Abstract
Haemophilus parasuis is a common opportunistic pathogen known for its ability to colonize healthy piglets and causes Glässer's disease. The lipooligosaccharide (LOS) of H. parasuis is a potential virulence-associated factor. In this study, two putative glycosyltransferases that might be involved in LOS synthesis in H. parasuis SC096 were identified (lgtB and lex-1). Mutants were constructed to investigate the roles of the lgtB and lex-1 genes. The LOS from the ΔlgtB or Δlex-1 mutant showed truncated structure on silver-stained SDS-PAGE gel compared to the wild-type strain. The ΔlgtB and Δlex-1 mutants were significantly more sensitive to 50% porcine serum, displaying 15.0 and 54.46% survival rates, respectively. Complementation of the lex-1 mutant restored the serum-resistant phenotype. Additionally, the ΔlgtB and Δlex-1 strains showed impaired ability to adhere to and invade porcine kidney epithelial cells (PK-15). The above results suggested that the lgtB and lex-1 genes of the H. parasuis SC096 strain participated in LOS synthesis and were involved in serum resistance, adhesion and invasion.
Keywords: Haemophilus parasuis, lipooligosaccharide, glycosyltransferase, serum resistance, adhesion and invasion
Introduction
Haemophilus parasuis is an important porcine pathogen and the etiological agent of Glässer's disease, which is characterized by fibrinous polyserositis, polyarthritis, and meningitis. It is a commensal organism found in the upper respiratory tract of swine that causes systemic symptoms in conditions with decreased resistance (Oliveira and Pijoan, 2004). The exact mechanisms by which H. parasuis invades internal organs to cause local and disseminated infection are not fully understood.
Lipooligosaccharide (LOS) has been identified as a potential H. parasuis virulence factor, however, only one investigation has analyzed the role of antigenic structure of the H. parasuis LOS (Xu et al., 2013). Most LOS molecules consist of two main components: lipid A and a nonrepeating core oligosaccharide. The core oligosaccharide components are typically 3-deoxy-D-manno-octulosonic acid (Kdo), heptose (Hep), glucose (Glu), galactose (Gal), and phosphate. The backbone of the lipid A moiety is substituted at position 6′ with a 2,4-linked Kdo disaccharide, which serves as an acceptor for the transfer of the first heptose residue to position 5 of the first Kdo residue; this transfer is accomplished by the heptosyltransferase family (Gronow et al., 2005). A lack of genes encoding heptosyltransferases often prevents the incorporation of the heptose residue and subsequently blocks the addition of other sugar moieties, resulting in truncated LOS in bacteria, including Haemophilus influenzae, Haemophilus ducreyi, and Campylobacter jejuni (Gibson et al., 1997; Gronow et al., 2005; Naito et al., 2010). In H. parasuis, deletion of the opsX, rfaF, and waaQ genes, which encode the three heptosyltransferases, produced severely truncated LOS structures, decreased resistance to complement-mediated killing in serum and a decreased ability to adhere to and invade porcine kidney epithelial (PK-15) and porcine umbilical vein-derived endothelial cells (PUVECs) (Xu et al., 2013). However, other glycosyltransferases associated with LOS biosynthesis and pathogenesis have yet to be investigated.
Glycosyltransferase family 25 (NCBI accession no. cd06532) has been reported to be involved in LOS biosynthesis (Jennings et al., 1995; Edwards et al., 2005; Masoud et al., 2008). Here, two putative glycosyltransferase family 25 genes (lgtB and lex-1) were identified in H. parasuis SC096 by sequencing analysis. The lgtB genes from Neisseria meningitidis and Neisseria gonorrhoeae encode the β-1,4-galactosyltransferase required for LOS core biosynthesis and show homology to the galactosyltransferases from Pasteurella haemolytica, H. ducreyi, Haemophilus sommnus (GenBank accession no. AF096997), and H. influenzae (High et al., 1993; Potter and Lo, 1995; Sun et al., 2000; Park et al., 2002). In H. influenzae type B, the lex-1 gene is involved in LOS biosynthesis and virulence. Genetic transformation using the cloned H. influenzae type b DNA fragment containing lex-1 increased the virulence in virulence-deficient LOS mutants (Cope et al., 1991; Ma et al., 1996). However, whether the lgtB or lex-1 gene of H. parasuis participates in LOS biosynthesis and disease pathogenesis is unknown. In this study, we generated ΔlgtB and Δlex-1 mutants of the H. parasuis SC096 strain to investigate their roles in serum resistance, host cell adherence, and invasion.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli plasmids were propagated in E. coli DH5α grown in Luria-Bertani medium (Oxoid) at 37°C. H. parasuis clinical strain SC096 was cultured on Trypticase Soy Agar (TSA) or Trypticase Soy Broth (TSB) (Oxiod) supplemented with 0.002% (w/v) nicotinamide adenine dinucleotide (NAD; Sigma) and 5% (v/v) inactivated bovine serum at 37°C in a 5% CO2-enriched atmosphere. For selection of the plasmid-containing strains, the medium were supplemented with 30 μg/mL of kanamycin or gentamycin.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristic(s) | Source |
|---|---|---|
| STRAINS | ||
| E. coli DH5α | F−, ϕ80d/lacZΔM15, Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 | Laboratory collection |
| H. parasuis SC096 | Serovar 4 clinical isolate | Zhang et al., 2012b |
| ΔlgtB | SC096 ΔlgtB::KanR | This study |
| Δlex-1 | SC096 Δlex-1::KanR | This study |
| ΔlgtB-c | SC096 complemented ΔlgtB strain, GmR KanR | This study |
| Δlex-1-c | SC096 complemented Δlex-1 strain, GmR KanR | This study |
| ΔlgtB-np | SC096 ΔlgtB::GmR, in-frame non polar deletion | This study |
| ΔlgtB-oc | SC096 complemented ΔlgtB strain, GmR KanR, original locus complement | This study |
| PLASMIDS | ||
| pMD-19T (simple) | T-vector, AmpR | Takara Inc. |
| pK18mobsacB | Suicide and narrow-broad-host vector, KanR | Schäfer et al., 1994 |
| pSF115 | Kan resistance cassette-carrying complement vector, KanR | Zou et al., 2013 |
| p34S-Gm | Gm resistance cassette-carrying vector, GmR | Laboratory collection |
| pSF116 | Gm resistance cassette-carrying complement vector, GmR | This study |
| pZQ001 | A 1937bp fragment containing KanR, the upstream and downstream sequences of the lgtB gene in pMD 19T(simple), KanR | This study |
| pZQ002 | A 2076bp fragment containing KanR, the upstream and downstream sequences of the lex-1 gene in pMD 19T(simple), KanR | This study |
| pZQ003 | A 1398bp fragment containing GmR and the lgtB gene in pSF116 | This study |
| pZQ004 | A 1460bp fragment containing GmR and the lex-1 gene in pSF116 | This study |
| pZQ005 | A 1786bp fragment containing GmR, the upstream and downstream sequences of the lgtB gene in pMD 19T(simple), GmR | This study |
| pZQ006 | A 2624 bp fragment containing GmR and the lgtB gene in pMD 19T(simple), GmR | This study |
Construction and complementation of the lgtB and lex-1 mutants
The oligonucleotides used for PCR are listed in Table 2. A DNA fragment encompassing the upstream region of the lgtB gene was amplified using the primer pair P1 and P2. The region downstream of the lgtB gene was amplified using the primer pair P3 and P4. A kanamycin resistance (KanR) cassette was amplified from pK18mobsacB using primers P9 and P10. These three fragments were connected by overlap PCR with primers P1 and P4 and then ligated into pMD-19T (simple) to obtain the plasmid pZQ001. Natural transformation was used to introduce pZQ001 into SC096 to obtain the lgtB mutant following a previously described method (Zhang et al., 2012a). The lex-1 mutant was constructed in the same manner with different primers. Primers P5 and P6 were used to amplify the upstream region of lex-1, primers P7 and P8 were used to amplify the downstream region, and primers P9 and P10 were used to amplify the KanR cassette. The three fragments were amplified using primers P5 and P8 by overlap PCR and then ligated into pMD-19T (simple) to obtain the plasmid pZQ002. Finally, the plasmid was introduced into SC096 using natural transformation then generated the lex-1 mutant. The mutants were confirmed by PCR and sequencing.
Table 2.
Sequences of the PCR primers used in this study.
| Primers | Primer sequences(5′-3′) |
|---|---|
| P1 (lgtB up-F) | ATACCGCTTGTGTGTGAGCGTCTTATATCAGCT |
| P2 (lgtB up-R) | ATGTCAATTCGGGATCCGCGTCTACTTCAGTAAGCGAA |
| P3 (lgtB down-F) | GATCGGCTTCGTCGACACGTTCGTATGTAGGAGCTGCTGGAT |
| P4 (lgtB down-R) | AGGGTAGAAGCACTCATATAG |
| P5 (lex-1 up-F) | ATACCGCTTGTGTCACCTAAGATAATATCATC |
| P6 (lex-1 up-R) | ATGTCAATTCGGGATCCGCGTATGTGAGCGTCTTATATCAG |
| P7 (lex-1 down-F) | GATCGGCTTCGTCGACACGTTCGCTCCTATTAATGGTAG |
| P8 (lex-1 down-R) | GTAGCTCAGAATGATTATCGCCA |
| P9 (Kan-F) | CGCGGATCCCGAATTGACAT TTTTATGGACAGCAAGCGAA |
| P10 (Kan-R) | ACGTGTCGACGAAGCCGATC TCAGAAGAACTCGTCAAGAA |
| P11 (lgtB comp-F) | GGTTCAAAAGAAGTTTCTATGTAAGAGTTAATTCATATTGAAGG |
| P12 (lgtB comp-R) | ATGTCAATTCGGGATCCCTATTTAAATTCAACAGTTC |
| P13 (lex-1 comp-F) | GGTTCAAAAGAAGTTTCTATGTAATATGCTATCTTAGCATAAAG |
| P14 (lex-1 comp-R) | ATGTCAATTCGGGATCC TTATTCAAAAGGAATAATAC |
| P15 (GmR-R) | GCGGTACTTGGGTCGATATC |
| P16 (GmR BamHI-F) | CGCGGATCCCGAATTGACATCGAATTGACATAAGCCTGTTC |
| P17 (GmR SalI-R) | ACGTGTCGACGAAGCCGATCTTAGGTGGCGGTACTTGGGTC |
| Expression studies by quantitative RT-PCR | |
| P18 (lgtB-F) | GACTGGTTTGAGCATTTAGATG |
| P19 (lgtB-R) | TCTAATACAGAATAGCGGG |
| P20 (rplM-F) | GTGACTGGTATGTAGTAG |
| P21 (rplM-R) | TGCCACCTACATAGCCAG |
| P22 (lgtB-F for test) | AATATCTTCTGCTTCCAAGG |
| P23 (lgtB-R for test) | CAATCAATCGGTGTTTTCTG |
| P24 (lgtB-up-F for non polar deletion) | ACCGCTTGTGTGCCGTACCATAATGTTTAG |
| P25 (lgtB-up-R for non polar deletion) | TATAATTTCCTTCAATATGAAT |
| P26 (lgtB-down-F for non polar deletion) | TGAAAAATATTACATATGTATTTG |
| P27 (lgtB-down-R for non polar deletion) | AATTGCGTTGCAGTACAAGC |
| P28 (GmR-F for non polar deletion) | ATTCATATTGAAGGAAATTATAATGTTACGCAGCAGCAACGA |
| P29 (GmR-R for non polar deletion) | CAAATACATATGTAATATTTTTCATTAGGTGGCGGTACTTGGGTC |
| P30 (lgtB-up-R for original locus complementation) | CTATTTAAATTCAACAGTTCT |
| P31 (GmR-F for original locus complementation) | AGAACTGTTGAATTTAAATAGCGAATTGACATAAGCCTGTTC |
The pSF116 vector was constructed as follows. A 554-bp DNA fragment including the gentamicin resistance (GmR) gene was amplified from p34S-Gm using the primer pair P16 and P17. Then the GmR gene and pSF115 were digested with BamHI and SalI. The two digested products were ligated to obtain the complement vector pSF116. To construct the complementing plasmids pZQ003 and pZQ004, the lgtB and lex-1 genes were amplified from SC096, then cloned into KpnI and BamHI-digested pSF116 using the In-fusion HD cloning kit (Clontech Laboratories, TaKaRa Bio Inc., Shiga, Japan) respectively.
To construct an in-frame non-polar lgtB mutant, A DNA fragment about 500-bp encompassing the upstream region of the lgtB was amplified using the primer pair P24 and P25, whereas the downstream DNA region of the lgtB was amplified using the primer pair P26 and P27. An intact gentamicin resistant (GmR) cassette (from ATG to TAA) was amplified by PCR from p34S-Gm using primers P28 and P29. These three fragments were connected by overlap PCR with the primers P24 and P27, then purified and ligated into pMD19-T to give plasmid pZQ005. Then the plasmid was introduced into SC096 using natural transformation to construct a mutant containing a non-polar, in frame mutation in lgtB, ΔlgtB-np. This mutant was confirmed by PCR and sequenced using primer P22 and P23.
To construct a strain in which the wild-type lgtB gene is restored in the lgtB::kan mutant, the fragment containing upstream region of the lgtB and the intact lgtB was amplified using the primer pair P24 and P30. The downstream region fragment was amplified using the primer pair P26 and P27. The gentamicin resistant (GmR) cassette was amplified by PCR from p34S-Gm using primers P31 and P29. The three fragments were connected using primers P24 and P27 by overlap PCR and then ligated into pMD-19T (simple) to obtain the plasmid pZQ006. The plasmid was transformed into lgtB insertion mutant (ΔlgtB::KanR) to get the lgtB original locus complement strain. This complement strain was confirmed by PCR and sequenced using primer P22 and P23.
Growth studies
To obtain growth curves for the wild-type SC096 strain, lgtB or lex-1 mutant, cultures of each strain were grown overnight in TSB supplemented with NAD and 5% serum. Then the cultures were inoculated into fresh TSB medium supplemented with NAD and 5% serum at a ratio of 1: 100 and incubated at 37°C. The optical density at 600 (OD600) was measured at 1 h intervals.
LOS preparation
Extraction of LOS with high purity was performed using a modified phenol-water extraction protocol accompanied by proteinase K digestion of the bacterial proteins and nuclease elimination of the nucleic acids (Hitchcock and Brown, 1983). The LOS preparations were treated with sodium dodecyl sulfate (SDS) loading buffer (100 mM Tris–HCl, pH 8.0, 2% β-mercaptoethanol, 4% SDS, 0.2% bromophenol blue, 0.2% xylene cyanole, and 20% glycerol), separated by SDS-polyacrylamide gel electrophoresis (PAGE; 4% stacking gel and 15% separating gel) at 100 V for 1 h and visualized by silver staining.
Serum bactericidal assays
Porcine serum was collected from four healthy piglets (3–4 weeks old) from a farm free of Glässer's disease and was stored at −80°C. Some serum aliquots were treated at 56°C for 30 min to inactivate complement. The serum bactericidal assay was performed as described by (Zhang et al., 2012a). Briefly, bacterial suspensions [107–108 colony-forming units (CFU)/mL] were cultured with either fresh porcine serum or heat-inactivated serum at 1:1 ratios or different serum concentrations for 1 h. After incubation, 10-fold serial dilutions of the samples were generated, spotted onto plates and incubated at 37°C for 48 h. Then, the bacterial numbers were counted, and the survival ratios were calculated. The results were expressed as the means of triplicates from three independent experiments.
Adhesion and invasion assays
PK-15 cells were used for the adhesion and invasion assays following the previously described method (Xu et al., 2013). The cells (5 × 105 CFU/mL) were seeded into 24-well tissue culture plates in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% heat-inactivated fetal bovine serum. The cells were cultured at 37°C in a humidified incubator with 5% CO2 for 24 h, washed twice with PBS and then infected with approximately 1 × 107 CFU of H. parasuis. The culture plates were incubated for up to 2 h at 37°C to allow bacterial adhesion. The cells were rigorously washed five times with PBS to eliminate non-specific bacterial attachment and were then incubated for 10 min at 37°C with 100 mL of 0.25% trypsin/EDTA. After incubation, 900 μL of ice-cold TSB was added, the cells were removed from the culture plates by scraping the bottoms of the wells. Bacterial enumeration was performed using serial 10-fold dilutions and plating on TSA plates. For the invasion assay, cell culture, bacterial infection, and bacterial counting were performed as described above for the bacterial adherence assay except that the extracellular bacteria were killed by incubation of the monolayer with DMEM containing chloromycetin (25 μg/mL) for another 2 h following the incubation with the bacteria and three washes with PBS. All of the above assays were performed in triplicate and repeated three times.
Quantitative real-time PCR
RNAs were isolated from the SC096 and ΔlgtB-c strain. The lgtB gene transcripts were analyzed by quantitative reverse transcription PCR (qRT-PCR). RNA was extracted using the bacterial RNA kit (Omega, USA) according to the manufacturer's instructions. The reactions were performed with the one-step SYBR® PrimeScript™ PLUS RT-PCR Kit (Clontech, USA). The 2(−ΔΔC(T)) method was used to relatively quantitate lgtB gene expression compared to the stably expressed rplM reference gene. The 7500 Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA) was used for the assay.
Statistical analysis
All experiments were repeated three times, and the results were expressed as means ± the standard deviations (SD). To determine significance of obtained results, comparison between groups was made using Student t-test. P < 0.01 were considered statistically significant.
Results
Analysis of the lgtB and lex-1 gene sequences
Two putative lipooligosaccharide biosynthesis genes (lgtB and lex-1) from H. parasuis SC096 were sequenced and identified using the BLAST program. The lgtB gene had 789 bp and showed 100% identity with a glycosyltransferase from H. parasuis ZJ0906 (NCBI accession no. AGO15727), whereas lex-1 had 837 bp and showed 96% identity with another glycosyltransferase from H. parasuis ZJ0906 (NCBI accession no. AGO15728).
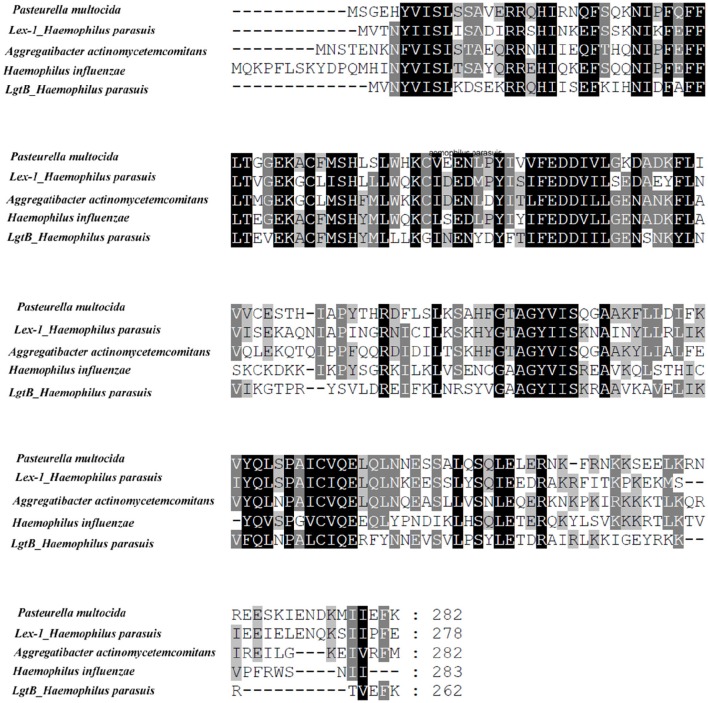
A global sequence comparison between the known lgtB and lex-1 proteins indicated that they belonged to glycosyltransferase family 25. lgtB showed 41, 42, and 41% identities with the β-1,4 galactosyltransferase from Pasteurella multocida (no. WP_014390682.1), the lipooligosaccharide biosynthesis protein lex-1 from Aggregatibacter actinomycetemcomitans (no. WP_053330004.1) and the lipooligosaccharide biosynthesis protein lic2B from H. influenzae (no. WP_048952714.1), respectively. lex-1 showed 45, 46, and 46% identities with the same sequences, respectively. Multiple alignments were created using CLUSTAL W (Figure 1).
Figure 1.
Multiple alignments of the lex-1 or lgtB amino acid sequences from H. parasuis SC096 with those from Pasteurella multocida (no. WP_014390682.1), Aggregatibacter actinomycetemcomitans (no. WP_053330004.1), Haemophilus influenza (no. WP_048952714.1). Shadowed letters indicated either identical residues or conservative changes.
Construction of the H. parasuis ΔlgtB/Δlex-1 mutants and complemented strains
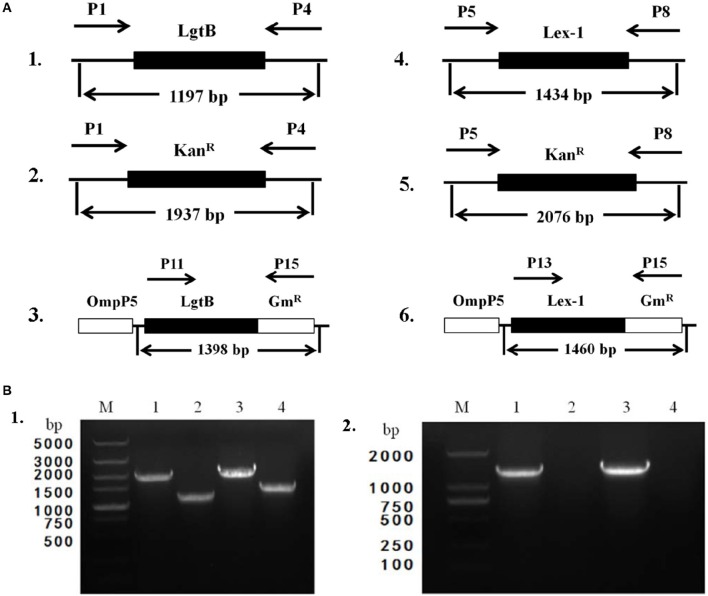
To obtain the ΔlgtB mutant, pZQ001, which contained the ΔlgtB::KanR insertion, was introduced into the SC096 strain by natural transformation. To obtain the Δlex-1 mutant, pZQ002, which contained the Δlex-1::KanR insertion, was introduced in the same manner. To determine the phenotypes of the lgtB or lex-1 strains due to the inactivation of the lgtB and lex-1 genes, pZQ003 was introduced into the ΔlgtB mutant by transformation to obtain a ΔlgtB-Comp strain, and pZQ004 was introduced to obtain a Δlex-1-Comp strain. In the complemented strains, the intact lgtB or lex-1 with a gentamycin resistance cassette was inserted immediately downstream of ompP5. Analysis of the transformants indicated that the lgtB and lex-1 genes plus a kanamycin resistance cassette were integrated into the homologous chromosomes of the mutant strains (Figure 2A). Colony PCR was used to confirm the transformants (Figure 2B). The ability of these mutants to grow under standard conditions was tested, but only negligible changes in the growth rates were detected (Figure S1).
Figure 2.
Construction and characterization of ΔlgtB and Δlex-1 mutants and complemented strains. (A) Parts 1–3 show the maps of the lgtB gene of wild-type SC096, the lgtB::KanR insertion mutant and its complemented strain. Parts 4–6 show the maps of the lex-1 gene of wild-type SC096, the lex-1::KanR insertion mutant and its complemented strain. (B) Part 1, Primers P1 and P4 were used to amplify upstream to downstream of lgtB from the ΔlgtB mutant strain (lane 1), the wild type SC096 (lane 2); Primers P5 and P8 were used to amplify upstream to downstream of lex-1 from the Δlex-1 mutant strain (lane 3), the wild type SC096 (lane 4). Part 2, Primers P11 and P15 were used to amplify lgtB and GmR from the ΔlgtB-c strain (lane 1), the wild type SC096 (lane 2); Primers P13 and P15 were used to amplify lex-1 and GmR from the Δlex-1-c strain (lane 3), the wild type SC096 (lane 4).
Denaturing gel electrophoresis of the lipooligosaccharides
To evaluate the variations in the LOS glycoforms, the LOSs of the wild-type SC096 strain, mutants and complemented strains were assessed by SDS-PAGE. Electrophoretic analysis of the LOS from the ΔlgtB or Δlex-1 mutant indicated that the profiles had changed compared to the profile of the parental strain SC096 (Figure 3). The SC096 strain had a LOS band at 20 kDa (Lane 1), whereas the ΔlgtB mutant had a LOS with a distinctly smaller molecular mass (Lane 2). Specifically, the LOS of the ΔlgtB or Δlex-1 mutant (Lanes 2 and 4) migrated faster than the LOS from the wild-type SC096 strain, and the LOS from the ΔlgtB mutant migrated even faster than the LOS from the Δlex-1 mutant. The LOSs of the complemented strains (Lanes 3 and 5) were similar or identical to the LOS of the parent strain, which suggested that LOS synthesis was restored in the complemented strains. These results suggested that lgtB and lex-1 might affect the LOS structures.
Figure 3.
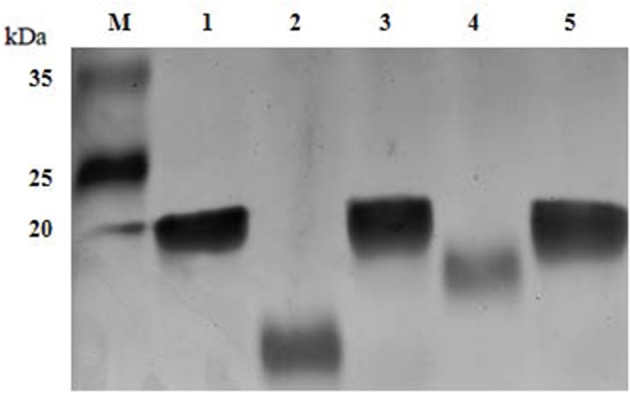
LOS profiles of H. parasuis SC096 strain, mutants, and complemented strains. Lane 1, wild-type SC096; Lane 2 and 3, ΔlgtB and its complemented strain ΔlgtB-c; Lane 4 and 5, Δlex-1 and its complemented strain Δlex-1-c.
Resistance to complement-mediated serum killing
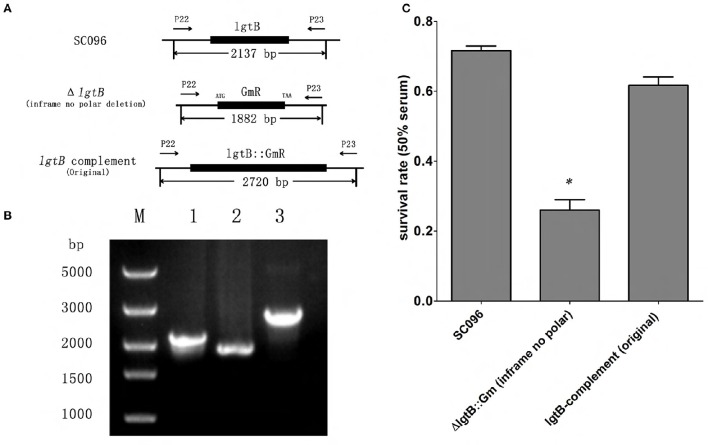
To investigate whether the lgtB and lex-1 genes were involved in serum resistance, the survival rates of ΔlgtB, Δlex-1, and their complemented strains were assessed in 50% porcine serum (Figure 4A). Compared to the wild-type SC096 strain, the ΔlgtB and Δlex-1 mutants were both significantly more sensitive to pig serum (p < 0.01), resulting in survival rates of 15.0 and 54.46% in 50% pig serum, respectively. The ΔlgtB mutant was more susceptible to pig serum than the Δlex-1 strain (p < 0.01). Furthermore, lex-1 mutant showed significantly increased susceptibility to serum of different concentrations compared with the wild type strain SC096 (p < 0.01; Figure S2). Interestingly, complementation of the lex-1 mutant restored the serum resistance phenotype, whereas the survival rate following complementation of the lgtB mutant was 8.51%, representing an approximately 10-fold reduction in 50% porcine serum susceptibility. A qRT-PCR analysis was performed to confirm the transcription level of the lgtB gene in the complemented strain (Figure 4B). The ΔlgtB-c strain exhibited lgtB transcription level that was increased 27.05-fold relative to the SC096 strain, indicating that lgtB was overexpressed in the complemented strain. In order to confirm the phenotype of lgtB mutant, a new in-frame deletion non-polar mutant (ΔlgtB::GmR) and an original locus complemented strain were constructed (Figures 5A,B). The new mutant was as sensitive to pig serum as the previous, whereas the original locus complemented strain restored the serum resistance phenotype (Figure 5C). The results suggested that lgtB and lex-1 may be associated with serum resistance.
Figure 4.
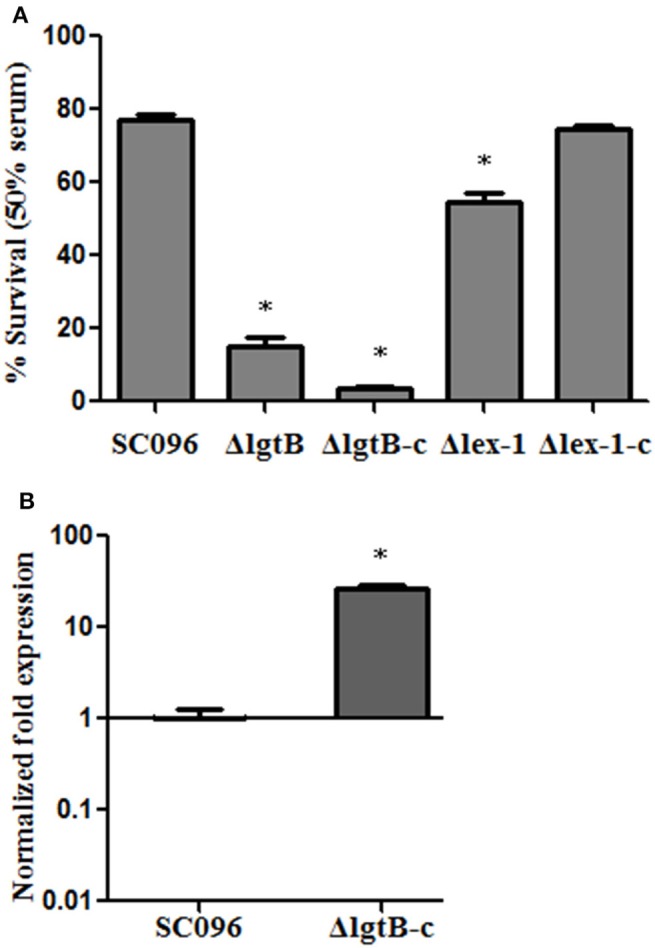
Survival of H. parasuis strains in 50% porcine sera (A). The survival percentage was calculated as the ratio of colonies in fresh serum to those in heat-treated serum. Error bars represent the standard deviation from three independent experiments. The asterisks indicate that the survival of bacteria in serum was statistically different (p < 0.01) from that of the wild-type SC096 strain as judged by the Student t-test. qRT-PCR analysis of mRNA levels of lgtB in ΔlgtB-c mutant compared with mRNA levels wild-type SC096 (B). The data represent means standard errors (n = 3). The asterisks indicate that mRNA levels of lgtB in the complemented strain was statistically different (p < 0.01) from that of the wild-type strain as judged by the Student t-test.
Figure 5.
Construction and verification of lgtB in-frame non polar mutant and the original complemented strain. (A) Schematic of lgtB in-frame non polar mutant and the original complemented strain. The lgtB in-frame non polar mutant was constructed by replacing lgtB with GmR gene from ATG to TAA codon. The original complemented strain was constructed by inserting the intact lgtB gene follow by GmR gene into previous constructed ΔlgtB::KanR strain in order to confirm the phenotype of the previous lgtB::KanR mutant. (B) PCR analysis verifying the lgtB in-frame non polar mutant and the original complemented strain. Primers P22 and P23 were used to amplify the locus region of lgtB from the wild type SC096 (lane 1), the lgtB in-frame non polar strain (lane 2), and the original complemented strain (lane 3); lane M shows a 5Kb DNA molecular marker. (C) Survival of lgtB in-frame non polar mutant and the original complemented strains treated with 50% porcine serum. The lgtB in-frame non polar mutant showed significantly increased susceptibility to serum compared with the wild type strain SC096 (p < 0.01) with 50% porcine serum, while the original complemented strain restored the serum resistant phenotype. Error bars represent the standard deviation of three independent experiments. The asterisks indicate that the survival of bacteria in serum was statistically different (p < 0.01) from that of the wild-type SC096 strain as judged by the Student t-test.
Adherence and invasion abilities
To assess the effects of the H. parasuis lgtB and lex-1 genes on host cell interactions, PK-15 cells were incubated with the wild-type, mutant, and complemented strains to compare the adherence and invasion abilities. As illustrated in Figure 6A, there was significantly less adhesion by the ΔlgtB and Δlex-1 mutants than the wild-type SC096 strain (p < 0.01). Similarly, the ΔlgtB and Δlex-1 mutants showed significantly less invasion efficiency (Figure 6B; p < 0.01). The adhesion and invasion levels were fully recovered in the complemented lex-1 strain. The complemented lgtB strain exhibited a 1.87-fold increase in adherence and a 1.64-fold increase in invasion. The results indicated that lgtB and lex-1 have an effect on the ability of the bacteria to interact with the host cells.
Figure 6.
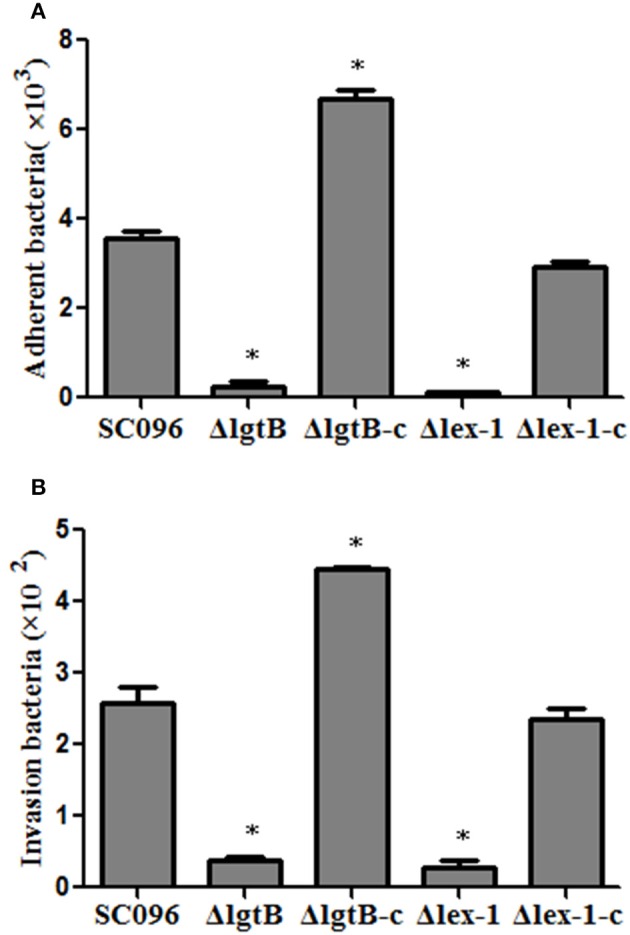
Adherence (A) and invasion (B) of H. parasuis wild-type SC096, ΔlgtB and Δlex-1 mutants and complemented strains in PK-15 cells. The data represented the number of bacteria that adhered to or invaded the cells in each well of a 24-well plate. Error bars represent the standard deviation from three independent experiments performed in triplicate. The asterisks indicate that the number of bacteria bound to the PK-15 cells was statistically different (p < 0.01) from that of the wild-type SC096 strain as judged by the Student t-test.
Discussion
Previous investigations have shown that the H. parasuis LOS has a significant influence on pathogenesis, including pathogen adherence, invasion, serum resistance, and endotoxicity (Amano et al., 1997; Tadjine et al., 2004; Zhang et al., 2014). Analysis of LOS biosynthesis and structures could contribute to explorations of the relationship between the LOS components and pathogenesis (Perry et al., 2013). One study presented evidence that the heptosyltransferases that transfer the heptose I and heptose II residues contributed to the virulence-associated properties of H. parasuis (Xu et al., 2013). Additionally, some galactosyltransferases are involved in the biosynthesis of LOS and virulence in certain pathogenic bacteria. For instance, β-1,4-galactosyltransferases encoded by the lgtB gene of N. meningitidis are required for the addition of least three sugars in the lacto-N-neotetraose chain (Park et al., 2002). In H. influenzae, the lex2 locus is identified as a phase-variable LPS biosynthetic locus and contributes to resistance of the bacteria to the killing effect of serum (Griffin et al., 2005; Deadman et al., 2009). Moreover, lic2B gene, which is required for addition of a galactose residue to the LOS outer core, is crucial for optimal survival of nontyeable H. influenzae in a mouse model of bacteremia and for evasion of serum complement (Wong et al., 2011). In H. ducreyi, the lbgAB genes are involved in lipooligosaccharide biosynthesis and serve as one indicator of the classification of H. ducreyi strains (Stevens et al., 1997; Tullius et al., 2002). To examine whether the lgtB and lex-1 of H. parasuis genes participate in LOS biosynthesis, we constructed chromosomal knockout ΔlgtB and Δlex-1 mutants of the H. parasuis SC096 strain. Deletion of either the lgtB or the lex-1 gene caused a truncated LOS profile on silver-stained SDS-PAGE gel, which demonstrated that lgtB and lex-1 were necessary for lipooligosaccharide biosynthesis in the SC096 strain. The phenotypic differences in the LOS patterns following the deletion of the lgtB and lex-1 genes in the SC096 strain could be attributed to the substrate specificity of the two glycosyltransferases.
Serum resistance is an important bacterial pathogenic mechanism. In H. influenzae, both lipooligosaccharide and capsular polysaccharide contribute to resistance against complement-mediated attacks and, hence, the increased survival of H. influenzae (Hallström and Riesbeck, 2010). In H. parasuis, LOS and the polysaccharide biosynthesis protein CapD have been reported to participate in resistance to complement-dependent bactericidal activity (Wang et al., 2013; Xu et al., 2013). Loss of heptose I or heptose II from LOS resulted in notable defects in serum resistance, and deletion of CapD significantly attenuated the serum resistance ability and pathogenicity of H. parasuis. Here, sensitivity to complement following the loss of the lgtB gene in the SC096 strain indicated that lgtB gene expression was associated with serum resistance. However, the lgtB complemented strain (lgtB located in the end of ompA) could not recover the serum-resistant phenotype and was even more sensitive than the lgtB mutant. In fact, the LOS pattern of the lgtB complemented strain was restored to the pattern observed for the wild-type strain. In order to confirm the phenotype of lgtB mutant, an in-frame non-polar deletion mutant (ΔlgtB::GmR) and an original locus complemented strain were constructed. Results obtained with the non-polar deletion mutant were consistent with those obtained with the previous insertion mutant which indicated that serum sensitive phenotype in lgtB mutant may be caused by lgtB gene deletion but not by mutation in other genes. The original locus complemented strain showed serum resistance phenotype, indicated that the original locus complementation of the mutation results in a return to the wild type phenotype. Therefore, the serum sensitive phenotype of the previous complemented strain ΔlgtB-c may be caused by lgtB over-expression.
Host cell invasion is a H. parasuis virulence mechanism and the LOS play specific roles in the process (Bouchet et al., 2008; Aragon et al., 2010). The truncated LOS in the ΔrfaE, ΔopsX, and ΔrfaF mutants reduced the adherence and invasion abilities in PUVEC and PK-15 cells (Xu et al., 2013; Zhang et al., 2014). Consistent with previous reports, this study showed impairments in adhesion and invasion of PK-15 cells for both the ΔlgtB and Δlex-1 mutants, indicating that the lgtB and lex-1 genes were also required for host cell interactions. Compared with the wild-type strain, the ΔlgtB-c strain exhibited increased attachment and invasion rates that appeared to be associated with up-regulation of the lgtB gene (Figure 6).
Conclusions
In conclusion, this study investigated the influences of the lgtB and lex-1 genes in the H. parasuis SC096 strain on LOS synthesis, serum resistance, adhesion, and invasion. The ΔlgtB and Δlex-1 mutants caused severe LOS truncations, significant sensitivity to complement-mediated serum and reductions in adherence to and invasion of PK-15 cells. Taken together, the data indicate that the lgtB and lex-1 genes are involved in lipooligosaccharide biosynthesis and may be novel pathogenicity-associated determinants in H. parasuis.
Author contributions
QZ: performed research, analyzed data, wrote paper. SF: analyzed data, wrote paper. JZ: analyzed data. AJ: helped with experiment. KY: helped with experiment. KX: helped with experiment. HF: funded research, analyzed data. ML: funded research, analyzed data.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Public Agriculture Specific Research Program (Grant No. 201303034).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00100
References
- Amano H., Shibata M., Takahashi K., Sasaki Y. (1997). Effects on endotoxin pathogenicity in pigs with acute septicemia of Haemophilus parasuis infection. J. Vet. Med. Sci. 59, 451–455. 10.1292/jvms.59.451 [DOI] [PubMed] [Google Scholar]
- Aragon V., Bouchet B., Gottschalk M. (2010). Invasion of endothelial cells by systemic and nasal strains of Haemophilus parasuis. Vet. J. 186, 264–267. 10.1016/j.tvjl.2009.08.013 [DOI] [PubMed] [Google Scholar]
- Bouchet B., Vanier G., Jacques M., Gottschalk M. (2008). Interactions of Haemophilus parasuis and its LOS with porcine brain microvascular endothelial cells. Vet. Res. 39, 42 10.1051/vetres:2008019 [DOI] [PubMed] [Google Scholar]
- Cope L. D., Yogev R., Mertsola J., Latimer J. L., Hanson M. S., McCracken G. H., Jr., et al. (1991). Molecular cloning of a gene involved in lipooligosaccharide biosynthesis and virulence expression by Haemophilus influenzae type B. Mol. Microbiol. 5, 1113–1124. 10.1111/j.1365-2958.1991.tb01884.x [DOI] [PubMed] [Google Scholar]
- Deadman M. E., Hermant P., Engskog M., Makepeace K., Moxon E. R., Schweda E. K., et al. (2009). Lex2B, a phase-variable glycosyltransferase, adds either a glucose or a galactose to Haemophilus influenzae lipopolysaccharide. Infect. Immun. 77, 2376–2384. 10.1128/IAI.01446-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. J., Allen S., Gibson B. W., Campagnari A. A. (2005). Characterization of a cluster of three glycosyltransferase enzymes essential for Moraxella catarrhalis lipooligosaccharide assembly. J. Bacteriol. 187, 2939–2947. 10.1128/JB.187.9.2939-2947.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B. W., Campagnari A. A., Melaugh W., Phillips N. J., Apicella M. A., Grass S., et al. (1997). Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J. Bacteriol. 179, 5062–5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin R., Bayliss C. D., Herbert M. A., Cox A. D., Makepeace K., Richards J. C., et al. (2005). Digalactoside expression in the lipopolysaccharide of Haemophilus influenzae and its role in intravascular survival. Infect. Immun. 73, 7022–7026. 10.1128/IAI.73.10.7022-7026.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronow S., Brabetz W., Lindner B., Brade H. (2005). OpsX from Haemophilus influenzae represents a novel type of heptosyltransferase I in lipopolysaccharide biosynthesis. J. Bacteriol. 187, 6242–6247. 10.1128/JB.187.17.6242-6247.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström T., Riesbeck K. (2010). Haemophilus influenzae and the complement system. Trends Microbiol. 18, 258–265. 10.1016/j.tim.2010.03.007 [DOI] [PubMed] [Google Scholar]
- High N. J., Deadman M. E., Moxon E. R. (1993). The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope alpha Gal(1-4)beta Gal. Mol. Microbiol. 9, 1275–1282. 10.1111/j.1365-2958.1993.tb01257.x [DOI] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. (1983). Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings M. P., Hood D. W., Peak I. R., Virji M., Moxon E. R. (1995). Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol. Microbiol. 18, 729–740. 10.1111/j.1365-2958.1995.mmi_18040729.x [DOI] [PubMed] [Google Scholar]
- Ma D., Alberti M., Lynch C., Nikaido H., Hearst J. E. (1996). The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19, 101–112. 10.1046/j.1365-2958.1996.357881.x [DOI] [PubMed] [Google Scholar]
- Masoud H., Moxon E. R., Richards J. C. (2008). Structural elucidation of lipopolysaccharide core oligosaccharides from lic1 and lic1/lic2 mutants of Haemophilus influenzae type b strain Eagan. Can. J. Microbiol. 54, 281–290. 10.1139/W08-009 [DOI] [PubMed] [Google Scholar]
- Naito M., Frirdich E., Fields J. A., Pryjma M., Li J., Cameron A., et al. (2010). Effects of sequential Campylobacter jejuni 81-176 lipooligosaccharide core truncations on biofilm formation, stress survival, and pathogenesis. J. Bacteriol. 192, 2182–2192. 10.1128/JB.01222-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira S., Pijoan C. (2004). Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet. Microbiol. 99, 1–12. 10.1016/j.vetmic.2003.12.001 [DOI] [PubMed] [Google Scholar]
- Park J. E., Lee K. Y., Do S. I., Lee S. S. (2002). Expression and characterization of beta-1,4-galactosyltransferase from Neisseria meningitidis and Neisseria gonorrhoeae. J. Biochem. Mol. Biol. 35, 330–336. 10.5483/BMBRep.2002.35.3.330 [DOI] [PubMed] [Google Scholar]
- Perry M. B., MacLean L. L., Gottschalk M., Aragon V., Vinogradov E. (2013). Structure of the capsular polysaccharides and lipopolysaccharides from Haemophilus parasuis strains ER-6P (serovar 15) and Nagasaki (serovar 5). Carbohydr. Res. 378, 91–97. 10.1016/j.carres.2013.04.023 [DOI] [PubMed] [Google Scholar]
- Potter M. D., Lo R. Y. (1995). Cloning and characterization of a gene from Pasteurella haemolytica A1 involved in lipopolysaccharide biosynthesis. FEMS Microbiol. Lett. 129, 75–81. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. 10.1016/0378-1119(94)90324-7 [DOI] [PubMed] [Google Scholar]
- Stevens M. K., Klesney-Tait J., Lumbley S., Walters K. A., Joffe A. M., Radolf J. D., et al. (1997). Identification of tandem genes involved in lipooligosaccharide expression by Haemophilus ducreyi. Infect. Immun. 65, 651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Schilling B., Tarantino L., Tullius M. V., Gibson B. W., Munson R. S., Jr. (2000). Cloning and characterization of the lipooligosaccharide galactosyltransferase II gene of Haemophilus ducreyi. J. Bacteriol. 182, 2292–2298. 10.1128/JB.182.8.2292-2298.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadjine M., Mittal K. R., Bourdon S., Gottschalk M. (2004). Production and characterization of murine monoclonal antibodies against Haemophilus parasuis and study of their protective role in mice. Microbiology 150(Pt 12), 3935–3945. 10.1099/mic.0.27443-0 [DOI] [PubMed] [Google Scholar]
- Tullius M. V., Phillips N. J., Scheffler N. K., Samuels N. M., Munson R. S., Jr., Hansen E. J., et al. (2002). The lbgAB gene cluster of Haemophilus ducreyi encodes a beta-1,4-galactosyltransferase and an alpha-1,6-DD-heptosyltransferase involved in lipooligosaccharide biosynthesis. Infect. Immun. 70, 2853–2861. 10.1128/IAI.70.6.2853-2861.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu X., Wu Y., Li L., Cao R., Cai X., et al. (2013). Polysaccharide biosynthesis protein CapD is a novel pathogenicity-associated determinant of Haemophilus parasuis involved in serum-resistance ability. Vet. Microbiol. 164, 184–189. 10.1016/j.vetmic.2013.01.037 [DOI] [PubMed] [Google Scholar]
- Wong S. M., St. Michael F., Cox A., Ram S., Akerley B. J. (2011). ArcA-regulated glycosyltransferase lic2B promotes complement evasion and pathogenesis of nontypeable Haemophilus influenzae. Infect. Immun. 79, 1971–1983. 10.1128/IAI.01269-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. G., Zhang L. Y., Zhang B., Feng S. X., Zhou S. M., Li J. Y., et al. (2013). Involvement of lipooligosaccharide heptose residues of Haemophilus parasuis SC096 strain in serum resistance, adhesion and invasion. Vet. J. 195, 200–204. 10.1016/j.tvjl.2012.06.017 [DOI] [PubMed] [Google Scholar]
- Zhang B., Feng S., Xu C., Zhou S., He Y., Zhang L., et al. (2012a). Serum resistance in Haemophilus parasuis SC096 strain requires outer membrane protein P2 expression. FEMS Microbiol. Lett. 326, 109–115. 10.1111/j.1574-6968.2011.02433.x [DOI] [PubMed] [Google Scholar]
- Zhang B., He Y., Xu C., Xu L., Feng S., Liao M., et al. (2012b). Cytolethal distending toxin (CDT) of the Haemophilus parasuis SC096 strain contributes to serum resistance and adherence to and invasion of PK-15 and PUVEC cells. Vet. Microbiol. 157, 237–242. 10.1016/j.vetmic.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Zhang B., Yu Y., Zeng Z., Ren Y., Yue H. (2014). Deletion of the rfaE gene in Haemophilus parasuis SC096 strain attenuates serum resistance, adhesion and invasion. Microb. Pathog. 74, 33–37. 10.1016/j.micpath.2014.07.006 [DOI] [PubMed] [Google Scholar]
- Zou Y., Feng S. X., Xu C. G., Zhang B., Zhou S. M., Zhang L. Y., et al. (2013). The role of galU and galE of Haemophilus parasuis SC096 in serum resistance and biofilm formation. Vet. Microbiol. 162, 278–284. 10.1016/j.vetmic.2012.08.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.