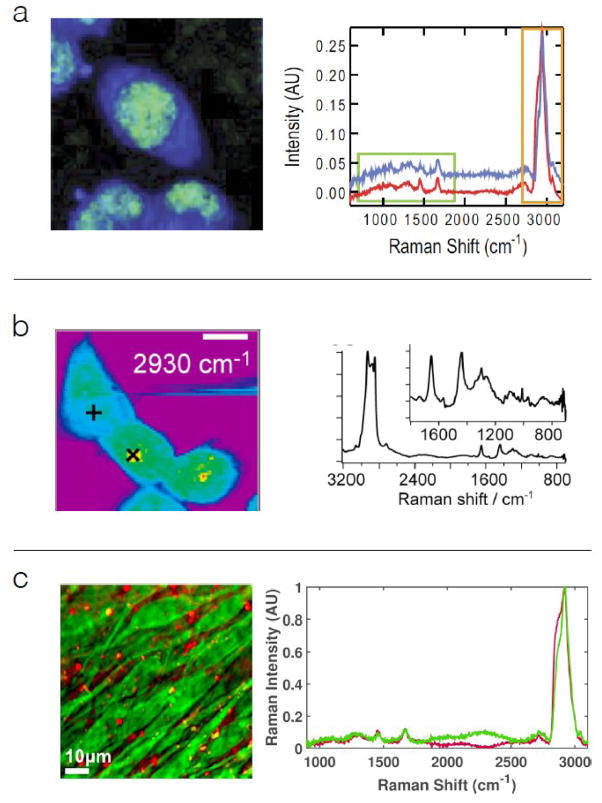
Spectroscopic CRI, primarily coherent anti-Stokes Raman scattering (CARS) techniques 22–25, have been developed to rapidly acquire signal from the fingerprint and CH stretch regions. These methods require spectrally broad pulsed light, or spectrally narrow but rapidly tunable pulsed light for excitation. Most spectroscopic CARS systems utilize continuum light generated in a photonic crystal fiber and collect signal in the spectral domain 26–28. Examples of spectra and images from such instruments are given in Figure 2. Generation of the fiber-based continuum light required for these approaches is now routine 26,29–31 but spectral domain signal detection creates challenging instrument constraints. Spectral domain detection requires focusing signal light onto the slit of a spectrometer so that it can be cleanly dispersed in the spectral domain onto a CCD camera. When signal light passes through biological samples, it is elastically scattered to some extent, and only a fraction of the signal can be properly focused onto the camera. Single-element detectors generally don’t require such high quality focusing of the signal light, and in this respect are easier to implement. Spectroscopic CARS techniques using single-element detectors include Fourier domain 32, spectral focusing 33, spectral scanning 34, or time domain 35 methods. Each of these spectroscopic CRI methods have been implemented almost exclusively as point-scanned approaches with spectral acquisition times typically in the range (25 to 50) ms / full Raman spectrum 36, which is somewhat faster than point-scanned SR scattering, but on par with line-scanned SR. Line-scan approaches can also be applied to spectroscopic CRI methods, with additional imaging speed improvements 37 of 5 to 10-fold, and no loss of spectral quality.
Figure 2.
CARS micro-spectroscopy: (a) Mouse fibroblast cells imaged with 50 ms spectral acquisition time, 42 mW laser power, and 830 nm probe 58 (b) Isotopically labeled surfactant uptake in Chinese hamster lung cells, using 50 ms spectral acquisition time, 10 mW laser power, and 1064 nm probe 50, (c) Confluent MC3T3 cells imaged at 9 ms spectral acquisition time, 50 mW laser power, and 830 nm probe 37.
In spite of increased spectral acquisition speed, the spectroscopic CRI methods have generally not been successful at producing fingerprint spectra on par with those of SR from biological systems. The spectral images in panels a and c, and to a lesser extent, panel b of Figure 2 exemplify this weakness. The relatively low signal to noise ratio in the CRI fingerprint spectra could be due to several factors. Some of the factors that are likely germane to Figure 2a and 2c are that the at 12 cm−1 spectral resolution of the probe pulse was wider than many natural peak widths, that the coherence generation process from pump and Stokes pulses was ~1 ps, so slightly shorter than the resonant buildup time 38, and there was no in-process noise filtering. The image and spectra in Figure 2c were obtained with high spectral resolution (< 0.1 cm−1 probe width), and coherence generation time was ~ 1 ns (much longer than the coherence buildup time). Also, the signal-processing algorithm included filters intended to reduce spectral noise. The predominant factor, common to all spectroscopic CRI approaches, is the necessity of spreading excitation pulse energy over a broad spectral range. As shown in Figure 2, most of the peaks in the fingerprint region (there are roughly 50 of them) are intrinsically quite weak, whereas the roughly five peaks of the CH stretch region (2800 to 3100 cm−1) are much stronger. For this reason, coherent Raman imaging studies (even spectroscopic CRI) have focused primarily on lipids and other species that have strong signals in the CH stretch region of the spectrum 39–41.
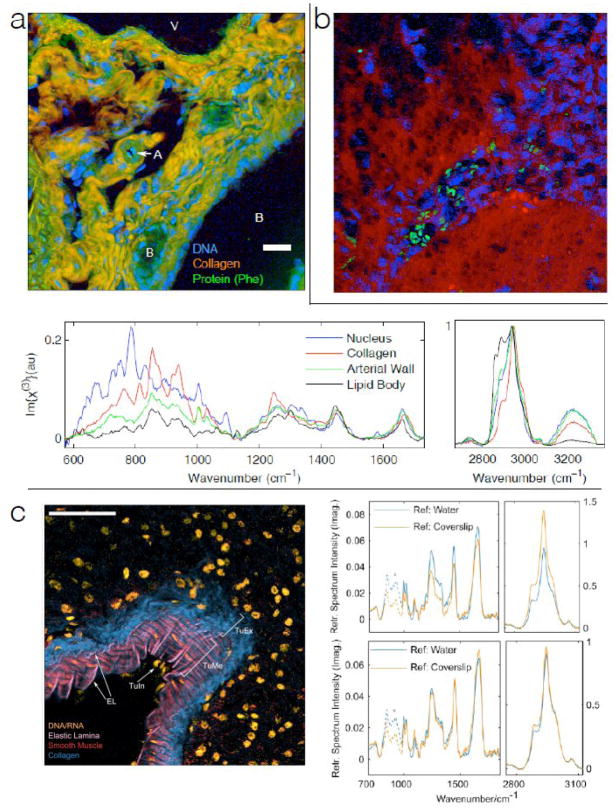
A major motivating factor for developing spectroscopic CRI has been to rapidly acquire high quality fingerprint spectra. This has recently become possible with application of a signal generation paradigm that is much more efficient than that previously used in spectroscopic CARS. Camp et al. recently showed that impulsive coherence generation can be used to generate high quality fingerprint spectra at 3.5 ms acquisition times 42. Figure 3 shows examples of spectra and images obtained with this highly efficient signal generation paradigm. The spectra shown are taken from individual image pixels, and are of sufficient quality to identify the presence of DNA, collagen, elastin, and other structural proteins. The detail in the fingerprint spectra is comparable to that obtained with SR at much longer acquisition times, and therefore will contain the same structural and functional information obtained from the fingerprint region in SR scattering (as shown in Figure 1). Impulsive coherence generation is key to rapidly obtaining these high quality fingerprint spectra. Fortunately, this coherence generation approach is compatible with spectral domain, Fourier domain, or time domain methods. It is also compatible with line-scan signal acquisition. A combination of impulsive coherence generation and line-scan detection would yield high quality Raman fingerprint spectra at sub-millisecond acquisition times.
Figure 3.
High-speed CARS micro-spectroscopy. All tissues imaged with 3.5 ms spectral acquisition time, 25 mW laser power, and 785 nm probe. Scale bars are 50 μm. (a) Mouse hepatic tissue, with contrast corresponding to collagen, DNA, and general protein. Representative spectra are shown from individual image pixels 42 (b) Mouse model of human glioblastoma, with tumor region identifiable through high density of (blue) nuclear regions. Green regions are red blood cells. 42 (c) Murine pancreatic artery tissue section, pseudocolor highlighting DNA/RNA, the elastic lamina, smooth muscle, and collagen. Arrows identify features of the arterial wall – EL: elastic lamina; TuIn: tunica intima; TuMe: tunica media; TuEx: tunica externa. The pairs of spectra plotted in the top and bottom panels to the right of the micrograph are a single CARS spectrum obtained from the TuME region, and processed using two different NRB approximations. Spectra in the top panel, showing significant differences, are processed without retrieved phase error correction, whereas spectra in the bottom panel are processed with retrieved phase error correction, and are nearly identical.
It is important that the correct Raman spectra are obtained from the overall coherent Raman signal. All CRI techniques yield a nonresonant background (NRB) component at the same wavelength(s) as the resonant signal of interest 38. For SRS 43 the nonresonant field is the laser excitation source, which is typically 106 to 108 times stronger than the resonant signal, but can typically rejected through signal modulation 43. For CARS, the NRB amplitude is typically 10 to 100 times larger than the resonant component in the fingerprint. However, since it is phase-locked with the resonant component, it can be used as a heterodyne amplifier for the weak peaks in the fingerprint region of the spectrum. In fact, without the NRB as a signal amplifier, the CARS signal originating from many low-concentration analytes in biological systems would be weaker than the corresponding SR signal 44, and it would be impossible to obtain fingerprint spectra with short acquisition times.
In order to extract the correct Raman spectrum, and to use the NRB as a heterodyne amplifier, the phase of the overall signal must be determined. There are a number of methods for accomplishing this 45–47, but these phase retrieval algorithms will always return small residual errors. Fortunately, these errors can be corrected analytically 48, and symmetry criteria peculiar to coherent Raman scattering can be used to ensure that retrieved peak height ratios are correct 48. Figure 3c shows reproducibility of retrieved spectra with and without the corrections to the retrieved phase.
Spectroscopic CRI methods can now generate full Raman spectra roughly 100 times faster than SR, and with little or no sample preparation. Both of these factors have and will lead to spectroscopic CRI being applied where SR imaging is not practical or possible. This includes applications providing chemo-structural information 49 and mechanistic information 50 that demand high spatial and chemical resolution, with limits on image acquisition time 51. Spectroscopically augmented histopathology (SHP) is a potentially important example of such an application. It is well established that interobserver diagnostic disagreement among pathologists varies from 10% to 40% depending on cancer type and severity 52–54. The lack of agreement can be traced back to subtly of diagnostically relevant features in tissue samples prepared with hematoxylin and eosin (H&E), the gold standard for hisopathology. One approach to reducing the diagnostic uncertainty is to add information about molecular species other than those highlighted by H&E. Vibrational spectroscopy provides a general, label-free way to provide relative abundance of a few diagnostically important tissue components. With spectra taken at just a few spots in the tissue, the agreement between Raman-based ranking and histopathologist panel assignment is typically found to be roughly 5% 7,55,56. These SR studies used single Raman spectra or relatively low resolution compositional maps of tissue, leading to possible sampling bias issues. Here, Spectroscopic CRI could add significant value by rapidly producing high-resolution spatial maps of these diagnostically important species.
For now, the benefit of significantly faster spectroscopic imaging through spectroscopic CRI comes at the price of significantly greater instrumental complexity. To date, these instruments are not commercially available. However, once the underlying technologies used to rapidly produce high quality CRI fingerprint spectra are well established, it is our expectation that the breadth of applications in biology and materials science will provide a favorable market for such instruments.
Research Highlights / Core Findings.
Raman spectroscopy can provide information of value to biological and biomedical researchers – primarily in the form of relative abundance of major molecular components
Spontaneous Raman spectral acquisition is sufficiently time consuming as to preclude its widespread use as an imaging technique
Spectroscopic coherent Raman imaging has been developed with an eye towards providing same spectroscopic information as spontaneous Raman, but at appreciably higher speed
Recent advances in coherent Raman spectroscopy have made it possible to acquire high-quality Raman spectra from typical biological cells and tissues on a ms timescale, fast enough to make the technique useful, but instrumentation is still in research phase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Klein K, Gigler A, Aschenbrenner T, Monetti R, et al. Label-Free Live-Cell Imaging with Confocal Raman Microscopy. Biophysical Journal. 2012;102:360–368. doi: 10.1016/j.bpj.2011.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthaus C, Chernenko T, Newmark JA, Warner CM, Diem M. Label-free detection of mitochondrial distribution in cells by nonresonant Raman microspectroscopy. Biophysical Journal. 2007;93:668–673. doi: 10.1529/biophysj.106.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YJ, Vega SL, Patel PJ, Aamer KA, et al. Quantitative, Label-Free Characterization of Stem Cell Differentiation at the Single-Cell Level by Broadband Coherent Anti-Stokes Raman Scattering Microscopy. Tissue Engineering Part C: Methods. 2014;20:562–569. doi: 10.1089/ten.tec.2013.0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Notingher I, Bisson I, Bishop AE, Randle WL, et al. In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal Chem. 2004;76:3185–3193. doi: 10.1021/ac0498720. [DOI] [PubMed] [Google Scholar]
- 5.Chan JW, Lieu DK, Huser T, Li RA. Label-Free Separation of Human Embryonic Stem Cells and Their Cardiac Derivatives Using Raman Spectroscopy. Analytical Chemistry. 2009;81:1324–1331. doi: 10.1021/ac801665m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haka AS, Volynskaya Z, Gardecki JA, Nazemi J, et al. In vivo Margin Assessment during Partial Mastectomy Breast Surgery Using Raman Spectroscopy. Cancer Research. 2006;66:3317–3322. doi: 10.1158/0008-5472.CAN-05-2815. [DOI] [PubMed] [Google Scholar]
- 7.Huang Z, McWilliams A, Lui H, McLean DI, et al. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. International Journal of Cancer. 2003;107:1047–1052. doi: 10.1002/ijc.11500. [DOI] [PubMed] [Google Scholar]
- 8.Chan JW, Taylor DS, Zwerdling T, Lane SM, et al. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophysical journal. 2006;90:648–656. doi: 10.1529/biophysj.105.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lloyd GR, Orr LE, Christie-Brown J, McCarthy K, et al. Discrimination between benign, primary and secondary malignancies in lymph nodes from the head and neck utilising Raman spectroscopy and multivariate analysis. Analyst. 2013 doi: 10.1039/c2an36579k. [DOI] [PubMed] [Google Scholar]
- 10.Gniadecka M, Philipsen PA, Sigurdsson S, Wessel S, et al. Melanoma diagnosis by Raman spectroscopy and neural networks: structure alterations in proteins and lipids in intact cancer tissue. J Invest Dermatol. 2004;122:443–449. doi: 10.1046/j.0022-202X.2004.22208.x. [DOI] [PubMed] [Google Scholar]
- 11.Gentleman E, Swain RJ, Evans ND, Boonrungsiman S, et al. Comparative materials differences revealed in engineered bone as a function of cell-specific differentiation. Nat Mater. 2009;8:763–770. doi: 10.1038/nmat2505. [DOI] [PubMed] [Google Scholar]
- 12.Notingher I, Jell G, Notingher PL, Bisson I, et al. Raman spectroscopy: Potential tool for in situ characterization of bone cell differentiation. Bioceramics. 2005;17:545–548. [Google Scholar]
- 13.Butler HJ, Ashton L, Bird B, Cinque G, et al. Using Raman spectroscopy to characterize biological materials. Nat Protoc. 2016;11:664–687. doi: 10.1038/nprot.2016.036. [DOI] [PubMed] [Google Scholar]
- 14.Matousek P, Towrie M, Stanley A, Parker AW. Efficient Rejection of Fluorescence from Raman Spectra Using Picosecond Kerr Gating. Applied Spectroscopy. 1999;53:1485–1489. [Google Scholar]
- 15.Schulze HG, Konorov SO, Piret JM, Blades MW, Turner RFB. Label-free imaging of mammalian cell nucleoli by Raman microspectroscopy. The Analyst. 2013;138:3416. doi: 10.1039/c3an00118k. [DOI] [PubMed] [Google Scholar]
- 16.Kong L, Navas-Moreno M, Chan JW. Fast Confocal Raman Imaging Using a 2-D Multifocal Array for Parallel Hyperspectral Detection. Anal Chem. 2016;88:1281–1285. doi: 10.1021/acs.analchem.5b03707. [DOI] [PubMed] [Google Scholar]
- 17.Schlücker S, Schaeberle MD, Huffman SW, Levin IW. Raman Microspectroscopy: A Comparison of Point, Line, and Wide-Field Imaging Methodologies. Anal Chem. 2003;75:4312–4318. doi: 10.1021/ac034169h. [DOI] [PubMed] [Google Scholar]
- 18.Ma J, Ben-Amotz D. Rapid Micro-Raman Imaging Using Fiber-Bundle Image Compression. Appl Spectrosc. 1997;51:1845–1848. [Google Scholar]
- 19.Hamada K, Fujita K, Smith NI, Kobayashi M, et al. Raman microscopy for dynamic molecular imaging of living cells. Journal of Biomedical Optics. 2008;13:044027–044027-4. doi: 10.1117/1.2952192. [DOI] [PubMed] [Google Scholar]
- 20.Saar BG, Freudiger CW, Reichman J, Stanley CM, et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science. 2010;330:1368–1370. doi: 10.1126/science.1197236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lei M, Winterhalder M, Selm R, Zumbusch A. Video-rate wide-field coherent anti-Stokes Raman scattering microscopy with collinear nonphase-matching illumination. JOURNAL OF BIOMEDICAL OPTICS. 2011;16 doi: 10.1117/1.3533707. [DOI] [PubMed] [Google Scholar]
- *22.Kee TW, Cicerone MT. Simple approach to one-laser, broadband coherent anti-Stokes Raman scattering microscopy. Optics Letters. 2004;29:2701–2703. doi: 10.1364/ol.29.002701. In these two papers, published in close time proximity, the authors first demonstrated the use of continuum light produced in highly nonlinear fiber for generating broadband (spectroscopic) coherent anti-Stokes Raman scattering in a microscope. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *23.Kano H, Hamaguchi H. Ultrabroadband (> 2500 cm(−1)) multiplex coherent anti-Stokes Raman scattering microspectroscopy using a supercontinuum generated from a photonic crystal fiber. Applied Physics Letters. 2005;86 In these two papers, published in close time proximity, the authors first demonstrated the use of continuum light produced in highly nonlinear fiber for generating broadband (spectroscopic) coherent anti-Stokes Raman scattering in a microscope. [Google Scholar]
- 24.Kano H, Hamaguchi H. Vibrationally resonant imaging of a single living cell by supercontinuum-based multiplex coherent anti-Stokes Raman scattering microspectroscopy. Optics Express. 2005;13:1322–1327. doi: 10.1364/opex.13.001322. [DOI] [PubMed] [Google Scholar]
- 25.Petrov GI, Yakovlev VV, Sokolov AV, Scully MO. Detection of Bacillus subtilis spores in water by means of broadband coherent anti-Stokes Raman spectroscopy. Optics Express. 2005;13:9537–9542. doi: 10.1364/opex.13.009537. [DOI] [PubMed] [Google Scholar]
- 26.Lee YJ, Parekh SH, Kim YH, Cicerone MT. Optimized continuum from a photonic crystal fiber for broadband time-resolved coherent anti-Stokes Raman scattering. Optics Express. 2010;18:4371–4379. doi: 10.1364/OE.18.004371. [DOI] [PubMed] [Google Scholar]
- 27.Kano H, Hamaguchi H-O. Characterization of a supercontinuum generated from a photonic crystal fiber and its application to coherent Raman spectroscopy. Optics Letters. 2003;28:2360–2362. doi: 10.1364/ol.28.002360. [DOI] [PubMed] [Google Scholar]
- 28.Yakovlev VV. Advanced instrumentation for non-linear Raman microscopy. Journal of Raman Spectroscopy. 2003;34:957–964. [Google Scholar]
- 29.Kieu KQ, Klein J, Evans A, Barton JK, Peyghambarian N. Ultrahigh resolution all-reflective optical coherence tomography system with a compact fiber-based supercontinuum source. Journal of Biomedical Optics. 2011;16:106004–106004-4. doi: 10.1117/1.3633340. [DOI] [PubMed] [Google Scholar]
- 30.Selm R, Winterhalder M, Zumbusch A, Krauss G, et al. Ultrabroadband background-free coherent anti-Stokes Raman scattering microscopy based on a compact Er:fiber laser system. Opt Lett. 2010;35:3282–3284. doi: 10.1364/OL.35.003282. [DOI] [PubMed] [Google Scholar]
- 31.Okuno M, Kano H, Leproux P, Couderc V, Hamaguchi H. Ultrabroadband (> 2000 cm(−1)) multiplex coherent anti-Stokes Raman scattering spectroscopy using a subnanosecond supercontinuum light source. Optics Letters. 2007;32:3050–3052. doi: 10.1364/ol.32.003050. [DOI] [PubMed] [Google Scholar]
- 32.Oron D, Dudovich N, Yelin D, Silberberg Y. Quantum control of coherent anti-Stokes Raman processes. Physical Review A. 2002;65 doi: 10.1103/PhysRevLett.88.063004. [DOI] [PubMed] [Google Scholar]
- 33.Hellerer T, Enejder AM, Zumbusch A. Spectral focusing: High spectral resolution spectroscopy with broad-bandwidth laser pulses. Applied Physics Letters. 2004;85:25–27. [Google Scholar]
- 34.Chimento PF, Jurna M, Bouwmans HSP, Garbacik ET, et al. High-resolution narrowband CARS spectroscopy in the spectral fingerprint region. Journal of Raman Spectroscopy. 2009;40:1229–1233. [Google Scholar]
- 35.Ideguchi T, Holzner S, Bernhardt B, Guelachvili G, et al. Coherent Raman spectro-imaging with laser frequency combs. Nature. 2013;502:355–358. doi: 10.1038/nature12607. [DOI] [PubMed] [Google Scholar]
- 36.Camp CH, Jr, Cicerone MT. Chemically sensitive bioimaging with coherent Raman scattering. Nat Photon. 2015;9:295–305. [Google Scholar]
- 37.Ryu IS, Camp CH, Jin Y, Cicerone MT, Lee YJ. Beam scanning for rapid coherent Raman hyperspectral imaging. Opt Lett. 2015;40:5826–5829. doi: 10.1364/OL.40.005826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eesley GL. Coherent raman spectroscopy. Journal of Quantitative Spectroscopy and Radiative Transfer. 1979;22:507–576. [Google Scholar]
- 39.Di Napoli C, Pope I, Masia F, Langbein W, et al. Quantitative Spatiotemporal Chemical Profiling of Individual Lipid Droplets by Hyperspectral CARS Microscopy in Living Human Adipose-Derived Stem Cells. Anal Chem. 2016 doi: 10.1021/acs.analchem.5b04468. [DOI] [PubMed]
- 40.Billecke N, Rago G, Bosma M, Eijkel G, et al. Chemical imaging of lipid droplets in muscle tissues using hyperspectral coherent Raman microscopy. Histochem Cell Biol. 2013 doi: 10.1007/s00418-013-1161-2. [DOI] [PubMed] [Google Scholar]
- 41.Billecke N, Bosma M, Rock W, Fleissner F, et al. Perilipin 5 mediated lipid droplet remodelling revealed by coherent Raman imaging. Integr Biol (Camb) 2015;7:467–476. doi: 10.1039/c4ib00271g. [DOI] [PubMed] [Google Scholar]
- **42.Camp CH, Jr, Lee YJ, Heddleston JM, Hartshorn CM, et al. High-speed coherent Raman fingerprint imaging of biological tissues. Nat Photon. 2014;8:627–634. doi: 10.1038/nphoton.2014.145. The authors of this paper combined the advantageous features of two signal excitation mechanisms to generate coherent Raman spectra at least 10-fold faster than had been previously achieved, and at much higher signal quality levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freudiger CW, Min W, Saar BG, Lu S, et al. Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Cui M, Bachler BR, Ogilvie JP. Comparing coherent and spontaneous Raman scattering under biological imaging conditions. Optics Letters. 2009;34:773–775. doi: 10.1364/ol.34.000773. [DOI] [PubMed] [Google Scholar]
- 45.Vartiainen EM. Phase retrieval approach for coherent anti-Stokes Raman scattering spectrum analysis. JOSA B. 1992;9:1209–1214. [Google Scholar]
- 46.Liu YX, Lee YJ, Cicerone MT. Broadband CARS spectral phase retrieval using a time-domain Kramers-Kronig transform. Optics Letters. 2009;34:1363–1365. doi: 10.1364/ol.34.001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Cicerone MT, Aamer KA, Lee YJ, Vartiainen E. Maximum entropy and time-domain Kramers–Kronig phase retrieval approaches are functionally equivalent for CARS microspectroscopy. Journal of Raman Spectroscopy. 2012;43:637–643. The authors demonstrate functional equivalence of the two widely used phase retrieval methods in spectroscopic CARS. They show that the Kramers-Kronig approach is computationally less demanding, although the Maximum Entropy approach could be recast in a less computationally demanding form. The author list includes inventors of both methods. [Google Scholar]
- *48.Camp CH, Lee YJ, Cicerone MT. Quantitative, comparable coherent anti-Stokes Raman scattering (CARS) spectroscopy: correcting errors in phase retrieval. Journal of Raman Spectroscopy. 2016;47:408–415. doi: 10.1002/jrs.4824. The authors derive equations for and demonstrate a protocol that allows one to objectively correct for phase errors that accumulate in spectral phase retrieval by Kramers-Kronig or Maximum Entropy methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gohad NV, Aldred N, Hartshorn CM, Jong Lee Y, et al. Synergistic roles for lipids and proteins in the permanent adhesive of barnacle larvae. Nat Commun. 2014;5 doi: 10.1038/ncomms5414. [DOI] [PubMed] [Google Scholar]
- *50.Okuno M, Kano H, Fujii K, Bito K, et al. Surfactant Uptake Dynamics in Mammalian Cells Elucidated with Quantitative Coherent Anti-Stokes Raman Scattering Microspectroscopy. PloS one. 2014;9:e93401. doi: 10.1371/journal.pone.0093401. The authors use spectroscopic CARS microscopy to show for the first time that surfactants are taken into Chinese hamster lung cells before lysis occurs. It was previously thought that surfactants directly associated with the membrane to induce lysis. This is an excellent example, (one of only a few) of using coherent Raman spectroscopic imaging to follow a live-cell process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hartshorn CM, Lee YJ, Camp CH, Liu Z, et al. Multicomponent Chemical Imaging of Pharmaceutical Solid Dosage Forms with Broadband CARS Microscopy. Anal Chem. 2013;85:8102–8111. doi: 10.1021/ac400671p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rhijn BW, van Leenders GJ, Ooms BC, Kirkels WJ, et al. The Pathologist’s Mean Grade Is Constant and Individualizes the Prognostic Value of Bladder Cancer Grading. European Urology. 2010;57:1052–1057. doi: 10.1016/j.eururo.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 53.Longacre TA, Ennis M, Quenneville LA, Bane AL, et al. Interobserver agreement and reproducibility in classification of invasive breast carcinoma: an NCI breast cancer family registry study. Mod Pathol. 2005;19:195–207. doi: 10.1038/modpathol.3800496. [DOI] [PubMed] [Google Scholar]
- 54.HUNNINGHAKE G, ZIMMERMAN MB, SCHWARTZ D, KING T, et al. Utility of a Lung Biopsy for the Diagnosis of Idiopathic Pulmonary Fibrosis. Am J Respir Crit Care Med. 2001;164:193–196. doi: 10.1164/ajrccm.164.2.2101090. [DOI] [PubMed] [Google Scholar]
- 55.Haka AS, Shafer-Peltier KE, Fitzmaurice M, Crowe J, et al. Diagnosing breast cancer by using Raman spectroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:12371. doi: 10.1073/pnas.0501390102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakker Schut TC, Maquelin K, van der Kwast T, Bangma CH, et al. Discrimination between Nontumor Bladder Tissue and Tumor by Raman Spectroscopy. Anal Chem. 2006;78:7761–7769. doi: 10.1021/ac061417b. [DOI] [PubMed] [Google Scholar]
- 57.Okada M, Smith NI, Palonpon AF, Endo H, et al. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proceedings of the National Academy of Sciences. 2012;109:28–32. doi: 10.1073/pnas.1107524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parekh SH, Lee YJ, Aamer KA, Cicerone MT. Label-Free Cellular Imaging by Broadband Coherent Anti-Stokes Raman Scattering Microscopy. Biophysical Journal. 2010;99:2695–2704. doi: 10.1016/j.bpj.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]