Abstract
Background
Blood pressure (BP) homeostasis involves complex interactions among genetic and non-genetic factors, providing major challenges to dissection of the genetic components that influence BP and hypertension. In this study, we examine the effects of interaction of genetic variants with physical activity on BP in a relatively genetically homogenous cohort of rural Chinese villagers.
Methods
Generalized estimating equations analysis was used to test for associations of systolic blood pressure (SBP) and diastolic blood pressure (DBP) with variants in 24 genes in BP pathways (196 SNPs) among 3,142 Chinese participants divided according to physical activity (active versus inactive groups).
Results
In the physically active group, 2 SNPs in NR3C2 were significantly associated with lower SBP, and a SNP in SCNN1B was significantly associated with lower SBP and DBP. In the physically inactive group, a SNP in APLNR was associated with lower SBP, a SNP in GNB3 was associated with higher SBP and DBP, and a SNP in BDKRB2 was associated with lower DBP. Cumulative effects in carriers of minor alleles of these SNPs showed reductions of SBP and DBP as large as 8 and 5 mmHg, respectively, in the active individuals compared to inactive individuals carrying the same number of minor alleles.
Conclusions
We found that physical activity modifies the effects of genetic variants on BP. However, our results also show that active individuals with specific genotypes always have lower BP than inactive individuals with the same genotypes, demonstrating the overall beneficial effects of physical activity on blood pressure.
Keywords: Blood pressure, Epidemiology, genetics, physical activity, gene by environment interaction
INTRODUCTION
High blood pressure (BP) or hypertension is a global health issue1 and a major risk factor for cardiovascular, cerebrovascular, renal2, and eye diseases3. Major risk factors for hypertension include physical inactivity, obesity, high sodium and low potassium diet, and alcohol consumption2, 4. Genetic factors also influence hypertension, likely via complex interactions among many genes, as well as interactions among genetic and environmental factors such as diet and physical activity.
Controlling high BP by medication and life style modification can have a substantial impact on avoiding or reducing its medical complications and end organ damage5-8. Increased physical activity is an important life style modification that is recommended for hypertension patients based on documented benefits in many randomized clinical trials and meta-analyses9, 10. Different physiological mechanisms have been proposed to explain the beneficial effects of physical activity on BP levels. Increases in cardiac output during physical activity to meet higher demands for oxygen cause increased arterial shear stress associated with enhanced release of nitric oxide, a potent vasorelaxant11, and reduced expression of endothelin 1, a potent vasoconstrictor12. Physical activity also reduces plasma levels of norepinephrine and renin that are associated with reduced sympathetic nervous system activities and systemic vascular resistance9. Improved endothelial function and reduced insulin resistance are other characteristics of physical activity that might account for its beneficial effects on BP9.
The search for genetic factors underlying hypertension and measures of BP has been difficult, in part due to the complexity of interactions with environmental factors such as physical activity. Previous studies have examined interactions of particular genetic variants with different forms of physical activity, but typically included limited numbers of variants and relatively small sample sizes11-19. In this study, we test for effects of interaction of genetic variation with physical activity on BP in large numbers of rural Chinese (n=3,142). The study population was comprised of Han Chinese families from six rural villages in Northern China that participated in the multicenter study “Genetic Epidemiology Network of Salt Sensitivity (GenSalt)”. We interrogated 24 genes from key metabolic and physiological pathways that regulate BP homeostasis using 196 SNPs selected from the International HapMap project to provide comprehensive gene coverage in the Chinese population20.
METHODS
Study population
The study subjects were Han Chinese families from six rural villages in Northern China that participated in the multicenter GenSalt study to identify genetic factors that influence BP response to sodium and potassium intake. Chinese families were recruited through 18-60 year old probands who were either prehypertensive or had stage 1 hypertension (SBP of 130-160 mmHg and/or DBP of 85-100 mmHg), but had never been on any hypertension medication. Family members were excluded if they had stage-2 hypertension, secondary hypertension, or were treated with antihypertensive medications or low sodium diets. Additional exclusions included history of cardiovascular disease, diabetes, pregnancy, or heavy alcohol drinkers. A total of 3,142 individuals in 631 families participated in the GenSalt study. A large number of demographic, anthropomorphic, and medical variables were measured in this population. More information about subject recruitment and study measurements are available elsewhere21. All the participants signed an informed consent, and the Institutional Review Board approvals for this study were obtained at all the participant institutions.
Measurements of Blood Pressure and physical activity
Prior to dietary interventions, GenSalt participants were measured for base line BP over a three-day period while consuming their usual diets. Three morning BP measurements on each of three days were taken by the same trained and certified technician using the same random-zero sphygmomanometer. The average of these 9 SBP measurements was used in this analysis. Information about the intensity and the duration of habitual physical activities during work and leisure was collected using a detailed questionnaire. A metabolic equivalent (MET) variable defined as the ratio of the metabolic rate while seated and resting to the metabolic rate while performing particular tasks, was calculated for each participant (weekday and weekend), according to the following formula:
The average daily MET for each person was calculated based on the following formula:
After adjustment for covariates (gender, age, age2, and age3), log transformed MET residuals were used to classify the participants as active (residual > 0) or inactive (residual ≤ 0).
SNP selection and genotyping
Lymphocyte DNA samples from GenSalt participants were genotyped for SNPs in genes in key pathways of BP regulation. Initially, 237 SNPs (26 genes) were selected based on linkage disequilibrium structure in the Chinese population in the international HapMap project20. High throughput SNP genotyping was performed using the ABI SNPlex platform according to the manufacturer’s protocol22. 41 SNPs were excluded from subsequent association analysis due to low call rate (<80%), low minor allele frequency (MAF <0.05), or deviations from Hardy-Weinberg Equilibrium (HWE) (p<0.001). Supplementary Table 1 shows detailed information about the 196 SNPs in 24 genes that were used for association analysis.
Statistical analysis
Plink and PedCheck were used to assess consistency of the SNP genotypes with Mendelian transmission in GenSalt pedigrees23, 24. ASPEX and GRR were used to check for potential pedigree misreported relationships25, 26. Haploview was used for SNP descriptive statistics27. Generalized Estimating Equation methods (GEE) were used to test for associations between BP and genetic variants in active, inactive, and total populations under the additive model28. BP was adjusted for age, age2, age3, gender, body-mass index (BMI), pedigree generation, BP measurement room temperature and field center, and standardized to ensure a mean of 0 and a SD of 1. All SNPs were coded additively for the minor allele, and GEE analysis was performed with SAS 9.1 (proc genmod), and exchangeable working correlation matrix. False Discovery Rate (FDR) was used to correct for the multiple testing in GEE analysis29. Examining the LD patterns between the 196 SNPs showed that 24 pairs of SNPs were in almost complete LD (r2 ≥0.90), yielding 172 independent tests for our analysis.
RESULTS
A total of 3,142 individuals from rural Chinese families that participated in the GenSalt multicenter project were included in this study. Table 1 shows their basic characteristics stratified according to physical activity status (active versus inactive). The average age in both groups was 50 years old and there was slightly, but not significantly, more males in the active group compared to the inactive group. By design, the active group had significantly higher MET than the inactive group (mean MET scores differed by 30.6). In addition, the inactive group had significantly higher BP and BMI, while there was no significant difference in cigarette smoking and alcohol consumption between the two groups.
Table 1.
Basic characteristics of the GenSalt participants (active and inactive groups).
| Active | Inactive | |
|---|---|---|
| N* | 1453 | 1689 |
| Male % | 53.6 | 49.2 |
| Age | 50.1 ± 16.8 | 50.0 ± 16.7 |
| BMI* | 22.8 ± 3.1 | 23.3 ± 3.3 |
| SBP* | 122.8 ± 20.4 | 126.1 ± 21.2 |
| DBP (mmhg)* | 73.0 ± 10.4 | 75.2 ± 11.1 |
| MET* | 73.1 ± 20.0 | 42.5 ± 10.0 |
| Smoke % | 16.3 | 18.2 |
| Alcohol % | 13.0 | 13.8 |
p<0.0001 for comparisons between physically active and inactive groups
BMI: Body mass index
SBP: Systolic blood pressure
DBP: Diastolic blood pressure
MET: Metabolic equivalent score
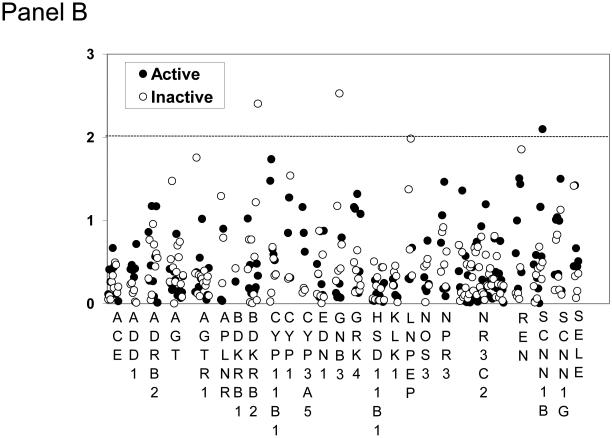
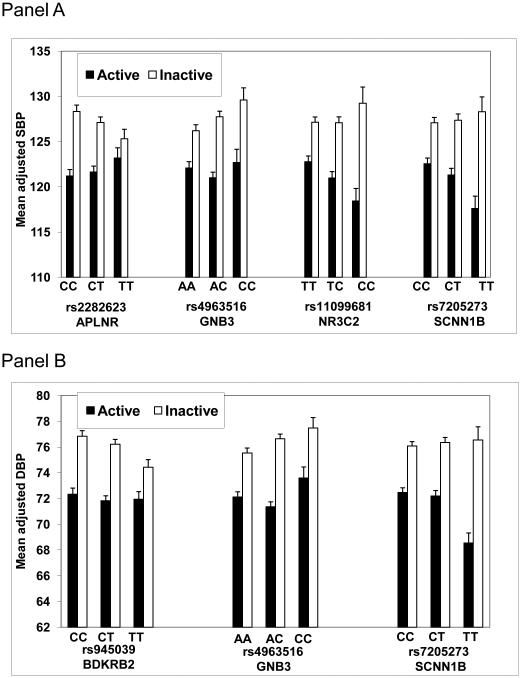
We genotyped the GenSalt subjects for DNA variants in 24 genes involved in metabolic and physiological pathways related to BP homeostasis. Figure 1 shows the results of association analysis in groups stratified according to physical activity status (complete list of 196 SNPs in Supplementary Table 1). We identified 6 SNPs in 5 genes (NR3C2, APLNR, GNB3, SCNN1B, BDKRB2) that were significantly associated in either the active or inactive groups (p < 0.01) (Table 2). Figure 2 shows mean adjusted effects on SBP and DBP for SNP genotypes that showed significant associations for active and inactive groups. SNP NR3C2-rs4835493 was not included in Figure 2 due to high LD with rs11099681 (r2 = 0.98). In the inactive group, the minor allele of APLNR-rs2282623 was associated with reduced SBP, the minor allele of BDKRB2-rs945039 was associated with reduced DBP, and the minor allele of GNB3-rs4963516 was associated with elevated SBP and DBP. In the active group, the minor allele of NR3C2-rs11099681 was associated with reduced SBP, and the minor allele of SCNN1B-rs7205273 was associated with reduced SBP and DBP. The FDR for all significant p-values was 0.37, meaning that less than 3 of the significant SNPs might be false positive results.
Figure 1.
−log p-values (Y axis) for associations of SBP (Panel A) and DBP (Panel B) with 196 individual SNPs separately for the physically active group and the physically inactive group of rural Chinese participants (n=3,142). The name of each of the 24 genes from blood pressure pathways is presented below graph (X axis). The dotted lines indicate p value <0.01.
Table 2.
Results of stratified analyses for individual SNPs that are significantly (p<0.01) associated with SBP (A) and DBP (B) in either physically active or inactive groups.
| (A) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Region | MAF | Maj/Min | β * | Active SE |
p | β | Inactive SE |
p | β | Combined SE |
p |
| APLNR | rs2282623 | 3' UTR | 0.40 | C/T | 0.043 | 0.044 | 0.320 | −0.102 | 0.038 | 0.008 | −0.040 | 0.030 | 0.185 |
| GNB3 | rs4963516 | 5' near gene | 0.33 | A/C | 0.024 | 0.044 | 0.591 | 0.113 | 0.040 | 0.004 | 0.077 | 0.031 | 0.012 |
| NR3C2 | rs11099681 | intron | 0.29 | T/C | −0.124 | 0.044 | 0.005 | 0.037 | 0.043 | 0.389 | −0.048 | 0.031 | 0.123 |
| NR3C3 | rs4835493 | intron | 0.29 | C/T | −0.120 | 0.044 | 0.006 | 0.037 | 0.043 | 0.385 | −0.047 | 0.032 | 0.138 |
| SCNN1B | rs7205273 | intron | 0.26 | C/T | −0.111 | 0.043 | 0.009 | 0.029 | 0.045 | 0.519 | −0.045 | 0.032 | 0.163 |
| (B) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Region | MAF | Maj/Min | β * | Active SE |
p | β | Inactive SE |
p | β | Combined SE |
p |
| BDKRB2 | rs945039 | intron | 0.43 | C/T | −0.029 | 0.039 | 0.462 | −0.108 | 0.037 | 0.004 | −0.072 | 0.027 | 0.008 |
| GNB3 | rs4963516 | 5' near gene | 0.33 | A/C | 0.024 | 0.043 | 0.576 | 0.126 | 0.043 | 0.003 | 0.086 | 0.030 | 0.005 |
| SCNN1B | rs7205273 | intron | 0.26 | C/T | −0.108 | 0.041 | 0.008 | 0.044 | 0.044 | 0.319 | −0.036 | 0.031 | 0.235 |
MAF: minor allele frequency
Maj/Min: Major/Minor allele
UTR: untranslated region
SE: standard error
p: p value.
β: beta coefficient
effect size shown as beta coefficient in SD units for the minor allele.
Figure 2.
Mean adjusted SBP (Panel A) and DBP (Panel B) (with standard errors) for each genotype for significantly associated SNPs (p<0.01) separately for the active group and the inactive group. Each gene, SNP number, and SNP genotype are presented below the graph.
Figure 3 shows combined genotypic effects on SBP and DBP in subjects carrying increasing numbers of minor alleles for the significant SNPs according to physical activity status. Within the inactive group, subjects did not show consistent effects on SBP and DBP with increasing numbers of minor alleles. In contrast, individuals within the active group showed reduced SBP and DBP with increasing numbers of minor alleles. Active individuals carrying >4 minor alleles have up to 8 mm lower SBP (Panel A), and those with >3 minor alleles have up to 5 mmHg lower DBP (Panel B) compared to inactive individuals carrying the same number of minor alleles (Figure 3).
Figure 3.
Cumulative effects of the minor alleles for all of the significant SNPs on the values of mean adjusted SBP (Panel A) and DBP (Panel B). The best fitting trend lines for each group and the p values interactions are presented. The numbers of individuals are shown next to each point.
DISCUSSION
The purpose of this study was to investigate effects of interactions between genetic variation and physical activity on BP in rural Chinese participants of the multicenter GenSalt study (n=3,142). Of 196 SNPs across 24 genes from physiological pathways that regulate BP homeostasis, we identified SNPs at five loci (NR3C2, APLNR, GNB3, SCNN1B, BDKRB2) that showed significant associations with BP in either the active or inactive groups (Figure 1, Table 2). Our results provide evidence for effects of interactions between SNPs and physical activity on BP, with demonstrable differences in individual SNP effects according to physical activity status (Figure 2).
In the physically active group, two correlated intronic SNPs (rs11099681 and rs4835493) in NR3C2 (nuclear receptor subfamily 3, group C, member 2) were significantly associated with SBP. NR3C2 encodes the mineralocorticoid receptor that is activated by binding with aldosterone and cortisol, leading to higher BP via alteration of renal sodium retention, expression of nitric oxide synthase and vascular endothelin 1, and vasoconstrictor sensitivity to catecholamines30. We also found a SNP (rs7205273) in the first intron of SCNN1B (sodium channel, nonvoltage-gated 1, beta subunit) that showed significant associations with SBP and DBP in the physically active group. Interestingly, the SNPs that showed significant associations in the physically active group are located in genes that also contain rare mutations that cause monogenic hypertension. Previous studies have found that rare NR3C2 variants cause monogenic forms of hypertension, including autosomal dominant pseudohypoaldosteronism31. Mutations in SCNN1B cause Liddle’s syndrome and pseudohypoaldosteronism type I (PHA I)32. Taken together, our results suggest that common variants in such genes that contain mutations causing monogenic disorders may also influence blood pressure homeostasis in the general population under particular contexts such as physical activity.
In the physically inactive group, we found significant association with SBP for rs2282623 in the 3’ untranslated region (potential regulatory region) of APLNR (apelin receptor) that encodes APJ, a G protein-coupled receptor. The apelin/APJ system plays a role in cardiovascular homeostasis, vascular tone, and cardiac contractile function. This system regulates BP via nitric oxide mediated mechanisms33, and is involved in energy balance and obesity34. In spontaneously hypertensive rats, exercise training decreased SBP and substantially lessened pathological cardiac hypertrophy via increased expression of the apelin/APJ system35. In addition, we found a SNP (rs4963516) in the 5’ flanking region (potential regulatory region) of GNB3 (guanine nucleotide binding protein, beta polypeptide 3) that was significantly associated with SBP and DBP in the physically inactive group. This subunit of the heterotrimeric G protein plays a role in sodium retention by altering activities of sodium exchange across cellular membranes leading to changes in BP36. Signal transduction via G proteins is also important for adipocyte formation37. Previous studies of GNB3 have reported associations of rs5443 with hypertension38 and obesity37. Interactions of GNB3 rs5443 and BP response to an exercise training program were identified in African American women18. This variant was also found to interact with obesity and physical activity in hypertension in African Americans19. However, we did not find association of GNB3 rs5443 with blood pressure measures and interaction with exercise in the GenSalt cohort. We also found an intronic SNP (rs945039) in BDKRB2 (bradykinin receptor B2) that was significantly associated with DBP in the physically inactive group. Bradykinin plays a central role in physiological pathways of vasodilation and insulin sensitivity. Other BDKRB2 variants have shown associations with physical performance in track athletes (efficiency of skeletal muscle contraction and endurance)39.
Many previous studies have investigated interactions between genetic variants and different forms of exercise or physical activity on BP traits11-19. Several such studies have used an intervention training program or physical activity monitoring to test how BP response differs based on an individual’s genotype at specific loci14, 15, 17, 18. Like these previous studies, our findings of significant genetic associations do not imply that physical activity is not useful for individuals with any particular genotype given the overall beneficial effects of physical activity on BP traits, as well as numerous other well-documented health benefits. We found that active individuals with specific genotypes always have lower BP than inactive individuals with the same genotypes (Figure 2). However, genetic information can help identify individuals where physical activity is not sufficient to control their BP, requiring other BP lowering interventions14. In addition, identification of particular genes and genetic variants that are associated with BP help further our understanding of the pathophysiology of BP regulation and the etiology of hypertension. Characterization of interactions between genetic variants and physical activity is also important to understand the mechanisms of how physical activity lowers BP measures, ultimately contributing to improved medications and better hypertension treatment modalities.
We recruited our study participants from a semi-isolated Han Chinese population to minimize genetic and environmental heterogeneity. No subjects were under any form of BP treatments to avoid all complications associated with controlling for drug response, and to ensure better BP measurements. The participants provided subjective assessments of their physical activity using questionnaires that are susceptible to recall bias. However, the effects should be minimal since this is a farming community with a routine and repetitive life style that should facilitate accurate recall. Future studies in other cohorts will be required to replicate these associations of genetic variants that interact with physical activity to influence SBP and DBP.
In conclusion, this study provides evidence that interactions between genetics variants and physical activity play a role in BP regulation. However, we found that effects of physical activity can be stronger than genetic effects, indicating that physical activity should continue to be recommended for hypertensive patients, regardless of their genetic makeup.
Supplementary Material
ACKNOWLEDGEMENTS
This report is from the Genetic Epidemiology Network of Salt Sensitivity (GenSalt) study that is supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health (U01HL072507). Upsher-Smith Laboratories, Inc. has provided Klor-Con M20 potassium tablets for the GenSalt study.
GenSalt Study Steering Committee
Dongfeng Gu, Jiang He (Chair), James E Hixson, Cashell E Jaquish, Depei Liu, DC Rao, Paul K Whelton, and Zhijian Yao.
GenSalt Collaborative Research Group
Tulane University Health Sciences Center, New Orleans, USA: Jiang He (PI), Lydia A Bazzano, Chung-Shiuan Chen, Jing Chen, Mei Hao, Lee Hamm, Tanika Kelly, Paul Muntner, Kristi Reynolds, Paul K Whelton, Wenjie Yang, and Qi Zhao.
Washington University School of Medicine, St Louis, USA: DC Rao (PI), Matthew Brown, Charles Gu, Hongyan Huang, Treva Rice, Karen Schwander, and Shiping Wang.
University of Texas Health Sciences Center at Houston: James E Hixson (PI) and Lawrence C Shimmin.
National Heart, Lung, and Blood Institute: Cashell E Jaquish.
Chinese Academy of Medical Sciences, Beijing, China: Dongfeng Gu (PI), Jie Cao, Jichun Chen, Jingping Chen, Zhenhan Du, Jianfeng Huang, Hongwen Jiang, Jianxin Li, Xiaohua Liang, Depei Liu, Xiangfeng Lu, Donghua Liu, Qunxia Mao, Dongling Sun, Hongwei Wang, Qianqian Wang, Xigui Wu, Ying Yang, and Dahai Yu.
Shandong Academy of Medical Sciences, Shandong, China: Fanghong Lu (PI), Zhendong Liu, Shikuan Jin, Yingxin Zhao, Shangwen Sun, Shujian Wang, Qengjie Meng, Baojin Liu, Zhaodong Yang, and Chuanrui Wei.
Shandong Center for Diseases Control and Prevention, Shandong, China: Jixiang Ma (PI), Jiyu Zhang, and Junli Tang.
Zhengzhou University: Dongsheng Hu, Hongwei Wen, Chongjian Wang, Minghui Shen, Jingjing Pan, and Liming Yang.
Xinle Traditional Chinese Medicine Hospital, Hebei, China: Xu Ji (PI), Rongyan Li, Haijun Zu, and Junwei Song.
Ganyu Center for Disease Control and Prevention: Delin Wu (PI), Xushan Wang, and Xiaofeng Zhang.
Xi’an Jiaotong University, Shanxi, China: Jianjun Mu (PI), Enrang Chen,Fuqiang Liu, and Guanji Wu.
Chinese National Human Genome Center at Beijing: Zhi-Jian Yao (PI), Shufeng Chen, Dongfeng Gu, Hongfan Li, Laiyuan Wang, and Penghua Zhang.
Support: NIH grant U01HL072507
Footnotes
Conflict of interest: None
REFERENCES
- 1.Mathers CD, Ezzati M, Lopez AD. Measuring the burden of neglected tropical diseases: the global burden of disease framework. PLoS Negl Trop Dis. 2007;1:e114. doi: 10.1371/journal.pntd.0000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Binder A. A review of the genetics of essential hypertension. Curr Opin Cardiol. 2007;22:176–184. doi: 10.1097/HCO.0b013e3280d357f9. [DOI] [PubMed] [Google Scholar]
- 3.van Leiden HA, Dekker JM, Moll AC, Nijpels G, Heine RJ, Bouter LM, et al. Risk factors for incident retinopathy in a diabetic and nondiabetic population: the Hoorn study. Arch Ophthalmol. 2003;121:245–251. doi: 10.1001/archopht.121.2.245. [DOI] [PubMed] [Google Scholar]
- 4.Whelton PK, He J, Appel LJ, Cutler JA, Havas S, Kotchen TA, et al. Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. JAMA. 2002;288:1882–1888. doi: 10.1001/jama.288.15.1882. [DOI] [PubMed] [Google Scholar]
- 5.Rigaud AS, Seux ML, Staessen JA, Birkenhager WH, Forette F. Cerebral complications of hypertension. J Hum Hypertens. 2000;14:605–616. doi: 10.1038/sj.jhh.1001118. [DOI] [PubMed] [Google Scholar]
- 6.Mancia G. Blood pressure reduction and cardiovascular outcomes: past, present, and future. Am J Cardiol. 2007;100:3J–9J. doi: 10.1016/j.amjcard.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 7.Wong TY, Mitchell P. Hypertensive retinopathy. N Engl J Med. 2004;351:2310–2317. doi: 10.1056/NEJMra032865. [DOI] [PubMed] [Google Scholar]
- 8.Cook NR, Cutler JA, Obarzanek E, Buring JE, Rexrode KM, Kumanyika SK, et al. Long term effects of dietary sodium reduction on cardiovascular disease outcomes: observational follow-up of the trials of hypertension prevention (TOHP) BMJ. 2007;334:885–888. doi: 10.1136/bmj.39147.604896.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagard RH, Cornelissen VA. Effect of exercise on blood pressure control in hypertensive patients. Eur J Cardiovasc Prev Rehabil. 2007;14:12–17. doi: 10.1097/HJR.0b013e3280128bbb. [DOI] [PubMed] [Google Scholar]
- 10.Whelton SP, Chin A, Xin X, He J. Effect of aerobic exercise on blood pressure: a meta-analysis of randomized, controlled trials. Ann Intern Med. 2002;136:493–503. doi: 10.7326/0003-4819-136-7-200204020-00006. [DOI] [PubMed] [Google Scholar]
- 11.Vimaleswaran KS, Franks PW, Barroso I, Brage S, Ekelund U, Wareham NJ, Loos RJ. Habitual energy expenditure modifies the association between NOS3 gene polymorphisms and blood pressure. Am J Hypertens. 2008;21:297–302. doi: 10.1038/ajh.2007.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rankinen T, Church T, Rice T, Markward N, Leon AS, Rao DC, et al. Effect of endothelin 1 genotype on blood pressure is dependent on physical activity or fitness levels. Hypertension. 2007;50:1120–1125. doi: 10.1161/HYPERTENSIONAHA.107.093609. [DOI] [PubMed] [Google Scholar]
- 13.Kimura T, Yokoyama T, Matsumura Y, Yoshiike N, Date C, Muramatsu M, Tanaka H. NOS3 genotype-dependent correlation between blood pressure and physical activity. Hypertension. 2003;41:355–360. doi: 10.1161/01.hyp.0000051500.02578.6d. [DOI] [PubMed] [Google Scholar]
- 14.Franks PW, Bhattacharyya S, Luan J, Montague C, Brennand J, Challis B, et al. Association between physical activity and blood pressure is modified by variants in the G-protein coupled receptor 10. Hypertension. 2004;43:224–228. doi: 10.1161/01.HYP.0000109319.63240.08. [DOI] [PubMed] [Google Scholar]
- 15.Delmonico MJ, Ferrell RE, Meerasahib A, Martel GF, Roth SM, Kostek MC, Hurley BF. Blood pressure response to strength training may be influenced by angiotensinogen A-20C and angiotensin II type I receptor A1166C genotypes in older men and women. J Am Geriatr Soc. 2005;53:204–210. doi: 10.1111/j.1532-5415.2005.53104.x. [DOI] [PubMed] [Google Scholar]
- 16.Rauramaa R, Kuhanen R, Lakka TA, Vaisanen SB, Halonen P, Alen M, et al. Physical exercise and blood pressure with reference to the angiotensinogen M235T polymorphism. Physiol Genomics. 2002;10:71–77. doi: 10.1152/physiolgenomics.00050.2002. [DOI] [PubMed] [Google Scholar]
- 17.Rankinen T, Gagnon J, Perusse L, Chagnon YC, Rice T, Leon AS, et al. AGT M235T and ACE ID polymorphisms and exercise blood pressure in the HERITAGE Family Study. Am J Physiol Heart Circ Physiol. 2000;279:H368–74. doi: 10.1152/ajpheart.2000.279.1.H368. [DOI] [PubMed] [Google Scholar]
- 18.Rankinen T, Rice T, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. G protein beta 3 polymorphism and hemodynamic and body composition phenotypes in the HERITAGE Family Study. Physiol Genomics. 2002;8:151–157. doi: 10.1152/physiolgenomics.00102.2001. [DOI] [PubMed] [Google Scholar]
- 19.Grove ML, Morrison A, Folsom AR, Boerwinkle E, Hoelscher DM, Bray MS. Gene-environment interaction and the GNB3 gene in the Atherosclerosis Risk in Communities study. Int J Obes (Lond) 2007;31:919–926. doi: 10.1038/sj.ijo.0803545. [DOI] [PubMed] [Google Scholar]
- 20.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 21.GenSalt Collaborative Research Group GenSalt: rationale, design, methods and baseline characteristics of study participants. J Hum Hypertens. 2007;21:639–646. doi: 10.1038/sj.jhh.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tobler AR, Short S, Andersen MR, Paner TM, Briggs JC, Lambert SM, et al. SNPlex genotyping system: a flexible and scalable platform for SNP genotyping. J Biomol Tech. 2005;16:398–406. [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinds DR. The ASPEX package:affected sib-pair exclusion mapping v1.88. 1999 M. [Google Scholar]
- 26.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Hanley JA, Negassa A, Edwardes MD, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157:364–375. doi: 10.1093/aje/kwf215. [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 30.Pippal JB, Fuller PJ. Structure-function relationships in the mineralocorticoid receptor. J Mol Endocrinol. 2008;41:405–413. doi: 10.1677/JME-08-0093. [DOI] [PubMed] [Google Scholar]
- 31.Riepe FG, Finkeldei J, de Sanctis L, Einaudi S, Testa A, Karges B, et al. Elucidating the underlying molecular pathogenesis of NR3C2 mutants causing autosomal dominant pseudohypoaldosteronism type 1. J Clin Endocrinol Metab. 2006;91:4552–4561. doi: 10.1210/jc.2006-1161. [DOI] [PubMed] [Google Scholar]
- 32.Rossi E, Farnetti E, Debonneville A, Nicoli D, Grasselli C, Regolisti G, et al. Liddle's syndrome caused by a novel missense mutation (P617L) of the epithelial sodium channel beta subunit. J Hypertens. 2008;26:921–927. doi: 10.1097/HJH.0b013e3282f85dfe. [DOI] [PubMed] [Google Scholar]
- 33.Chandrasekaran B, Dar O, McDonagh T. The role of apelin in cardiovascular function and heart failure. Eur J Heart Fail. 2008;10:725–732. doi: 10.1016/j.ejheart.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Rayalam S, Della-Fera MA, Krieg PA, Cox CM, Robins A, Baile CA. A putative role for apelin in the etiology of obesity. Biochem Biophys Res Commun. 2008;368:815–819. doi: 10.1016/j.bbrc.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Ren CX, Qi YF, Lou LX, Chen L, Zhang LK, et al. Exercise training promotes expression of apelin and APJ of cardiovascular tissues in spontaneously hypertensive rats. Life Sci. 2006;79:1153–1159. doi: 10.1016/j.lfs.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 36.Siffert W, Dusing R. Sodium-proton exchange and primary hypertension. An update. Hypertension. 1995;26:649–655. doi: 10.1161/01.hyp.26.4.649. [DOI] [PubMed] [Google Scholar]
- 37.Siffert W, Naber C, Walla M, Ritz E. G protein beta3 subunit 825T allele and its potential association with obesity in hypertensive individuals. J Hypertens. 1999;17:1095–1098. doi: 10.1097/00004872-199917080-00008. [DOI] [PubMed] [Google Scholar]
- 38.Bagos PG, Elefsinioti AL, Nikolopoulos GK, Hamodrakas SJ. The GNB3 C825T polymorphism and essential hypertension: a meta-analysis of 34 studies including 14,094 cases and 17,760 controls. J Hypertens. 2007;25:487–500. doi: 10.1097/HJH.0b013e328011db24. [DOI] [PubMed] [Google Scholar]
- 39.Williams AG, Dhamrait SS, Wootton PT, Day SH, Hawe E, Payne JR, et al. Bradykinin receptor gene variant and human physical performance. J Appl Physiol. 2004;96:938–942. doi: 10.1152/japplphysiol.00865.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.