Significance
In nematodes, acyl-CoA oxidases function not only in primary metabolism, where they participate in the breakdown of long-chain fatty acids, but also in secondary metabolism, where they participate in the biosynthesis of the ascarosides, a family of pheromones that regulate development and behavior. Here, we provide a molecular basis for the diverse substrate specificities of the acyl-CoA oxidases. Unlike their homologs in other organisms, the acyl-CoA oxidases of nematodes bind ATP at the dimer interface, leading to a conformational change that promotes FAD cofactor binding and enzyme activity. Our results suggest a mechanism by which the biosynthesis of most ascarosides is increased under conditions that promote higher cellular ATP concentrations in the worm.
Keywords: ascarosides, beta-oxidation, ATP, crystal structure, dauer pheromone
Abstract
Caenorhabditis elegans secretes ascarosides as pheromones to communicate with other worms and to coordinate the development and behavior of the population. Peroxisomal β-oxidation cycles shorten the side chains of ascaroside precursors to produce the short-chain ascaroside pheromones. Acyl-CoA oxidases, which catalyze the first step in these β-oxidation cycles, have different side chain-length specificities and enable C. elegans to regulate the production of specific ascaroside pheromones. Here, we determine the crystal structure of the acyl-CoA oxidase 1 (ACOX-1) homodimer and the ACOX-2 homodimer bound to its substrate. Our results provide a molecular basis for the substrate specificities of the acyl-CoA oxidases and reveal why some of these enzymes have a very broad substrate range, whereas others are quite specific. Our results also enable predictions to be made for the roles of uncharacterized acyl-CoA oxidases in C. elegans and in other nematode species. Remarkably, we show that most of the C. elegans acyl-CoA oxidases that participate in ascaroside biosynthesis contain a conserved ATP-binding pocket that lies at the dimer interface, and we identify key residues in this binding pocket. ATP binding induces a structural change that is associated with tighter binding of the FAD cofactor. Mutations that disrupt ATP binding reduce FAD binding and reduce enzyme activity. Thus, ATP may serve as a regulator of acyl-CoA oxidase activity, thereby directly linking ascaroside biosynthesis to ATP concentration and metabolic state.
In mammals, peroxisomal β-oxidation plays a critical role in primary metabolism, catabolizing the CoA-thioesters of long/very long-chain fatty acids, branched-chain fatty acids, and bile acid intermediates (1, 2). In the nematode C. elegans, peroxisomal β-oxidation not only functions in primary metabolism, but also has been coopted in the biosynthesis of a class of secondary metabolites known as the ascarosides (3–7). These ascarosides, which are secreted as pheromones, are derivatives of the 3,6-dideoxy-l-sugar ascarylose and are modified with fatty acid-derived side chains of various lengths (SI Appendix, Fig. S1 A and B) (8–13). C. elegans makes ascarosides with long side chains and then shortens these side chains through β-oxidation to make the short-chain ascaroside pheromones (3–7). These pheromones enable C. elegans to coordinate the development and behavior of the population. Different subsets of these short-chain ascarosides are used to induce the stress-resistant dauer larval stage, to attract males to hermaphrodites, to attract hermaphrodites to males, to induce aggregation, and to induce dispersal (6, 9–12, 14–16).
Acyl-CoA oxidases (ACOXs) catalyze the rate-limiting step in peroxisomal β-oxidation cycles, using a catalytic glutamate residue to install a double-bond α-β to the CoA-thioester of their substrates (SI Appendix, Fig. S2A) (17, 18). These enzymes oxidize their substrates by passing the electrons to FAD and, subsequently, to molecular oxygen to make hydrogen peroxide. Thus, the enzymatic mechanism of the ACOXs is distinct from that of the acyl-CoA dehydrogenases that catalyze the first step in mitochondrial β-oxidation cycles and oxidize fatty acyl-CoA substrates by passing electrons to the electron transport chain (19). Different mammalian ACOXs are specialized to process different types of substrates (1, 2, 20–22). Palmitoyl-CoA oxidase is specifically active toward the CoA-thioesters of straight-chain fatty acids, pristanoyl-CoA oxidase toward the CoA-thioesters of 2-methyl-branched-chain fatty acids and long/very long straight-chain fatty acids, and cholestanoyl-CoA oxidase toward the CoA-thioesters of bile acids (20–22). The crystal structures of a few ACOXs have been determined, including that of rat liver palmitoyl-CoA oxidase homodimer (ACO-II) bound to a fatty acid and FAD cofactor (17, 18, 23–26).
In C. elegans, an ACOX enzyme also catalyzes the first step in each peroxisomal β-oxidation cycle. The remaining three enzymes in each cycle include an enoyl-CoA hydratase, a (3R)-hydroxyacyl-CoA dehydrogenase, and a 3-ketoacyl-CoA thiolase (3–6). Specific ACOX enzymes have evolved to process ascarosides with specific side-chain lengths, and thus, these enzymes help to determine the mixture of short-chain ascarosides that are produced by the worm (5–7). The ascarosides are divided into two main classes: the (ω-1)-ascarosides and the ω-ascarosides, which are biosynthesized through two parallel β-oxidation pathways, each involving different ACOX enzymes (SI Appendix, Fig. S2 B and C) (7). The side chain of the (ω-1)-ascarosides is attached at its penultimate (or ω-1) carbon to the ascarylose sugar, whereas the side chain of the ω-ascarosides is attached at its terminal (or ω) carbon to the ascarylose sugar (SI Appendix, Fig. S1A). Although some of the ACOX enzymes in C. elegans process both fatty acyl-CoA substrates and ascaroside-CoA substrates, others are specialized for processing only specific ascaroside-CoA substrates (SI Appendix, Fig. S2 B and C) (7). An ACOX-1 homodimer processes fatty acyl-CoA substrates, as well as specific ascaroside-CoA substrates, including an (ω-1)-ascaroside with a 9-carbon side chain (7). In contrast, an ACOX-1/ACOX-3 heterodimer is specific to an (ω-1)-ascaroside substrate with a 7-carbon side chain, and an ACOX-2 homodimer is specific to an ω-ascaroside substrate with a 5-carbon side chain (7). By regulating the expression of these ACOXs, C. elegans can control the production of specific ascarosides (7). For example, C. elegans regulates the expression of ACOX-2 to influence the production of an ascaroside with a short ω-side chain, the dauer pheromone asc-ωC3 (C3; ascr#5), and it regulates the expression of ACOX-3 to influence the production of ascarosides with shorter (ω-1)-side chains, such as the dauer pheromone asc-C6-MK (C6; ascr#2) (SI Appendix, Figs. S1B and S2 B and C).
Here, we provide the structural basis for the substrate specificities of the ACOX enzymes in C. elegans by obtaining the crystal structures of an ACOX-1 homodimer and an ACOX-2 homodimer bound to its substrate. Site-directed mutagenesis confirms the importance of key residues in the active sites of the enzymes for substrate recognition. We identify an ATP binding pocket that is present at the dimer interface of many of the ACOX enzymes and show how ATP may regulate pheromone production.
Results and Discussion
Comparison of the Structures of ACOX-1 and ACOX-2.
In initial efforts to crystallize ACOX-1, we were only able to generate crystals of ACOX-1 without bound FAD, even when FAD was added to all buffers, as had been done for the purification and crystallization of certain plant ACOX enzymes (27). Serendipitously, we discovered that the ACOX-1(E434A) mutant can be readily crystallized with bound FAD. In this catalytically inactive mutant, a conserved glutamate that removes the α-proton in the substrate has been changed to an alanine (SI Appendix, Fig. S2A). Unfortunately, we could not obtain enzyme-substrate cocrystals either by crystallizing ACOX-1 in the presence of asc-C9-CoA (1), asc-C9, nonanoyl-CoA, or nonanoic acid, or by soaking ACOX-1 crystals in the presence of these substrates or substrate mimics. Similar to our experience with ACOX-1, making the catalytic mutant of ACOX-2 facilitated our work with that enzyme. Although wild-type ACOX-2 expresses relatively poorly in E. coli (∼0.1 mg per L of E. coli culture), the ACOX-2(E432A) catalytic mutant expresses at much higher levels (∼1 mg per L). Furthermore, we were able to crystallize this ACOX-2 mutant bound to its FAD cofactor and asc-ωC5-CoA (2) substrate. The asc-ωC5-CoA (2) substrate was used in cocrystallization experiments because ACOX-2 is specifically active toward this substrate and cannot process (ω-1)-ascarosides (7) or longer-chain ω-ascarosides (SI Appendix, Fig. S3).
The structure of wild-type apo-ACOX-1, which does not have a bound FAD cofactor, was solved in two different lattices (SI Appendix, Table S1) by molecular replacement, using rat liver peroxisomal ACO-II as a search model (17). The structures of ACOX-1(E434A) and ACOX-2(E432A) were solved by molecular replacement, using the wild-type ACOX-1 structure as a search model (see SI Appendix, Table S1 for crystallographic data and refinement statistics). ACOX-1 and ACOX-2 crystallize as dimers as the biological unit with a similar overall fold (root-mean-square deviation = 0.64 Å) (SI Appendix, Fig. S4). For both ACOX-1(E434A) and ACOX-2(E432A), an FAD cofactor is bound at two twofold symmetric sites that lie at the interface between the two subunits of the dimer (Fig. 1 A and B). Whereas most of the FAD cofactor is encased by protein, the adenine portion of the FAD is largely exposed to solvent. Surprisingly, for both proteins, electron density that is consistent with ATP is observed at two twofold symmetric sites that lie at the interface between the two subunits of the dimer (Fig. 1 A and B). In the ACOX-2 structure, the asc-ωC5-CoA (2) substrate is bound in a cleft that is adjacent to the FAD cofactor (Fig. 1 A and B). The electron density for the ascaroside and CoA portion of the substrate, as well as the FAD cofactor, is well resolved (Fig. 2A). Thus, although the CoA group is near the surface of the protein and is exposed to solvent, it is bound in a specific manner.
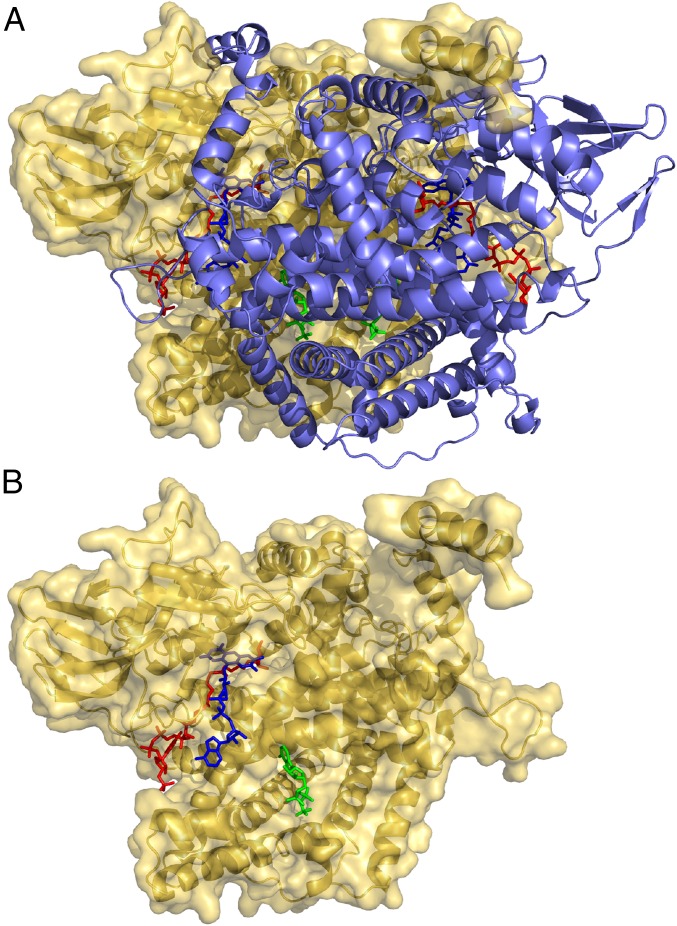
Fig. 1.
Structure of the ACOX-2 homodimer and bound small molecules. (A) The ACOX-2(E432A) homodimer (with one subunit shown in yellow and one in purple) bound to two molecules of the FAD cofactor (blue), the asc-ωC5-CoA (2) substrate (red), and ATP (green). The structure of asc-ωC5-CoA (2) is shown in Fig. 2C and SI Appendix, Fig. S2C. (B) One of the subunits of the ACOX-2(E432A) homodimer and the FAD cofactor (blue), asc-ωC5-CoA (2) substrate (red), and ATP (green) associated with that subunit.
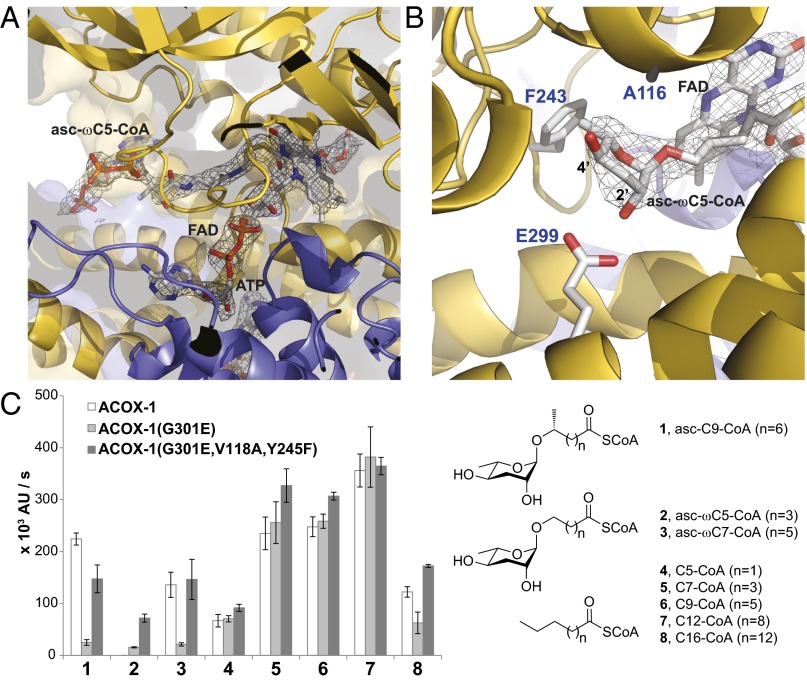
Fig. 2.
Comparison of the active sites of ACOX-1 and ACOX-2. (A) Active site of ACOX-2(E432A) with bound asc-ωC5-CoA (2) substrate and FAD cofactor. The electron density associated with the substrate and cofactor is shown (2Fo-Fc map, σ=1.0). (B) Close up of the asc-ωC5-CoA (2) substrate in the active site of ACOX-2(E432A), shown from the opposite perspective as shown in A. Glu-299 hydrogen bonds to the 2′-hydroxyl of the ascarylose ring of the substrate. Phe-243 and Ala-116 help to accommodate the ascarylose ring. (C) In vitro enzyme activity assay showing the effect of mutating active site residues (Gly-301, Val-118, and Tyr-245) of ACOX-1 to the corresponding residues in ACOX-2. The structures of the (ω-1)-ascaroside-CoA (1), ω-ascaroside-CoA (2–3), and fatty acyl-CoA (4–8) substrates that were tested are shown. Data represent the mean ± SD of three independent experiments.
Structural Basis for Substrate Specificity.
In the ACOX-2 active site, Glu-299 is positioned to hydrogen bond with the 2′-hydroxyl of the ascarylose ring of the asc-ωC5-CoA (2) substrate (Fig. 2B). The homologous position in the ACOX-1 active site has a glycine (Gly-301) instead of a Glu, possibly reducing the binding of ACOX-1 to asc-ωC5-CoA (2). To implicate this residue in conferring substrate specificity, we made the Gly-301 to Glu mutation in the ACOX-1 active site to make it resemble the ACOX-2 active site. This mutation greatly reduced the activity of ACOX-1 toward its ascaroside substrate, asc-C9-CoA (1), but maintained its activity toward fatty acyl-CoA substrates (Fig. 2C). Thus, this mutation does not affect enzyme folding or catalysis, but only recognition and accommodation of a specific ascaroside substrate. Furthermore, the ACOX-1(G301E) mutant also demonstrated some minor activity toward the ACOX-2 substrate, asc-ωC5-CoA (2), suggesting a glutamate in this position in the ACOX-2 active site does play a role in binding this substrate (Fig. 2C).
Phe-243 and Ala-116 in ACOX-2 (Fig. 2B) are replaced by Tyr-245 and Val-118, respectively, in ACOX-1, and these residues may further reduce the affinity of ACOX-1 for an asc-ωC5-CoA (2) substrate. Alignment of the ACOX-1 structure with the ACOX-2 structure with bound asc-ωC5-CoA (2) suggests that if ACOX-1 were to bind ACOX-2’s substrate, the Tyr-245 hydroxyl would be within 2.1 Å of the ascarylose methyl group, and the Val-118 side chain would be within 2.3 Å of the ascarylose 4′-hydroxyl. To implicate these residues in substrate selection, we mutated ACOX-1(G301E) further with the Y245F and V118A mutations. The ACOX-1 triple mutant demonstrates a level of activity toward the ACOX-2 substrate, asc-ωC5-CoA (2), that approaches that of ACOX-2 (Fig. 2C) (7). This triple mutant, however, has similar levels of activity as wild-type ACOX-1 toward most other substrates. Thus, although Glu-299, Phe-243, and Ala-116 are important for ACOX-2’s activity toward its asc-ωC5-CoA (2) substrate, additional residues in ACOX-2 must account for its extremely limited substrate range.
Comparison of the ACOX-1 and ACOX-2 active sites shows that the ACOX-1 active site is much larger than that of ACOX-2 (Fig. 3 A and B and SI Appendix, Fig. S5 A and B). Modeling of the asc-ωC5-CoA (2) substrate into the ACOX-1 structure (based on its position in the ACOX-2 structure) shows that the ACOX-1 active site opens to two channels that run along the surface of the protein (indicated by arrows in Fig. 3B). These channels may enable the ACOX-1 enzyme to accommodate longer ascaroside-CoA and fatty acyl-CoA substrates in its active site than asc-ωC5-CoA (2). The ACOX-2 active site has limited access to the outer surface of the protein (Fig. 3A). Lys-291 and Glu-93 form a salt bridge, and Gln-295 and Arg-101 form a hydrogen bond. These residues, along with Leu-97, greatly reduce the size of the active site. In the ACOX-1 active site, in contrast, these residues are replaced by a number of hydrophobic and aromatic residues that would interact favorably with the alkyl chain of substrates and may enable them to extend outward into one of the two outer surface channels (Fig. 3B).
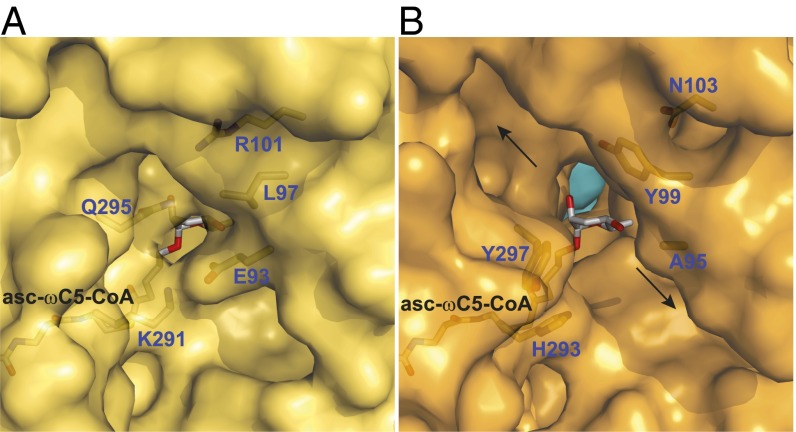
Fig. 3.
Amino acid residues that determine whether the active site is closed or open to the external surface. (A) A region of the outer surface of ACOX-2(E432A) (yellow), showing that its bound substrate asc-ωC5-CoA (2) is largely encased by the protein. The FAD cofactor located behind the substrate is not shown for clarity. A hydrogen bond between Gln-295 and Arg-101 and a salt bridge between Lys-291 and Glu-93 close the active site to the outer surface. (B) A region of the outer surface of ACOX-1(E434A) (orange, with the other subunit in light blue). The substrate of ACOX-2 (asc-ωC5-CoA, 2) has been placed into the ACOX-1 active site to show that the active site is open to the outer surface. The FAD cofactor is not shown for clarity. Hydrophobic and aromatic residues (Ala-95, Tyr-99, His-293, and Tyr-297) line the opening to the outer surface, which leads to two channels (marked by arrows) that run along the outer surface. The location of the region shown in A and B on the full structures is shown in SI Appendix, Fig. S5 A and B.
Comparison of the active site residues across ACOX enzymes in C. elegans helps to explain the substrate specificities of biochemically characterized ACOXs and could potentially enable the substrate specificities of uncharacterized ACOX enzymes to be predicted (Fig. 4). For example, Glu-299 in ACOX-2, which hydrogen bonds to the 2′-hydroxyl of the ascarylose ring of the substrate (Fig. 2B), corresponds to Asp in ACOX-3 and to Gly in ACOX-1 and ACOX-4. From these data, one might predict that ACOX-3 would prefer ascaroside substrates that are intermediate in size between the preferred substrates of ACOX-1 and ACOX-2 (i.e., asc-C7-CoA; SI Appendix, Fig. S2), as has been shown (7). One might also predict that ACOX-4 would prefer ascarosides substrates that are similar in size to the preferred substrates of ACOX-1 [e.g., asc-C9-CoA (1)]. In terms of the residues that might determine whether the active site is open or closed to the outer surface (i.e., those highlighted in Fig. 3), ACOX-3 resembles ACOX-2, suggesting that, similar to ACOX-2, it has a closed active site and processes shorter substrates (i.e., asc-C7-CoA; SI Appendix, Fig. S2), as has been shown (7). ACOX-6, in contrast, resembles ACOX-1, suggesting that, like ACOX-1, it has an open active site and processes longer substrates.
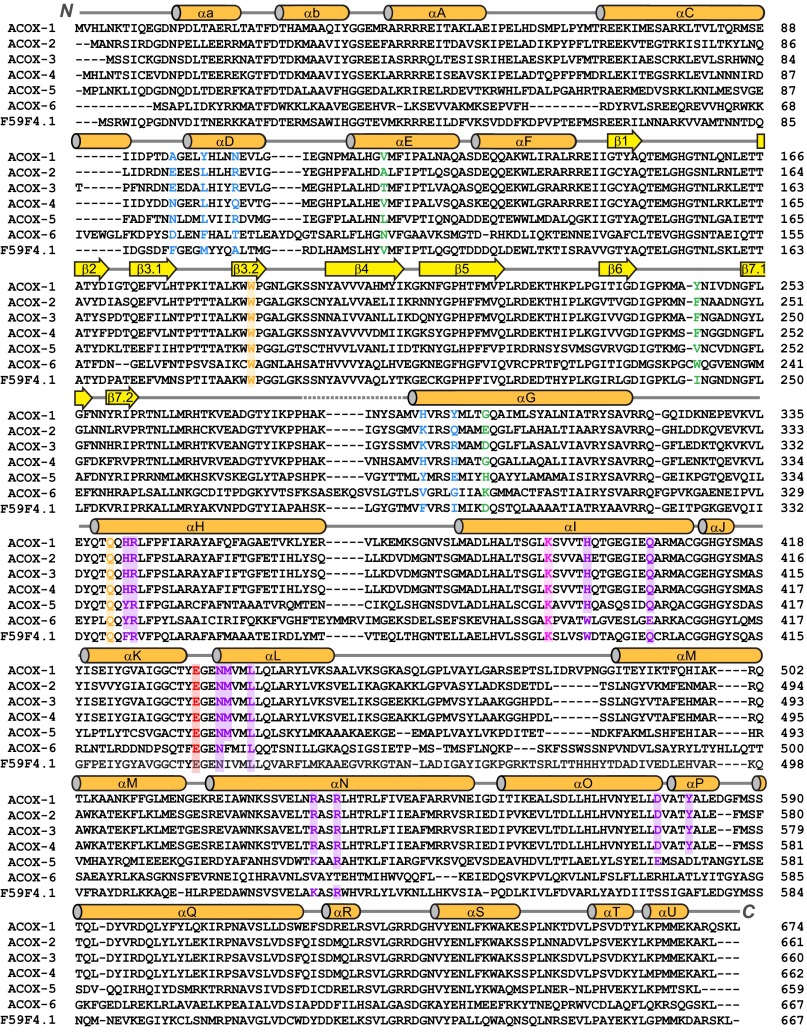
Fig. 4.
Protein sequence alignment of ACOX homologs in C. elegans. The secondary structure in ACOX-1 is shown across the top of the sequences, and the nomenclature used for the α helices and β sheets is based on that of Rattus norvegicus ACO-II (23). The conserved catalytic glutamate found in all C. elegans ACOX enzymes is highlighted in red. Residues (from Fig. 2B) that are predicted to play a key role in substrate binding in ACOX-1 and ACOX-2 are highlighted in green. Residues (from Fig. 3 A and B) that likely play a key role in determining the size of the active site and whether it is open to the outer surface of the protein are highlighted in blue. Residues in the ATP-binding site (shown in Fig. 5A) are highlighted in purple. A conserved lysine that hydrogen bonds to Asn-437 in the ATP-binding pocket may stabilize the loop between αK and αL and is highlighted in pink. A conserved Trp that stacks with the isoalloxazine ring of FAD and a conserved Gln that hydrogen bonds to the ribose ring of FAD are highlighted in orange.
Unusual ATP Binding Domain in the ACOX Enzymes.
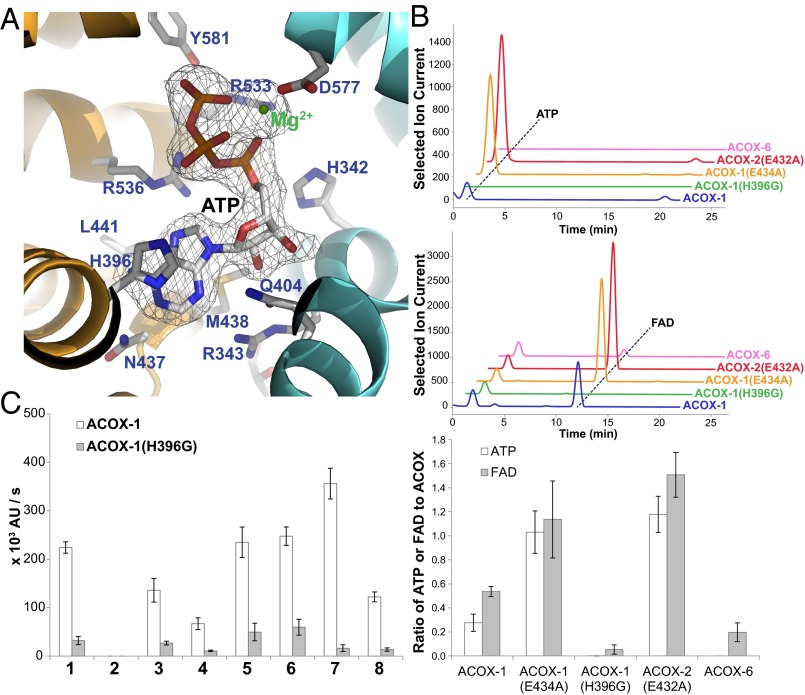
During the refinement of the ACOX-1(E434A) and ACOX-2(E432A) structures, electron density was observed at two twofold symmetric sites at the dimer interface that was consistent with a bound ATP molecule coordinating a Mg2+ ion (Fig. 5A and SI Appendix, Fig. S6 A and B). To confirm that the ACOX enzymes bind ATP, we extracted small molecules from purified ACOX-1, ACOX-1(E434A), and ACOX-2(E432A) and used LC-MS to analyze the extracts. These data demonstrate that approximately two ATP molecules are bound per dimer for ACOX-1(E434A) and ACOX-2(E432A) (Fig. 5B). A smaller amount of bound ATP is seen for wild-type ACOX-1 (Fig. 5B). Each ATP binding site contains conserved residues contributed by both dimer subunits (Fig. 5A). To confirm these residues are involved in ATP binding, we made several ACOX-1 mutants, including ACOX-1(H396G), ACOX-1(R533E), and ACOX-1(R536E), that should not be able to bind ATP as well as wild-type ACOX-1. We were able to confirm that these mutants do not bind ATP (Fig. 5B). The ATP-binding site is conserved in all of the ACOX enzymes in C. elegans, except ACOX-6 and perhaps F59F4.1 (see purple highlighting in Fig. 4). Correspondingly, recombinantly expressed ACOX-6 was not associated with any bound ATP (Fig. 5B). This ATP-binding site may be unique to C. elegans and closely related nematodes, as the site residues are not strictly conserved in the ACOX enzymes from other organisms (SI Appendix, Fig. S7). Furthermore, the crystal structure of rat liver ACO-II bound to a fatty acid and FAD, as well as the crystal structures of Arabidopsis and Tomato ACX1 bound to FAD, did not show any electron density at the dimer interface consistent with ATP (17, 24, 26).
Fig. 5.
Role of ATP-binding site in ACOX enzymes. (A) ATP-binding site in ACOX-1 located at the dimer interface (between the orange and light blue subunits). Conserved residues are indicated in purple. The electron density for ATP (2Fo-Fc map, σ=1.0) is also shown. (B) LC-MS traces showing the amount of associated ATP (Upper) and FAD (Middle) for recombinantly expressed wild-type and mutant ACOX enzymes, and the ratio of ATP or FAD to ACOX subunits determined from the LC-MS traces (Lower). In the bottom panel, data represent the mean ± SD of two independent experiments. (C) In vitro enzyme activity assay showing the effect of mutating a histidine in the ATP-binding site of ACOX-1 (His-396). The structures of the (ω-1)-ascaroside-CoA (1), ω-ascaroside-CoA (2–3), and fatty acyl-CoA (4–8) substrates that were tested are shown in Fig. 2C. Data represent the mean ± SD of three independent experiments.
The enzymatic activity of the ACOX-1(H396G) mutant enzyme was studied further. The mutation did not disrupt protein folding, as judged by CD spectroscopy (SI Appendix, Fig. S8). Although the mutant expressed well as a dimer, it lost its FAD cofactor during purification; much more so than did wild-type ACOX-1 (Fig. 5B). Furthermore, the ATP-binding mutant was not as active as wild-type ACOX-1 toward ascaroside-CoA and fatty acyl-CoA substrates (Fig. 5C). Wild-type ACOX-1 is still catalytically active even when FAD is not included in the in vitro activity assay, presumably because it remains associated with its FAD cofactor during purification (SI Appendix, Fig. S9). The ATP-binding mutant ACOX-1(H396G), in contrast, shows virtually no activity when FAD is not included in the assay, presumably because it lost any associated FAD during purification (SI Appendix, Fig. S9).
In general, association of the ACOX enzymes with FAD and with ATP appear to be correlated, with the levels of associated FAD and ATP high for the catalytic mutants ACOX-1(E434A) and ACOX-2(E432A), lower for wild-type ACOX-1, and lowest for the ATP-binding mutant ACOX-1(H396G) (Fig. 5B). One possibility is that the presence of ATP at the ACOX dimer interface leads to a conformational change that further stabilizes the association of ACOX with FAD. Given that the ATP binding site is buried in the dimer interface, it is unlikely that ATP can bind after dimer formation. In fact, when either ACOX-1 or ACOX-1(H396G) was incubated with a 10 molar excess of FAD and ATP, no additional FAD or ATP became bound to the proteins (SI Appendix, Fig. S10). Thus, ATP binding likely occurs during ACOX protein folding and/or dimer formation.
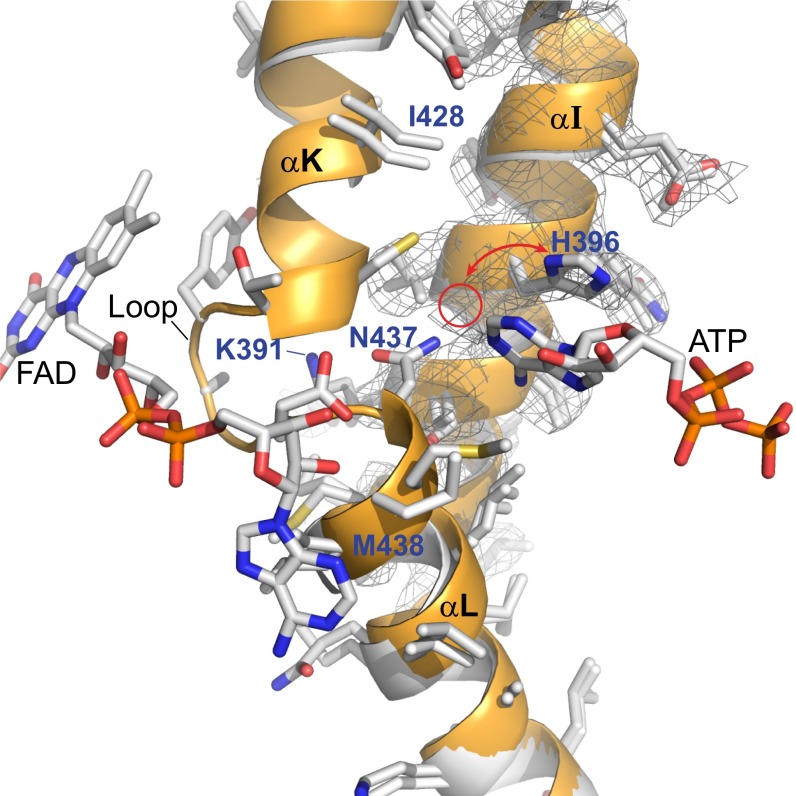
To investigate whether there is a structural basis for the correlation between ATP and FAD binding, we compared the crystal structure of apo-ACOX-1, which is not bound to either ATP or FAD, and the crystal structure of ACOX-1(E434A), which is bound to both ATP and FAD (Fig. 6 and SI Appendix, Figs. S11 and S12). In the ACOX-1(E434A) structure (shown in orange in Fig. 6 and SI Appendix, Figs. S11 and S12), the loop between αK and αL is ordered and interacts with FAD. Asn-437 on αL hydrogen bonds to the adenine ring of ATP, as well as Lys-391 on αI, and the interaction between Asn-437 and Lys-391 appears to stabilize the loop between αK and αL. Conversely, in the apo-ACOX-1 structure (shown in white in Fig. 6 and SI Appendix, Figs. S11 and S12), this loop is disordered. The electron density of the apo-ACOX-1 structure shows that His-396 can adopt two different conformations: one similar to its conformation in the ACOX-1(E434A) structure and one that might disrupt an interaction between with Asn-437 and Lys-391. Unlike other residues in αI, Lys-391 cannot be modeled in the apo-ACOX-1 structure, suggesting it is flexible in the absence of ATP and/or FAD binding. Consistent with the role of Asn-437 and Lys-391 in coordinating the binding of ATP and FAD, the ACOX-1(N437A) and ACOX-1(K391A) mutants bind little to no ATP and FAD (SI Appendix, Fig. S13). ACOX-1(W189A), which is lacking a Trp residue that stacks with the isoalloxazine ring of FAD, fails to bind FAD, as might be expected, but also fails to bind to ATP, even though the Trp residue is located on β3.2 and is quite distant from the ATP binding site (SI Appendix, Fig. S13). ACOX-1(Q340A), which is lacking a Gln located on αH that hydrogen bonds to the ribose ring of FAD, also fails to bind both FAD and ATP (SI Appendix, Fig. S13). Thus, binding of FAD and ATP appear to be dependent on one another.
Fig. 6.
Interaction between the ATP and FAD binding sites. Overlay of apo-ACOX-1 structure (no bound ATP or FAD) in white with holo-ACOX-1(E434A) structure (with bound ATP and FAD) in orange. α-helices αI, αK, and αL, as well as the electron density (2Fo-Fc map, σ=1.0) associated with αI of apo-ACOX-1, are shown. The loop between αK and αL is disordered between Ile-428 and Met-438 in apo-ACOX-1 and is ordered in ACOX-1(E434A) and interacts with FAD. The electron density of the apo-ACOX-1 structure shows that His-396 can adopt two different conformations, one of which is shown and one of which is indicated with a red circle. The back side of this image is shown in SI Appendix, Fig. S11, and a stereoview of this image and its back side is shown in SI Appendix, Fig. S12 A and B.
Conclusions
The ACOX enzymes play a central role in controlling ascaroside-based chemical communication in C. elegans, and likely in other nematode species as well. Here, we have provided a molecular understanding of the substrate specificities of these enzymes. ACOX-2 has a smaller, closed active site with specific amino acid residues that recognize the ascarylose ring of its preferred short-chain substrate, asc-ωC5-CoA (2). ACOX-1, in contrast, has a larger active site that opens to two hydrophobic channels on the outer surface of the protein, enabling the enzyme to process medium-chain ascaroside-CoA and fatty acyl-CoA substrates. Furthermore, we have shown that most of the ACOX enzymes in C. elegans have a conserved ATP-binding pocket at the dimer interface, suggesting ATP levels may regulate the function of these enzymes in ascaroside biosynthesis. Because the ATP binding site is buried, it is unlikely that the ATP is rapidly exchanged. It is more likely that ATP binding occurs during the process of ACOX protein folding and/or dimer formation, and that once the ACOX dimer forms, the ATP becomes trapped with no further opportunities for exchange. Although it remains to be demonstrated whether the ACOX enzymes bind ATP in C. elegans in vivo, the ability of heterologously expressed ACOX enzymes to bind ATP is correlated with their ability to bind FAD, which is required for their enzymatic activity. Thus, ATP levels may affect the overall level of fully functional ACOX complexes in the worm. We previously showed that most ascarosides are produced at higher levels in well-fed worms (7). Thus, the ATP-binding site in the ACOX enzymes may enable the worm to produce more ascaroside pheromones under well-fed conditions when cellular ATP levels are high (SI Appendix, Fig. S14). The chemical message of C. elegans may be further fine-tuned by the transcriptional regulation of specific ACOX enzymes to increase or decrease the relative production of certain ascarosides (SI Appendix, Fig. S14). For example, we have shown that food down-regulates the transcription of acox-3 in certain larval stages, thereby suppressing the increased production of shorter chain (ω-1)-ascarosides (7). We suggest that C. elegans may produce dauer-inducing ascaroside pheromones not necessarily to protect itself but, rather, to protect its progeny. The well-fed conditions of the moment might presage a population boom that will eventually deplete food resources and endanger future progeny. Thus, the linking of ascaroside production levels to metabolic state and ATP levels may result in an evolutionary advantage.
Methods
Expression, Purification, Crystallization, and Structure Determination of ACOX-1 and ACOX-2.
Expression vectors for ACOX-1a (the longest splice variant of ACOX-1) and ACOX-2 were described previously (7). Point mutations were made using the Q5 site-directed mutagenesis kit (New England Biolabs). Enzymes were expressed as described previously (7), with modifications (SI Appendix, SI Methods). Details regarding expression, purification, crystallization and structure determination can be found in the SI Appendix, SI Methods. Crystallographic data and refinement statistics are shown in SI Appendix, Table S1.
Synthesis of CoA-Thioesters and ACOX Activity Assay.
Ascarosides were synthesized as described previously (28). Pentanoic, heptanoic, and nonanoic acid were purchased (Sigma). CoA-thioesters of ascarosides and fatty acids were synthesized according to a previously described method (7), except that C12-CoA (7) was purchased from Sigma and C16-CoA (8) was purchased from ChemImpex. The activity of wild-type and mutant ACOXs enzymes were determined using a previously published method (7), except that substrates were tested at 24 μM and reactions were run at room temperature.
Supplementary Material
Acknowledgments
We thank the staff at the LS-CAT 21-ID-F/G beamline at Argonne National Laboratory, Advanced Photon Source, for X-ray access and data collection. This work was supported by a CAREER award from the National Science Foundation (1555050), the Research Corporation for Science Advancement, the Ellison Medical Foundation, and the Alfred P. Sloan Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5K3G, 5K3H, 5K3I, and 5K3J).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608262113/-/DCSupplemental.
References
- 1.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 2.Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK. Peroxisomal β-oxidation--a metabolic pathway with multiple functions. Biochim Biophys Acta. 2006;1763(12):1413–1426. doi: 10.1016/j.bbamcr.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 3.Butcher RA, et al. Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc Natl Acad Sci USA. 2009;106(6):1875–1879. doi: 10.1073/pnas.0810338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joo HJ, et al. Caenorhabditis elegans utilizes dauer pheromone biosynthesis to dispose of toxic peroxisomal fatty acids for cellular homoeostasis. Biochem J. 2009;422(1):61–71. doi: 10.1042/BJ20090513. [DOI] [PubMed] [Google Scholar]
- 5.Joo HJ, et al. Contribution of the peroxisomal acox gene to the dynamic balance of daumone production in Caenorhabditis elegans. J Biol Chem. 2010;285(38):29319–29325. doi: 10.1074/jbc.M110.122663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Reuss SH, et al. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small-molecule signals in C. elegans. J Am Chem Soc. 2012;134(3):1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, et al. Acyl-CoA oxidase complexes control the chemical message produced by Caenorhabditis elegans. Proc Natl Acad Sci USA. 2015;112(13):3955–3960. doi: 10.1073/pnas.1423951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong PY, et al. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature. 2005;433(7025):541–545. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- 9.Butcher RA, Fujita M, Schroeder FC, Clardy J. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol. 2007;3(7):420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- 10.Butcher RA, Ragains JR, Kim E, Clardy J. A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proc Natl Acad Sci USA. 2008;105(38):14288–14292. doi: 10.1073/pnas.0806676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasan J, et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454(7208):1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butcher RA, Ragains JR, Clardy J. An indole-containing dauer pheromone component with unusual dauer inhibitory activity at higher concentrations. Org Lett. 2009;11(14):3100–3103. doi: 10.1021/ol901011c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pungaliya C, et al. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106(19):7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivasan J, et al. A modular library of small molecule signals regulates social behaviors in Caenorhabditis elegans. PLoS Biol. 2012;10(1):e1001237. doi: 10.1371/journal.pbio.1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izrayelit Y, et al. Targeted metabolomics reveals a male pheromone and sex-specific ascaroside biosynthesis in Caenorhabditis elegans. ACS Chem Biol. 2012;7(8):1321–1325. doi: 10.1021/cb300169c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artyukhin AB, et al. Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J Biol Chem. 2013;288(26):18778–18783. doi: 10.1074/jbc.C113.477000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokuoka K, et al. Three-dimensional structure of rat-liver acyl-CoA oxidase in complex with a fatty acid: Insights into substrate-recognition and reactivity toward molecular oxygen. J Biochem. 2006;139(4):789–795. doi: 10.1093/jb/mvj088. [DOI] [PubMed] [Google Scholar]
- 18.Arent S, Pye VE, Henriksen A. Structure and function of plant acyl-CoA oxidases. Plant Physiol Biochem. 2008;46(3):292–301. doi: 10.1016/j.plaphy.2007.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Kim JJ, Wang M, Paschke R. Crystal structures of medium-chain acyl-CoA dehydrogenase from pig liver mitochondria with and without substrate. Proc Natl Acad Sci USA. 1993;90(16):7523–7527. doi: 10.1073/pnas.90.16.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osumi T, Hashimoto T, Ui N. Purification and properties of acyl-CoA oxidase from rat liver. J Biochem. 1980;87(6):1735–1746. doi: 10.1093/oxfordjournals.jbchem.a132918. [DOI] [PubMed] [Google Scholar]
- 21.Van Veldhoven PP, Van Rompuy P, Vanhooren JC, Mannaerts GP. Purification and further characterization of peroxisomal trihydroxycoprostanoyl-CoA oxidase from rat liver. Biochem J. 1994;304(Pt 1):195–200. doi: 10.1042/bj3040195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Veldhoven PP, Van Rompuy P, Fransen M, De Béthune B, Mannaerts GP. Large-scale purification and further characterization of rat pristanoyl-CoA oxidase. Eur J Biochem. 1994;222(3):795–801. doi: 10.1111/j.1432-1033.1994.tb18926.x. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima Y, et al. Three-dimensional structure of the flavoenzyme acyl-CoA oxidase-II from rat liver, the peroxisomal counterpart of mitochondrial acyl-CoA dehydrogenase. J Biochem. 2002;131(3):365–374. doi: 10.1093/oxfordjournals.jbchem.a003111. [DOI] [PubMed] [Google Scholar]
- 24.Pedersen L, Henriksen A. Acyl-CoA oxidase 1 from Arabidopsis thaliana. Structure of a key enzyme in plant lipid metabolism. J Mol Biol. 2005;345(3):487–500. doi: 10.1016/j.jmb.2004.10.062. [DOI] [PubMed] [Google Scholar]
- 25.Mackenzie J, Pedersen L, Arent S, Henriksen A. Controlling electron transfer in Acyl-CoA oxidases and dehydrogenases: A structural view. J Biol Chem. 2006;281(41):31012–31020. doi: 10.1074/jbc.M603405200. [DOI] [PubMed] [Google Scholar]
- 26.Powers RA, Rife CL, Schilmiller AL, Howe GA, Garavito RM. Structure determination and analysis of acyl-CoA oxidase (ACX1) from tomato. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 6):683–686. doi: 10.1107/S0907444906014107. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen L, Henriksen A. Expression, purification and crystallization of two peroxisomal acyl-CoA oxidases from Arabidopsis thaliana. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 6):1125–1128. doi: 10.1107/S0907444904007577. [DOI] [PubMed] [Google Scholar]
- 28.Hollister KA, et al. Ascaroside activity in Caenorhabditis elegans is highly dependent on chemical structure. Bioorg Med Chem. 2013;21(18):5754–5769. doi: 10.1016/j.bmc.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.