Abstract
MicroRNAs (miRNAs), which are small (~21 nucleotides) non-coding RNAs, are important players in endochondral ossification, articular cartilage homeostasis, and arthritis pathogenesis. Comprehensive and genetic analyses of cartilage-specific or cartilage-related miRNAs have provided new information on cartilage development, homeostasis, and related diseases. State-of-the-art combinatorial approaches, including transcription-activator like effector nuclease (TALEN)/clustered regularly interspaced short palindromic repeats (CRISPR) technique for targeting miRNAs and high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation for identifying target messenger RNAs, should be used to determine complex miRNA networks and miRNA-dependent cartilage regulation. Use of advanced drug delivery systems involving cartilage-specific miRNAs will accelerate the application of these new findings in arthritis therapy.
Keywords: Cartilage, Chondrocytes, MicroRNAs, Osteoarthritis
INTRODUCTION
MicroRNAs (miRNAs) are small (~21 nucleotides) non-coding RNAs that play critical roles in various biological events such as development; homeostasis; and pathogenesis of diseases, including cancer and arthritis. To date, approximately 2,000 miRNAs have been identified in humans and mice. However, only a small number of these miRNAs have been examined in detail. This is partly because mammalian miRNAs were identified only approximately a decade ago and because experimental approaches for examining miRNAs need some skill and information. Nevertheless, growing evidence on the functional roles of miRNAs in various human diseases have prompted us to obtain insights on tissue- and/or disease-specific miRNAs of interest. Functions of several miRNAs in cartilage development and arthritis pathogenesis were confirmed in the first decade of miRNA research in arthritis. Results of these early studies have increased our chances of determining new miRNAs and of using these miRNAs for medical diagnosis and as therapeutic targets. This review provides a relatively short historical background of miRNA discovery and highlights cutting-edge research on miRNAs involved in cartilage development and arthritis pathogenesis.
HISTORY OF miRNA DISCOVERY
Although non-coding RNAs such as transfer RNA and ribosomal RNA have been known for decades, miRNAs have been identified recently in Caenorhabditis elegans by performing advanced genetic experiments.
Lin-4, a gene encoding an miRNA that was first identified by Dr. Ambros, regulates developmental timing in C. elegans and therefore is called a "heterochronic gene".[1] Mutations in C. elegans lin-4 induce incorrect cell division at specific developmental stages because of the upregulation of LIN-14 protein.[1,2]
For a while, this epoch-making discovery of miRNAs was thought to be an exception to the "central dogma," in which gene function appears as protein through messenger RNA (mRNA) transcription. However, after the identification of another miRNA (let-7) in 2000 in C. elegans and other species, including flies and humans, significant number of miRNAs were identified, which were transcribed from novel conserved genes.
MOLECULAR MECHANISMS UNDERLYING miRNA EXPRESSION AND FUNCTIONS
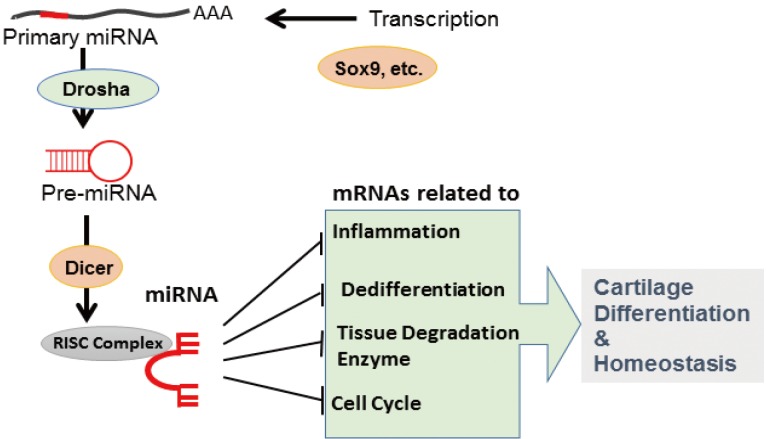
In general, miRNAs are generated from large transcripts called primary miRNA transcripts that are transcribed by Pol-II and processed by RNases Drosha in the nucleus and Dicer in the cytosol. Pre-miRNAs, which are transient products obtained from primary miRNAs during miRNA maturation, are transported from the nucleus to the cytosol by exportin 5, a dsRNA-binding protein. The final products, i.e., miRNAs, are integrated into RNA-induced silencing complex (RISC) to mediate their function, i.e., recognition of target mRNAs and repression of their expression (Fig. 1).
Fig. 1. MicroRNA (miRNA) synthesis and functions in chondrocytes. miRNAs are transcribed by specific transcription factors and processed by Drosha- and Dicer-mediated editing. A functional miRNA complex targets multiple messenger RNAs to regulate cartilage differentiation and homeostasis.
Spatio-temporal miRNA expression patterns have been examined by performing microarray analysis, next-generation sequencing, systematic and quantitative reverse transcription-polymerase chain reaction (RT-PCR), and whole-mount in situ hybridization. Results of these analyses have indicated that expression of some miRNAs is tissue and signal dependent. This specific miRNA expression is regulated at the level of Pol-II-dependent primary miRNA transcription and maturation, such as let-7 family by Lin-28a.
ROLE OF miRNAs IN SKELETAL DEVELOPMENT
Chondrocytes play a major role in skeletal development [3,4] and articular cartilage homeostasis.[5] The precise patterning underlying the development of skeletal framework relies on the appropriate control of chondrogenesis during which mesenchymal cells differentiate into chondrocytes.[3,4] During endochondral ossification, chondrocyte differentiation is divided into several stages, including proliferation, maturation, hypertrophy, and apoptosis. Finally, a cartilaginous scaffold formed by chondrocytes is replaced by the bone.[3,4] These differentiation and growth stages are tightly regulated by various molecules, including growth factors, cell signaling molecules, transcription factors, and miRNAs.[6]
Critical functions of miRNAs in endochondral ossification were determined by generating chondrocyte-specific Dicer-deleted mice that showed skeletal growth defects.[7] As stated above, Dicer is essential for synthesizing most miRNAs. Loss of Dicer produces diverse miRNA types in chondrocytes. Cartilage-specific Dicer-knockout mice develop growth defects because of the reduced proliferation of chondrocytes and because of the differentiation of these chondrocytes into their hypertrophic stages.
Based on this finding, which indicated the critical role of miRNAs in chondrogenesis, researchers next examined miRNA types involved in the generation of this phenotype. Expression of some miRNAs is ubiquitous, whereas that of some miRNAs is spatially and temporally restricted. Various miRNAs specifically expressed in chondrocytes were identified by performing microarray analysis, next-generation sequencing, etc., and their functions in cartilage development were examined.
A study involving in situ hybridization-based screening by using zebrafish development was the first to survey miRNAs expressed in chondrocytes.[8] This study identified 10 miRNAs (miR-23a, miR-27a, miR-27b, miR-140, miR-140*, miR-199, miR-199*, and miR-214) in the pharyngeal arches of zebrafish, suggesting their potential role in chondrocytes.[8] Cartilage-specific miRNAs were examined in various species, and miR-140 and miR-199 were identified as chondrocyte-specific miRNAs.[9] More comprehensive approaches such as next-generation sequencing by using various tissues from different species and in different developmental stages can be used to identify cartilage-specific miRNAs.[10]
Functions of specific miRNAs have been examined by generating miRNA-knockout mice.[11,12] Targeted deletion of miR-140 induces a mild defect in skeletal development. miR-140-null mice show disturbed endochondral ossification and reduced longitudinal bone growth.[11,12] Different target genes, including 1-(4,5-dimethoxy-2-nitrophenyl)ethyl ester (Dnpe), platelet derived growth factor receptor alpha (Pdgfra) and histone deacetylase-4 (HDAC4), are involved in the development of this phenotype.[11,12]
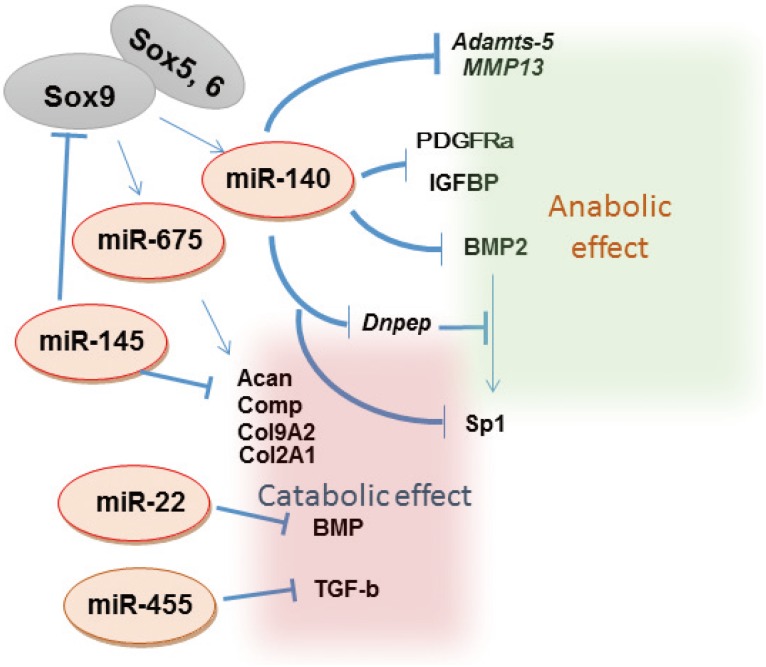
Although genetic evidence of the role of chondrocyte-specific miRNAs in bone development is limited, in vitro studies suggest that multiple miRNAs are involved in chondrogenesis. Use of mesenchymal stem cell (MSC) chondrogenesis as a model of cartilage development indicated that miR-199a, miR-124a, miR-130b, miR-152, miR28, miR26b, miR-455, miR192b, and miR-140 were upregulated and that miR-96 and miR-145 were downregulated during chondrogenesis (Fig. 2).[6,13]
Fig. 2. MicroRNA (miRNA) molecular networks involved in chondrocyte differentiation and homeostasis. Each miRNA has specific target messenger RNAs and affect anabolic or catabolic signals in chondrocytes. MMP, matrix metalloproteinase; PDGFRα, platelet-derived growth factor receptors-alpha; IGFBP, insulin-like growth factor binding protein; Dnpep, aspartyl aminopeptidase; BMP, bone morphogenetic protein; TGF-β, transforming growth factor-beta.
ARTICULAR CARTILAGE HOMEOSTASIS AND OSTEOARTHRITIS (OA) PATHOGENESIS
Articular cartilage, which lines the ends of bones in synovial joints, facilitates load transfer and frictionless movement.[5] Chondrocytes in the articular cartilage regulate cartilage homeostasis partly by synthesizing an extracellular matrix rich in type II collagen, proteoglycans, and related macromolecules.[14]
The articular cartilage is a target tissue for catabolic and inflammatory processes in joint diseases such as OA and rheumatoid arthritis.[15,16] Although precise mechanisms underlying the pathogenesis of arthritis are unknown, abnormal gene regulation in chondrocytes is suggested to be the main cause.[16]
OA is the most prevalent disorder of the synovial joints. It is characterized pathologically by focal articular cartilage degradation centered on load-bearing areas and is associated with new bone formation at joint margins (osteophytes), changes in the subchondral bone, and variable degrees of synovitis.[17] OA-associated changes in the articular cartilage include gradual proteolytic degradation of extracellular matrix along with increased synthesis of normal matrix components and of proteins that are normally absent in the articular cartilage, such as type X collagen or type IIA procollagen. These molecular events result in early morphological changes, cartilage surface fibrillation, cleft formation, and cartilage volume loss. Collectively, these changes in OA cartilage are attributed to the abnormal activation or differentiation of articular chondrocytes and possibly of resident adult MSCs.[17] Studies on the molecular control of the normal function of these cells and changes in arthritic joints have the potential to identify new disease mechanisms and therapeutic targets (Fig. 2).
miRNAs in OA
More than 30 miRNAs, including miR-455s,[18] miR-145, [10,13,19] miR-27b,[20] miR-146a,[21,22] miR-199a,[23,24] and miR-125b,[25] are abnormally expressed in human OA samples, suggesting the potential implications of cartilage homeostasis and OA development.[26,27,28,29,30]
miR-140 is persistently expressed in chondrocytes of the adult articular cartilage. Expression levels of miR-140 are significantly altered in human chondrocytes in the articular cartilage of patients with arthritis. Several studies performed to date indicate that miR-140 expression is downregulated in chondrocytes of patients with OA because of interleukin-1 beta (IL-1β) stimulation, which is suggested to be an in vitro model of arthritis pathogenesis.
Expression level of miR-140 may be upregulated in OA cartilage. This discrepancy may be explained as follows. OA may promote the regeneration of cartilage in some areas where chondrocyte proliferation is upregulated, resulting in the overexpression of these miRNAs. However, chondrocytes in severely damaged cartilage may lose their cartilage-specific gene expression, including cartilage-specific miRNA expression, and undergo dedifferentiation.
Knockdown of miR-140 in chondrocytes induces various changes in the expression profiles of genes involved in anabolic and catabolic phases. Importantly, miR-140-null mice show early onset of osteoarthritic changes in various articular cartilages. Consistently, miR-140 transgenic mice are resistant to arthritis induction compared with wild-type mice. Multiple potential miR-140 targets have been reported, including ADAMTS5 and MMP13 mRNAs, which are critical mediators of OA pathogenesis. The 3´-untranslated region (3´-UTR) of these mRNAs contain a miR-140 target sequence called "seed sequence," which is well conserved among various species, indicating that miR-140 expression strongly affects chondrocyte homeostasis and that consistent miR-140 expression in chondrocytes protects against arthritis progression (Fig. 2).
Sox9-DEPENDENT REGULATION OF CARTILAGE-SPECIFIC miRNAs
Sox9 is the key transcription factor in chondrogenesis. Sox proteins are homologous to proteins encoded by sex-determining region of Y chromosome (Sry) [31] and represent high-mobility group domain transcription factors. Sox9 plays an essential role in establishing precartilaginous condensations and in initiating chondroblast differentiation.[4,32,33] Sox5 and Sox6 cooperatively activate transcription along with Sox9 during chondrogenic differentiation. Mutations in SOX9 are observed in patients with a rare congenital dwarfism syndrome called campomelic dysplasia.[34,35,36] Analysis of mouse chimeras by using Sox9-/- embryonic stem cells indicates that Sox9-/- cells are excluded from cartilage tissues and that conditional deletion of Sox9 in cartilage leads to severe cartilage phenotype, with the downregulation of chondrocyte-specific extracellular matrix genes such as Col2a1.[37,38] Several studies have shown that Sox9 is downregulated in OA cartilage and that IL-1- and tumor necrosis factor-alpha (TNF-α) inhibit the expression of cartilage-specific genes such as Col2a1, Col11a2, Col9a2, and aggrecan,[39] suggesting that IL-1- and TNF-α-dependent downregulation of Sox9 contributes to OA pathogenesis.[39]
Primary miR-140 expression pattern are shadowing Sox9 and Col2 expression. Detailed analysis of potential tissue-specific enhancers of miR-140 by performing in vivo reporter assay have identified a non-coding sequence close to the gene encoding miR-140, which contains putative Sox9-binding sequences,[40,41,42] indicating cartilage-specific enhancer activity. Further studies indicate that Sox9 along with Sox5 and Sox6, which act together during Col2 expression, synergistically promote enhancer-dependent transcriptional activity, suggesting that miR-140 expression in cartilage is tightly regulated by the tissue-specific Sox trio at the level of primary miR-140 transcription.[42]
Sox9 expression is regulated by miR-145.[10,43] This regulation may inhibit chondrogenesis by repressing Sox9 expression. Therefore, reduced miR-145 expression may trigger chondrocyte differentiation (Fig. 2).[10,43]
NEW STRATEGIES FOR EXAMINING CARTILAGE-SPECIFIC miRNAs
Although analysis of miRNA functions is more difficult than that of mRNA functions, recent technological advances have identified the exact and comprehensive miRNA molecular networks in chondrocytes. The following sites are frequently used for bioinformatics approaches: Target–Scan, http://www.targetscan.org/; miRanda, http://cbio.mskcc.org/research/sander/data/miRNA2003/miranda_new.html; and PicTar, http://pictar.bio.nyu.edu/.
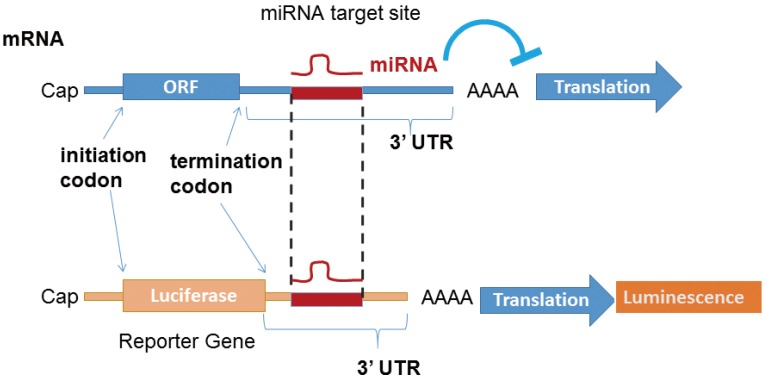
To identify the target mRNA of each miRNA, the potential target site of 3´-UTR sequence is subcloned into a luciferase reporter cassette, and the effect of the overexpression of a specific miRNA on luminescence activity is monitored (Fig. 3).
Fig. 3. MicroRNA (miRNA) target site validation. Reporter assay is routinely performed to determine whether an messenger RNA is a direct functional target of an miRNA.
High-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP) is a powerful strategy to determine the direct targets of each miRNA.[44] In HITS-CLIP, chondrocytes are UV cross-linked and an enriched mRNA–miRNA–Ago2 complex obtained by Ago2 immunoprecipitation is treated with RNase to completely degrade unmasked mRNAs. The remaining masked mRNAs, which escape RNase treatment, are examined by performing next-generation sequencing. To further validate the potential targets identified by HITS-CLIP, RNA-Seq is performed.[45]
Clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein-9 nuclease (Cas9) and transcription-activator like effector nuclease (TALEN), which introduce small deletions in a targeted sequence in vivo, are recently developed editing techniques for miRNA research.[46] We found that CRISPR-Cas9 and TALEN could be used to delete specific miRNAs and generate miRNA-knockout mice.[47,48] Application of these techniques allows us to delete various cartilage-related miRNAs rapidly and easily without affecting host gene expression.
THERAPEUTIC POTENTIAL OF miRNAs
Despite substantial recent progress in understanding OA pathogenesis, disease-modifying therapies for treating this prevalent joint disease are unavailable. miRNAs are a type of regulatory non-coding RNAs. Our and other recent studies suggest that miRNAs are important regulators of cartilage development, homeostasis, and OA.[29,30]
Accumulating evidence on the role of miRNAs in cancer, metabolic diseases, viral infections, and arthritis have suggested their use as novel therapeutic agents. However, efficient and precise delivery of miRNAs for therapeutic purposes is very challenging because of multiple obstacles such as methodologies for targeting various organs or tissues, protection against degradation by RNases, escape from Toll-like receptor-mediated immune system activation, appropriate incorporation into endogenous RISC, and off-target effects.[49,50]
Although recent advances for stabilizing anti-miRNAs, such as 2'-sugar modification, including 2'-O-methoxyethyl and locked nucleic acid, allow the knockdown of specific miRNAs, appropriate delivery systems are necessary to efficiently apply miRNA mimics in vivo.[49,50]
Footnotes
This work was supported by the Core Research for the Evolutionary Science and Technology (CREST) funding from the Japan Science and Technology Agency, JSPS KAKENHI (Grant Number: 26113008, 15H02560, and 15K15544), Bristol-Myers K.K. RA Clinical Investigation Grant, and grants from the NIH (grant numbers: AR050631 and AR065379).
No potential conflict of interest relevant to this article was reported.
References
- 1.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 2.Euling S, Ambros V. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell. 1996;84:667–676. doi: 10.1016/s0092-8674(00)81045-4. [DOI] [PubMed] [Google Scholar]
- 3.Cancedda R, Descalzi Cancedda F, Castagnola P. Chondrocyte differentiation. Int Rev Cytol. 1995;159:265–358. doi: 10.1016/s0074-7696(08)62109-9. [DOI] [PubMed] [Google Scholar]
- 4.de Crombrugghe B, Lefebvre V, Behringer RR, et al. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000;19:389–394. doi: 10.1016/s0945-053x(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 5.Hunziker EB, Kapfinger E, Geiss J. The structural architecture of adult mammalian articular cartilage evolves by a synchronized process of tissue resorption and neoformation during postnatal development. Osteoarthritis Cartilage. 2007;15:403–413. doi: 10.1016/j.joca.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Hong E, Reddi AH. MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering. Tissue Eng Part B Rev. 2012;18:445–453. doi: 10.1089/ten.TEB.2012.0116. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi T, Lu J, Cobb BS, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci U S A. 2008;105:1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 9.Ason B, Darnell DK, Wittbrodt B, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Sanchez A, Dudek KA, Murphy CL. Regulation of human chondrocyte function through direct inhibition of cartilage master regulator SOX9 by microRNA-145 (miRNA-145) J Biol Chem. 2012;287:916–924. doi: 10.1074/jbc.M111.302430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura Y, Inloes JB, Katagiri T, et al. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31:3019–3028. doi: 10.1128/MCB.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyaki S, Sato T, Inoue A, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong E, Reddi AH. Dedifferentiation and redifferentiation of articular chondrocytes from surface and middle zones: changes in microRNAs-221/-222, -140, and -143/145 expression. Tissue Eng Part A. 2013;19:1015–1022. doi: 10.1089/ten.TEA.2012.0055. [DOI] [PubMed] [Google Scholar]
- 14.Mow VC, Ratcliffe A, Poole AR. Cartilage and diarthrodial joints as paradigms for hierarchical materials and structures. Biomaterials. 1992;13:67–97. doi: 10.1016/0142-9612(92)90001-5. [DOI] [PubMed] [Google Scholar]
- 15.Månsson B, Carey D, Alini M, et al. Cartilage and bone metabolism in rheumatoid arthritis. Differences between rapid and slow progression of disease identified by serum markers of cartilage metabolism. J Clin Invest. 1995;95:1071–1077. doi: 10.1172/JCI117753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felson DT, Lawrence RC, Dieppe PA, et al. Osteoarthritis: new insights. Part 1: the disease and its risk factors. Ann Intern Med. 2000;133:635–646. doi: 10.7326/0003-4819-133-8-200010170-00016. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto M, Nakasa T, Hikata T, et al. Molecular network of cartilage homeostasis and osteoarthritis. Med Res Rev. 2008;28:464–481. doi: 10.1002/med.20113. [DOI] [PubMed] [Google Scholar]
- 18.Swingler TE, Wheeler G, Carmont V, et al. The expression and function of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum. 2012;64:1909–1919. doi: 10.1002/art.34314. [DOI] [PubMed] [Google Scholar]
- 19.Yang B, Kang X, Xing Y, et al. Effect of microRNA-145 on IL-1beta-induced cartilage degradation in human chondrocytes. FEBS Lett. 2014;588:2344–2352. doi: 10.1016/j.febslet.2014.05.033. [DOI] [PubMed] [Google Scholar]
- 20.Akhtar N, Rasheed Z, Ramamurthy S, et al. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis Rheum. 2010;62:1361–1371. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamasaki K, Nakasa T, Miyaki S, et al. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis Rheum. 2009;60:1035–1041. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones SW, Watkins G, Le Good N, et al. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis Cartilage. 2009;17:464–472. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Lin EA, Kong L, Bai XH, et al. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. J Biol Chem. 2009;284:11326–11335. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akhtar N, Haqqi TM. MicroRNA-199a* regulates the expression of cyclooxygenase-2 in human chondrocytes. Ann Rheum Dis. 2012;71:1073–1080. doi: 10.1136/annrheumdis-2011-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsukawa T, Sakai T, Yonezawa T, et al. MicroRNA-125b regulates the expression of aggrecanase-1 (ADAMTS-4) in human osteoarthritic chondrocytes. Arthritis Res Ther. 2013;15:R28. doi: 10.1186/ar4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirzamohammadi F, Papaioannou G, Kobayashi T. MicroRNAs in cartilage development, homeostasis, and disease. Curr Osteoporos Rep. 2014;12:410–419. doi: 10.1007/s11914-014-0229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papaioannou G, Mirzamohammadi F, Kobayashi T. MicroRNAs involved in bone formation. Cell Mol Life Sci. 2014;71:4747–4761. doi: 10.1007/s00018-014-1700-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barter MJ, Young DA. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Curr Rheumatol Rep. 2013;15:353. doi: 10.1007/s11926-013-0353-z. [DOI] [PubMed] [Google Scholar]
- 29.Gibson G, Asahara H. microRNAs and cartilage. J Orthop Res. 2013;31:1333–1344. doi: 10.1002/jor.22397. [DOI] [PubMed] [Google Scholar]
- 30.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nat Rev Rheumatol. 2012;8:543–552. doi: 10.1038/nrrheum.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarkson MJ, Harley VR. Sex with two SOX on: SRY and SOX9 in testis development. Trends Endocrinol Metab. 2002;13:106–111. doi: 10.1016/s1043-2760(01)00541-0. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka K, Matsumoto Y, Nakatani F, et al. A zinc finger transcription factor, alphaA-crystallin binding protein 1, is a negative regulator of the chondrocyte-specific enhancer of the alpha1(II) collagen gene. Mol Cell Biol. 2006;26:5202. doi: 10.1128/MCB.00845-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Lefebvre V, Huang W, Harley VR, et al. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–2346. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner T, Wirth J, Meyer J, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 35.Foster JW, Dominguez-Steglich MA, Guioli S, et al. Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature. 1994;372:525–530. doi: 10.1038/372525a0. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Q, Eberspaecher H, Lefebvre V, et al. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377–386. doi: 10.1002/(SICI)1097-0177(199708)209:4<377::AID-AJA5>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 37.Bi W, Deng JM, Zhang Z, et al. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 38.Bi W, Huang W, Whitworth DJ, et al. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami S, Lefebvre V, de Crombrugghe B. Potent inhibition of the master chondrogenic factor Sox9 gene by interleukin-1 and tumor necrosis factor-alpha. J Biol Chem. 2000;275:3687–3692. doi: 10.1074/jbc.275.5.3687. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura Y, He X, Kato H, et al. Sox9 is upstream of microRNA-140 in cartilage. Appl Biochem Biotechnol. 2012;166:64–71. doi: 10.1007/s12010-011-9404-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, Qin S, Yi C, et al. MiR-140 is co-expressed with Wwp2-C transcript and activated by Sox9 to target Sp1 in maintaining the chondrocyte proliferation. FEBS Lett. 2011;585:2992–2997. doi: 10.1016/j.febslet.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita S, Miyaki S, Kato Y, et al. L-Sox5 and Sox6 proteins enhance chondrogenic miR-140 microRNA expression by strengthening dimeric Sox9 activity. J Biol Chem. 2012;287:22206–22215. doi: 10.1074/jbc.M112.343194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang B, Guo H, Zhang Y, et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Licatalosi DD, Mele A, Fak JJ, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takada S, Asahara H. Current strategies for microRNA research. Mod Rheumatol. 2012;22:645–653. doi: 10.1007/s10165-011-0583-8. [DOI] [PubMed] [Google Scholar]
- 46.Matsubara Y, Chiba T, Kashimada K, et al. Transcription activator-like effector nuclease-mediated transduction of exogenous gene into IL2RG locus. Sci Rep. 2014;4:5043. doi: 10.1038/srep05043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takada S, Sato T, Ito Y, et al. Targeted gene deletion of miRNAs in mice by TALEN system. PLoS One. 2013;8:e76004. doi: 10.1371/journal.pone.0076004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inui M, Miyado M, Igarashi M, et al. Rapid generation of mouse models with defined point mutations by the CRISPR/Cas9 system. Sci Rep. 2014;4:5396. doi: 10.1038/srep05396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Shushan D, Markovsky E, Gibori H, et al. Overcoming obstacles in microRNA delivery towards improved cancer therapy. Drug Deliv Transl Res. 2014;4:38–49. doi: 10.1007/s13346-013-0160-0. [DOI] [PubMed] [Google Scholar]