Abstract
Eudistomin U is a member of the β-carboline class of heterocyclic amine-containing molecules that are capable of binding to DNA. The structure of eudistomin U is unique since it contains an indole ring at the 1-position of the pyridine ring. While simple β-carbolines are reported to intercalate DNA, an examination of the mode of binding of eudistomin U has been lacking. We report preliminary spectroscopic (UV-Vis, thermal denaturation, CD) and calorimetric (DSC) data on the binding of eudistomin U to DNA, which suggest that eudistomin U binds weakly according to a mechanism that is more complicated than other members of its class.
Keywords: β-carboline, DNA, eudistomin U, UV-Vis, thermal denaturation, circular dichroism, differential scanning calorimetry
Graphical Abstract
Targeting DNA as a macromolecular receptor is an attractive route for perturbing biological processes with small molecules.1,2 DNA is an appealing target since its inherent chirality makes possible the rational design of molecular probes. In addition to the carbon-centered chirality of the nucleotide building blocks, DNA also possesses helicity that is necessary for molecular recognition. Of the many small molecules that bind to DNA, polyamides,2 octahedral metal complexes,3 cis-platin,4 peptide nucleic acid,5 polyimides,6 and natural products like mitomycin7 and daunorubicin,8 are the most studied. Investigations into their binding thermodynamics, binding kinetics, and sequence specificities have enhanced our understanding of ligand–DNA interactions and provide further motivation for the search of new architectural motifs with similar potential as probes for chemical biology.
β-carbolines are naturally occurring or synthetic small molecules that also bind DNA noncovalently.9 These molecules are indolo[3,2-b]pyridines, which are flat, aromatic amines capable of inserting themselves in between base pairs of duplex DNA. Simple β-carbolines such as harman (1) and norharman (2, Figure 1) are widely known to intercalate into prokaryotic and eukaryotic DNA, resulting in loss of replication fidelity and impaired repair processes.10,11 They have also been shown to inhibit DNA transcription in vitro and induce DNA strand breaks.12,13 There are other more complex β-carbolines whose DNA binding properties are less well understood. For example, the marine metabolite eudistomin U (3) has been shown to bind strongly to GC-rich regions of DNA, but an in depth investigation of this interaction has been lacking.8 The interaction between eudistomin U and DNA is particularly intriguing since the natural product possesses an aromatic ring attached to the 1-position of the β-carboline, which is unique in this family of molecules.
Figure 1.
Structures of natural and synthetic β-carbolines
In order to evaluate eudistomin U as a novel molecular probe, we recently disclosed the synthesis of 3 in five steps through a key palladium-catalyzed Suzuki cross coupling reaction.14 A broad cytotoxicity screen of eukaryotic and prokaryotic cells established that the natural product inhibited the growth of gram-positive bacteria at low micromolar concentrations. Given the evidence for DNA binding in the literature,10–13 we hypothesized that binding to DNA may be responsible for the observed cytotoxicity and sought to characterize this interaction biochemically. In this Letter, we describe the DNA binding behavior of eudistomin U using spectroscopic and calorimetric techniques and further comparison to simple β-carbolines and known intercalators.
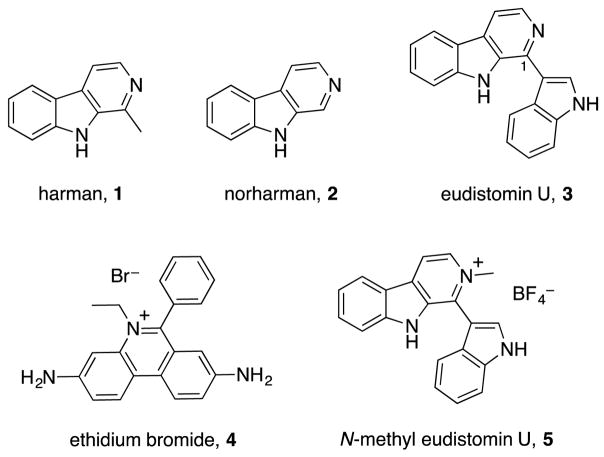
We first investigated the binding of β-carbolines to DNA using ultraviolet-visible spectroscopy (UV-Vis). When a small molecule binds to DNA through any of a variety of mechanisms including intercalation or groove binding, spectral changes will occur that are concentration-dependent.15 Figure 2 shows the UV-Vis spectrum of eudistomin U with increasing concentrations of calf thymus DNA. As DNA is titrated into a sample of eudistomin U at pH 7, a dramatic hyperchromic shift occurred at 257 nm, indicative of a simple interaction. Furthermore, the Soret peak around 400 nm shifted to longer wavelengths upon titration of DNA. This red-shifting of almost 20 nm is a further sign of a complexation between DNA and the natural product. The known intercalators harman and norharman displayed similar spectral changes in neutral phosphate buffer (see Supporting Information).16 Finally, since eudistomin U possesses a basic amine functional group, we investigated the pH-dependence of this binding through changes in the UV-Vis spectrum. Similar hyperchromicity at 257 nm and red-shifting above 400 nm were observed in both basic and acidic buffer. This suggests that protonation of the pyridine had no effect on binding, and that electrostatic interactions with the phosphate backbone of DNA were unlikely.
Figure 2.
UV-Vis spectra of eudistomin U and calf thymus DNA
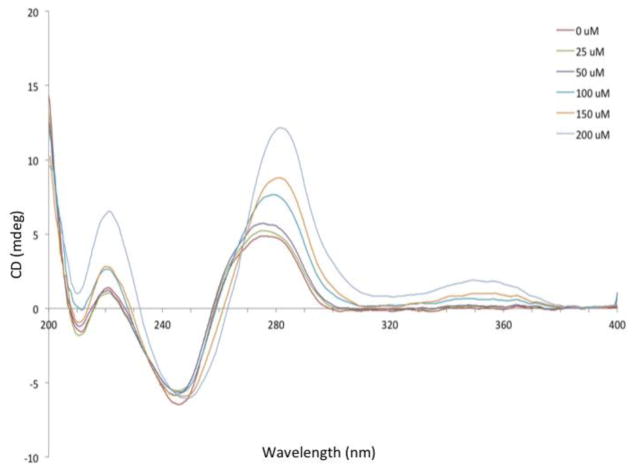
We next investigated the thermal denaturation of calf thymus DNA in the presence of eudistomin U. It is known that binding of a small molecule to DNA causes stabilization/destabilization of the double helix.17 For example, stabilization will increase the energy required to unravel the double helix into single stranded DNA. The melting temperature (Tm) is a measure of the stability of the double helix to temperature-induced denaturation and is useful in determining small molecule binding.18–19 Table 1 shows the Tm values for harman and eudistomin U at 50 μM concentration, as well as the known intercalator ethidium bromide. These results indicate a much weaker interaction between eudistomin U and double-stranded DNA than the UV-Vis spectra suggest. Harman produced a slightly larger increase in Tm than eudistomin U. Not surprisingly, the well-known intercalator ethidium bromide 4 (Figure 1) produced strong increases in the melting temperature at much lower concentrations, which indicates strong intercalation to DNA.
Table 1.
Thermal denaturation of calf thymus DNA
| compound | Tm (°C) | change |
|---|---|---|
| control | 66.8 | -- |
| harman, 1a | 70.8 | 4.0 |
| eudistomin U, 3a | 69.1 | 2.3 |
| ethidium bromide, 4b | 75.7 | 8.9 |
50 μM;
2.5 μM
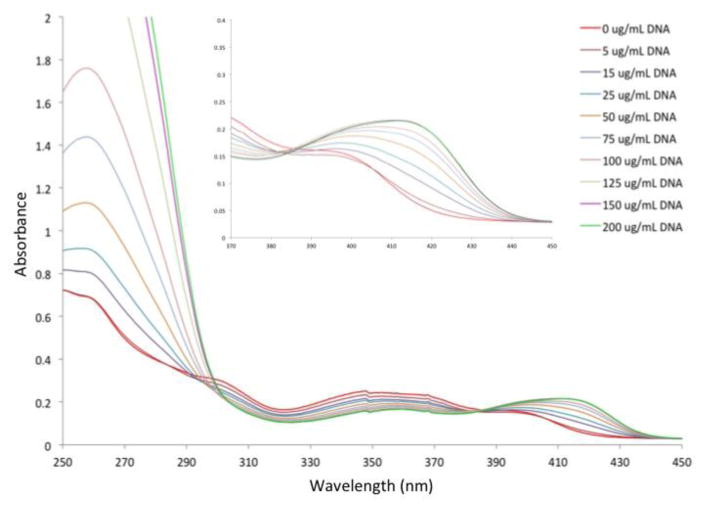
Next, we investigated the ability of these small molecules to perturb the secondary structure of DNA. When small molecules bind to DNA via specific mechanisms like intercalation or groove binding, the ability of the native double helix to absorb circularly polarized light will change.20 One can measure these changes using circular dichroism (CD). The helicity of DNA is very sensitive to small molecules and even weak binding can produce large changes in the CD spectrum. Ethidium bromide, for example, has been shown to have concentration-dependent hyperchromic effects on the helicity of DNA, but also induces ellipticity in the 300–400 nm region.
Figure 3 shows the CD spectrum of DNA upon titration with eudistomin U. Hyperchromicity and red-shifting occurred with increasing concentrations around 280 nm, indicating changes to the B-DNA structure. Induced ellipticity was observed at 349 nm, but this signal was much weaker than known intercalators like ethidium bromide. Nearly identical results were obtained under high salt conditions (0.1 M NaCl, see Supporting Information). Under basic conditions (pH = 8.9), there were no concentration-dependent changes to the CD spectrum (see Supporting Information). Under acidic conditions (pH = 5.4), increases in ellipticity were similar to neutral conditions, demonstrating a pH-dependence for this interaction.
Figure 3.
Circular dichroism spectra of eudistomin U and calf thymus DNA
In order to further investigate this pH-dependence, we synthesized N-methyl eudistomin U (5, Figure 1). We hypothesized that the positive formal charge on the β-carboline may more closely resemble the structure of ethidium bromide 4. This change might direct the small molecule to DNA through stronger electrostatic interactions with the phosphate backbone, where it can then associate with DNA more easily via intercalation or groove binding. Unfortunately, we observed no induced ellipticity around 350 and only weak hyperchromicity at the B-DNA maximum. We were also surprised that both harman and norharman did not produce induced ellipticity at 350 nm, which directly challenges the conclusion that these molecules solely intercalate between the base pairs of DNA.18 Previous CD studies of the interaction between these simple β-carbolines and bulk DNA suggested that they bind via intercalation, but the observed changes in ellipticity were obtained only at high concentrations of ligand.21 We were unable to reproduce this effect due to the insolubility of the ligands at these concentrations.
We also investigated the sequence-dependence of the eudistomin U-DNA interaction, which was suggested in the literature to be GC-specific. Thus, we measured the CD signal using AT- or GC-rich 16-mer nucleotides. Surprisingly, we observed no induced ellipticity nor hyperchromicity, even at concentrations as high as 200 μM. This result indicates that the interaction of DNA with eudistomin U is likely not sequence-specific.
Finally, through differential scanning calorimetry (DSC), we were able to quantify the effects on thermal denaturation of calf thymus DNA in the presence of eudistomin U (3), harman (1), and ethidium bromide (4). DSC is a microcalorimetry technique that measures Tm and enthalpy associated with melting and has been used successfully to screen the binding thermodynamics of nitrogen-containing small molecules.22–24 The DSC melting curves shown in Figure 4 confirm the results obtained by optical spectroscopy. All three small molecules increased the thermal stability of calf thymus DNA as evidenced by a strong increase in Tm when compared to control. While eudistomin U (3) stabilized duplex DNA with a Tm of 67 °C at 5 mM, roughly equal or stronger effects were observed for harman (1) and ethidium bromide (4) at much lower concentrations. An increase in the binding enthalpy was observed for the interaction between each small molecule and calf thymus DNA, which is concentration dependent for eudistomin U (3).
Figure 4.
Differential scanning calorimetry of small molecules and calf thymus DNA: a) melting profile of eudistomin U + calf thymus DNA; b) thermodynamic binding parameters: Tm and ΔH;c) melting profile of harman + calf thymus DNA; d) melting profile of ethidium bromide + calf thymus DNA
In conclusion, we have described spectroscopic data for the interaction of DNA with the natural product eudistomin U. Our UV-Vis, thermal denaturation, and CD studies all suggest a weak binding between eudistomin U and DNA. Furthermore, the lack of any specific interaction with small oligomers of DNA indicates that a more complicated binding mechanism is involved. Finally, we have evidence that challenges literature reports that harman and norharman are intercalators, suggesting that further studies into their mechanism of binding is needed.
Supplementary Material
Acknowledgments
This research was supported in part by Providence College, the Rhode Island Foundation Medical Research Funds, and an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under the grant number P20GM103430. The authors would also like to acknowledge Dr. Tun-Li Shen at Brown University for HR-MS measurements.
Footnotes
Supplementary data associated with this article, including all experimental details, spectroscopic data, and the full characterization of new compounds can be found in the online version.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Palchaudhuri R, Hergenrother P. J Curr Opin Biotechnology. 2007;18:497. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB. Bioorg Med Chem. 2001;9:2215. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 3.Barton JK, Danishefsky AT, Goldberg JM. J Am Chem Soc. 1984;106:2172. [Google Scholar]
- 4.Peyrone M. Annalen Chemie Pharmazie. 1844;51:1–29. [Google Scholar]
- 5.Nielsen PE. Acc Chem Res. 1999;32:624. [Google Scholar]
- 6.Lokey RS, Kwok Y, Guelev V, Pursell CJ, Hurley LH, Iverson BL. J Am Chem Soc. 1997;119:7202. [Google Scholar]
- 7.Tomasz M. Chem Biol. 1995;2:575. doi: 10.1016/1074-5521(95)90120-5. [DOI] [PubMed] [Google Scholar]
- 8.Wilson WD, Jones RL. Nucleic Acids Res. 1982;10:1399. doi: 10.1093/nar/10.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao R, Peng W, Wang Z, Xu A. Curr Med Chem. 2007;14:479. doi: 10.2174/092986707779940998. [DOI] [PubMed] [Google Scholar]
- 10.de Meester C. Mutation Research. 1995;339:139. doi: 10.1016/0165-1110(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 11.Funayama Y, Nishio K, Wakabayashi K, Nagao M, Shimoi K, Ohira T, Hasegawa S, Saijo N. Mutation Research. 1996;349:183. doi: 10.1016/0027-5107(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 12.Madle E, Obe G, Hansen J, Ristow H. Mutation Research. 1981;90:433. doi: 10.1016/0165-1218(81)90065-3. [DOI] [PubMed] [Google Scholar]
- 13.Mita S, Kamataki T, Kato R. Carcinogenesis. 1984;5:715. doi: 10.1093/carcin/5.6.715. [DOI] [PubMed] [Google Scholar]
- 14.Roggero CM, Giulietti JM, Mulcahy SP. Bioorg Med Chem Lett. 2014;24:3549. doi: 10.1016/j.bmcl.2014.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu LC, Chen CS, Hsiao IC, Chern JW, Lin CH, Shen YC, Yeh SF. Chem Biol. 2005;12:1317. doi: 10.1016/j.chembiol.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, Cao R, Shi B, Yu L, Song H, Ren Z. Chem Pharm Bull. 2010;58:901. doi: 10.1248/cpb.58.901. [DOI] [PubMed] [Google Scholar]
- 17.Suh D, Chaires JB. Bioorg Med Chem. 1995;3:723. doi: 10.1016/0968-0896(95)00053-j. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi K, Nagao M, Sugimura T. Nucleic Acids Res. 1977;4:3679. doi: 10.1093/nar/4.11.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puglisi JD, Tinoco I., Jr Methods in Enzymol. 1989;180:304. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 20.Brahms J, Mommaerts WF. J Mol Biol. 1964;10:73. doi: 10.1016/s0022-2836(64)80029-2. [DOI] [PubMed] [Google Scholar]
- 21.Taira Z, Kanzawa S, Dohara C, Ishida S, Matsumoto M, Sakiya Y. Japanese Journal of Toxicology and Environmental Health. 1997;43:83. [Google Scholar]
- 22.Guthrie KM, Parenty ADC, Smith LV, Cronin L, Cooper A. Biophys Chem. 2007;126:117. doi: 10.1016/j.bpc.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Del Vecchio P, Esposito D, Ricchi L, Barone G. Int J Biol Macromol. 1999;24:361. doi: 10.1016/s0141-8130(99)00058-6. [DOI] [PubMed] [Google Scholar]
- 24.Paul P, Hossain M, Yadav RC, Kumar GS. Biophys Chem. 2010;148:93. doi: 10.1016/j.bpc.2010.02.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.