Significance
Extant vertebrates include jawless and jawed species. Jawless vertebrates, such as lamprey and hagfish, do not possess paired fins, whereas jawed vertebrates have two pairs of appendages. Although paired appendages are important in performing complex movements, including swimming, burrowing, and flying, their evolutionary origin remains elusive. In this study, we compare jawless and jawed vertebrate embryos and identify fundamental differences in the expression and regulation of a gene that is essential for the pectoral fin and girdle formation. Our data suggest that modification of the expression and regulation of this gene is coincident with the origin of paired appendages.
Keywords: paired fins, evolution, development, Tbx5 enhancer
Abstract
The diversification of paired appendages has been a major factor in the evolutionary radiation of vertebrates. Despite its importance, an understanding of the origin of paired appendages has remained elusive. To address this problem, we focused on T-box transcription factor 5 (Tbx5), a gene indispensable for pectoral appendage initiation and development. Comparison of gene expression in jawless and jawed vertebrates reveals that the Tbx5 expression in jawed vertebrates is derived in having an expression domain that extends caudal to the heart and gills. Chromatin profiling, phylogenetic footprinting, and functional assays enabled the identification of a Tbx5 fin enhancer associated with this apomorphic pattern of expression. Comparative functional analysis of reporter constructs reveals that this enhancer activity is evolutionarily conserved among jawed vertebrates and is able to rescue the finless phenotype of tbx5a mutant zebrafish. Taking paleontological evidence of early vertebrates into account, our results suggest that the gain of apomorphic patterns of Tbx5 expression and regulation likely contributed to the morphological transition from a finless to finned condition at the base of the vertebrate lineage.
Paired appendages are one of the fundamental novelties of vertebrates. Having emerged in Paleozoic taxa, they have been associated with major patterns of phylogenetic, ecological, and functional diversification ever since. An understanding of the origin of paired appendages is a problem that links multiple approaches—from paleontology to genomics (1–7). The main challenge to progress in the field derives from understanding the similarities and differences between taxa with and without paired fins: How did the mechanisms that pattern paired appendages arise from taxa that lack them altogether?
Whereas jawed vertebrates have two sets of paired fins—pectoral and pelvic—their outgroups, jawless vertebrates, have a range of conditions. Extant jawless fish such as the lamprey and hagfish do not have any paired appendages. The fossil record, however, reveals extinct jawless fish that have pectoral appendages but lack pelvic ones (8). The phylogenetic distribution of extant and extinct species supports the notion that pectoral fins arose before the pelvics (7–12). Therefore, an understanding of pectoral fin development looms large in analyses of the origin of paired appendages.
Tbx5, and its homolog in jawless fish, Tbx4/5, have emerged as attractive candidates to explore the origin of pectoral fins. Phylogenetic analysis reveals that Tbx4/5 of an ancestral jawless vertebrate split into two functional paralogs in species with paired appendages, Tbx4 and Tbx5. These paralogs are involved in the initiation of the pectoral and pelvic appendages, respectively (6, 13–17). Of particular interest is Tbx5 because of its role in pectoral fin development (16–22). The expression pattern of Tbx5 has been studied in multiple chordate species, including amphioxus Tbx4/5. However, data on the expression of Tbx4/5 is sparse in jawless vertebrates (13–16, 18, 23, 24), and detailed comparative expression analysis in taxa with and without paired appendages is lacking. Moreover, whereas it has been hypothesized that cis-regulatory changes of Tbx5 have played an important role in pectoral fin evolution, an understanding of Tbx5 regulation is limited to amniotes and, therefore, lacking in more basal outgroups (15, 24–28).
Here, we performed embryological and genomic analyses of Tbx5 and Tbx4/5 in vertebrates lacking paired fins as well as diverse finned and limbed vertebrates. First, we assessed lamprey Tbx4/5 expression and compared it with that of vertebrates with paired appendages. Subsequently, we explored Tbx5 enhancer activity through a combination of phylogenetic footprinting, functional genomics, and transgenic reporters in zebrafish. Our data reveal phylogenetic patterns of Tbx5 expression and regulation that provide a window into the origin of mechanisms that pattern paired appendages.
Results
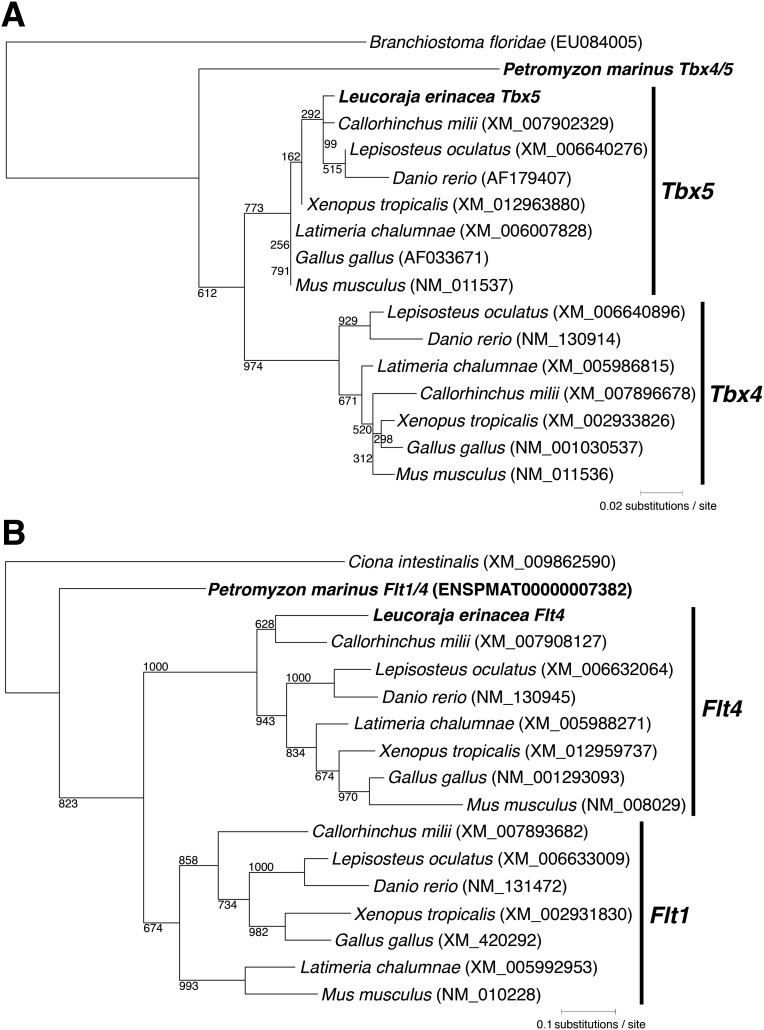
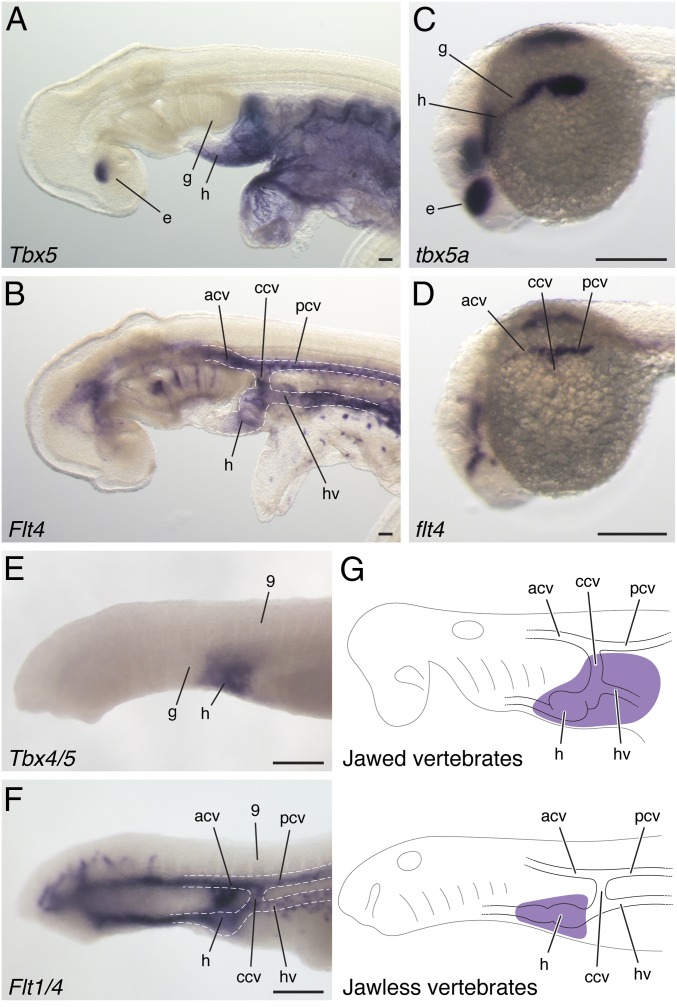
To compare Tbx5 expression in jawless and jawed vertebrates, we cloned sea lamprey Tbx4/5, skate Tbx5, and zebrafish tbx5a and performed whole-mount in situ hybridization. The phylogenetic relationship of sea lamprey Tbx4/5 and skate Tbx5 was confirmed by using the maximum-likelihood method (Fig. S1). In skate and zebrafish embryos, Tbx5 was expressed in the dorsal portion of the eye, heart, and LPM (lateral plate mesoderm) of the pectoral fin field (Fig. 1 A and C). In embryos of skate and zebrafish, expression in the heart and pectoral fin formed a continuous domain as in tetrapods (Fig. 1 A and C) (16, 18, 23). In sea lamprey, Tbx4/5 expression was found in the heart (Fig. 1E and Fig. S2) (24) but, lacking molecular markers, it is difficult to compare the posterior border of expression with jawed vertebrates. To delineate the posterior limit of the heart field, we isolated Flt (fms-like tyrosine kinase/vascular endothelial growth factor receptor) and observed its expression (Fig. 1 B, D, and F and Figs. S1 and S2). Flt is a marker of angioblasts and prefigures the formation of blood vessels. Flt4 and Flt1/4 genes marked anterior and posterior cardinal veins (acv and pcv), which extended in both rostral and caudal directions in the embryos. These two veins were confluent and continued vertically toward the common cardinal vein (ccv, also known as ductus Cuvieri), posterior to the gills. The vein ran further ventrally, bifurcated the hepatocardiac vein, and reached to the posterior extremity of the heart (Fig. 1 B, D, F, and G and Fig. S2). This circulatory pattern, and its topological relationships with surrounding structures, is conserved in vertebrates (5, 29). When the patterns of Tbx5 and Flt4 expression were compared by using these morphological landmarks, we found that the Tbx5 domain included the dorsal part of eye, heart, ccv, and LPM posterior to this vein in jawed vertebrates. In contrast, the Tbx4/5 domain resides exclusively in the heart in sea lamprey with no posterior extension (Fig. 1 and Figs. S2 and S3). Because Tbx5 expression posterior to the heart and gills is limited to jawed vertebrates and Tbx5 is essential for pectoral fin/limb development (16–22), these results imply that the expansion of Tbx5 expression in the LPM posterior to the heart might be associated with the evolution of pectoral fins.
Fig. S1.
Molecular phylogenetic trees of Tbx4/5 (A) and Flt1/4 (B) genes. (A) One hundred eighty-six amino acid sites of Tbx4/5 cognate genes in chordates are used to construct the maximum-likelihood tree. Skate Tbx5 is included in Tbx5 gene cluster, whereas sea lamprey Tbx4/5 is placed outside of Tbx4 and Tbx5 gene clusters. (B) Two hundred seventy-one amino acid sites are used in Flt gene phylogenetic analysis. The maximum-likelihood tree shows the orthology of skate Flt4 with other Flt4 genes. Sea lamprey Flt1/4 is located outside of Flt1 and Flt4 gene clusters. The clear orthology of sea lamprey genes (Tbx4/5 and Flt1/4) to jawed vertebrate genes (Tbx5 and Flt4) is uncertain due to the incomplete sequencing of the sea lamprey genome and hidden paralogy (51). Amphioxus (Branchiostoma floridae) and tunicate (Ciona intestinalis) genes are applied for the out-group. Numbers near the branches represent boot-strap values of 1,000 replicates.
Fig. 1.
Comparison of Tbx5 cognate genes in vertebrates. Skate Tbx5 (A) and Flt4 (B) expression at stage 23. Zebrafish tbx5a (C) and flt4 (D) expression at 22 hpf. Sea lamprey Tbx4/5 (E) and Flt1/4 (F) expression at stage 24. (G) A depiction of the Tbx5 expression domain (purple) relative to embryonic components in jawed and jawless vertebrates. acv, anterior cardinal vein; ccv, common cardinal vein; e, eye; g, gills; h, heart; hv, hepatocardiac vein; pcv, posterior cardinal vein; 9, somite 9. (Scale bars: 200 μm.)
Fig. S2.
Tbx4/5 and Flt1/4 expression in sea lamprey embryos. Tbx4/5 (A−L) and Flt1/4 (M−P) expression in developing sea lampreys. (A) Tbx4/5 was undetectable at stage 21. (B) Tbx4/5 is initially detected in the heart primordium at stage 22. (C−F) Tbx4/5 is expressed in the heart. The posterior border of Tbx4/5 expression and heart domain are not clear due to the premature development of heart and cardinal veins at these stages. (G and H) The heart expression of Tbx4/5 becomes barely detectable at stages 27 and 28. (I−L) Tbx4/5 is undetectable in the heart at stages 29 and 30. The expression is never found in the median fin (J and L). Tbx4/5 expression was not found in the dorsal portion of the eye in sea lamprey embryos. (M) Flt1/4 was detectable in the dorsal and ventral aortas, but not in the cardinal veins at stage 23. (N and O) Flt1/4 was detected in the cardinal veins at stages 24 and 25. (P) Flt1/4 was weakly expressed in the cardinal veins at stage 26. (Scale bars: 200 μm.)
Fig. S3.
tbx5a and flt4 expression in zebrafish. tbx5a (A and B) and flt4 (C and D) expression at 24 hpf of zebrafish embryos. Dorsolateral views (A and C) and dorsal views (B and D). tbx5a is expressed next to somites 1–4. The posterior half of the common cardinal vein labeled by flt4 is located next to somite 1. acv, anterior cardinal vein; ccv, common cardinal vein; pcv, posterior cardinal vein; s1, somite 1; s3, somite 3. (Scale bars: 200 μm.)
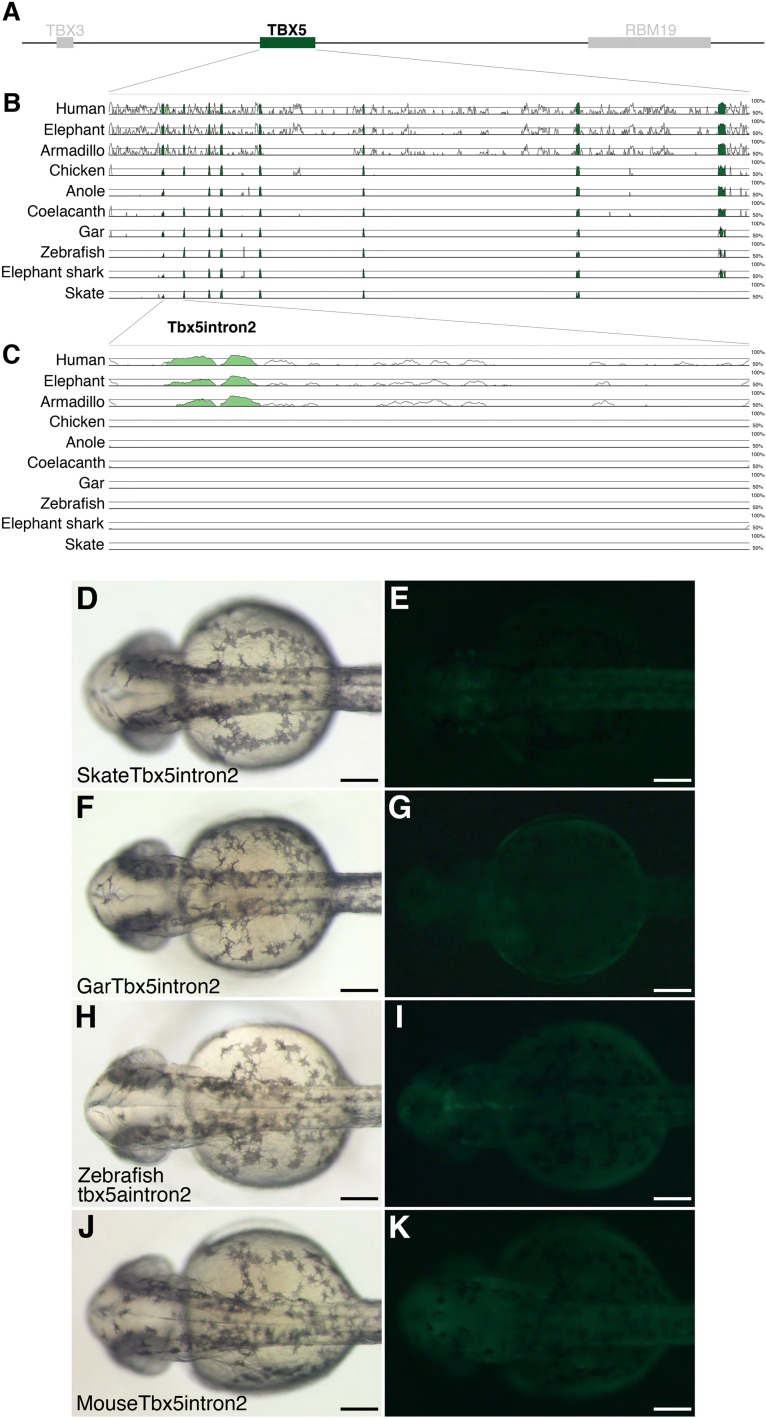
The regulation of Tbx5 in limbs has been explored in mouse, revealing one forelimb enhancer in the second intron (Tbx5 intron2) and its regulation by Hox genes (15, 25, 30). To assess the phylogenetic diversity of Tbx5 regulation by this enhancer, we first analyzed the evolutionary conservation of Tbx5 intron2 activity. By comparing vertebrate genomic sequences adjacent to the Tbx5 locus, we found that Tbx5 intron2 was conserved in mammals, but not in other vertebrates (Fig. S4 A–C). Because enhancers can retain function without displaying sequence similarity (31), the regulatory potential of Tbx5 intron2 from skate, gar, and zebrafish was tested in transgenic zebrafish. Several independent stable lines were established in zebrafish, but none could recapitulate the endogenous expression pattern of Tbx5 (five lines with skate sequence, three lines with gar sequence, and seven lines with zebrafish sequence) (Fig. S4 D–I). Enhancers can change their position in the genome (32, 33), and if zebrafish retains a Tbx5 intron2-like enhancer somewhere else in the genome along with the similar trans environment to mouse, transgenic zebrafish carrying mouse Tbx5 intron2 could express GFP in the pectoral fin. To test this possibility, we assayed mouse Tbx5 intron2 activity in transgenic zebrafish. Six stable lines were established, but conspicuous GFP signal was never detected in the pectoral fin (Fig. S4 J and K). Altogether, these results do not support the notion that Tbx5 intron2 and its regulation are evolutionarily conserved.
Fig. S4.
Sequence alignment of Tbx5 coding region and Tbx5 intron2 transgenic zebrafish. (A) A schematic representation of human TBX5 locus. (B) mVISTA analysis around Tbx5 using the mouse sequence as base. (C) Enlargement of Tbx5 intron2. Conserved sequences are indicated as peaks. Brightfield (D, F, H, and J) and fluorescent (E, G, I, and K) images of Tbx5 intron2 transgenic zebrafish. All images are from dorsal view at 36 hpf. Skate (D and E), gar (F and G), zebrafish (H and I) and mouse (J and K) Tbx5 intron2 never drive GFP signal in the pectoral fin bud. (Scale bars: 200 μm.)
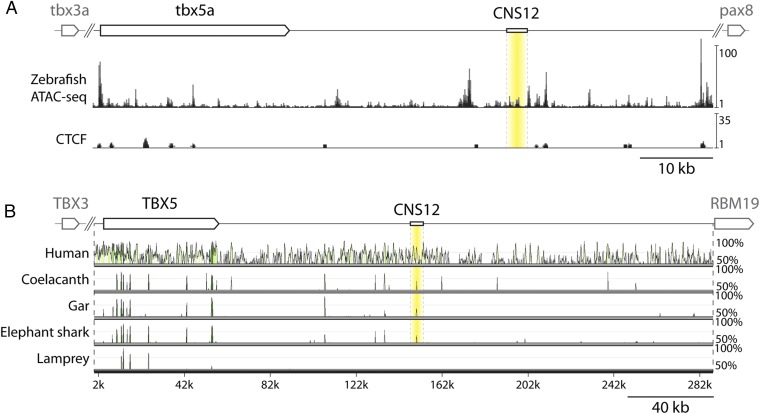
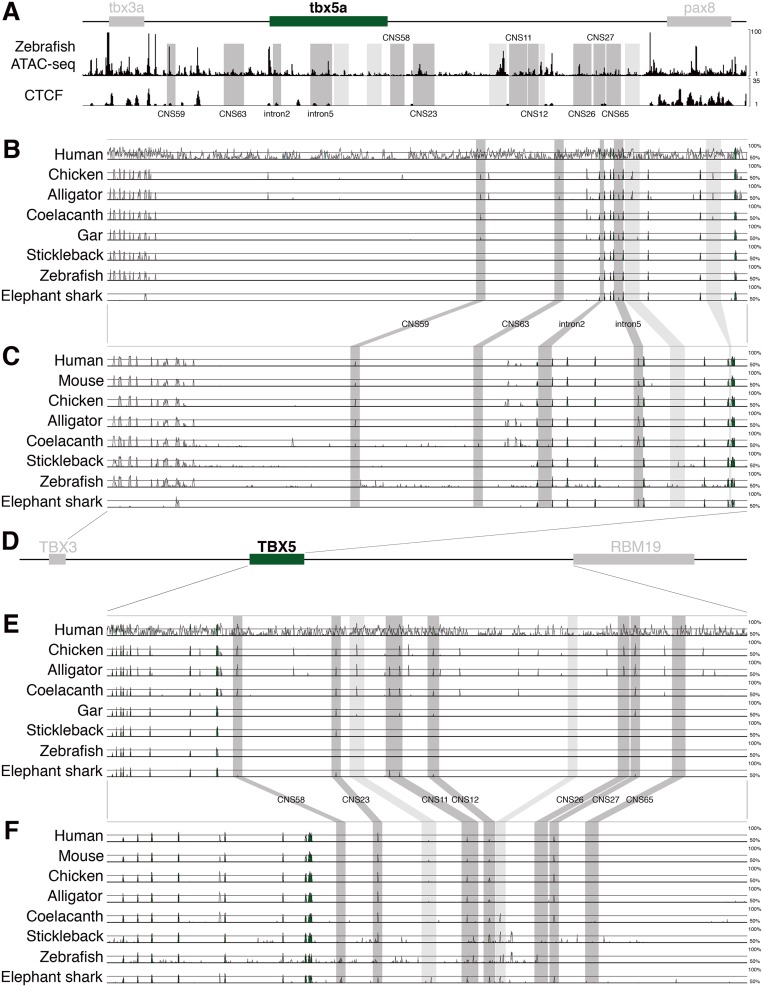
Next, we assayed for Tbx5 fin-specific enhancers. We used recently published ATAC-seq (Assay for Transposase-Accessible Chromatin) data from whole zebrafish embryos at 24 h after fertilization (hpf) to assess open chromatin regions as sites of putative enhancers (34). In addition, we inspected predicted CTCF (CCCTC-binding factor) sites in the open chromatin regions (35). Because these sites are involved in the formation of chromatin loops, we prioritized open chromatin regions without CTCF sites. Using these criteria, more than 30 open chromatin regions were detected within a 120-kb genome sequence between tbx3a and pax8, two flanking genes of tbx5a in the zebrafish genome (Fig. 2A and Fig. S5). Subsequently, we assessed sequence conservation of diverse vertebrate species to reveal CNSs (conserved noncoding sequences) around the Tbx5 locus (Fig. 2B and Fig. S5). This approach revealed more than 30 CNSs. The intersection of these two criteria (open chromatin regions and CNSs) winnowed the pool to 10 candidate enhancer regions to assay by using functional tests in transgenic reporter constructs. The gar genome was used to engineer these constructs because it is more similar to the tetrapod genome than zebrafish one, being basal to the teleost-specific whole genome duplication (34, 36). Gar DNA fragments were integrated into the GFP reporter vector and injected into zebrafish eggs to establish stable transgenic lines (Fig. S6). With these lines, we identified CNS12, a 3,108-bp sequence located downstream of the Tbx5 coding region, as a fin enhancer. CNS12 drove GFP expression in the dorsal part of eye and pectoral fin (Figs. 2 and 3, three independent transgenic stable lines), in a spatial pattern identical to that of Tbx5 ortholog in jawed vertebrates. Importantly, GFP signals in the LPM driven by the enhancer reside posterior to the heart and gills in the same apomorphic pattern detected in jawed vertebrates (Figs. 1 and 3 and Figs. S3 and S7) (16, 18, 23).
Fig. 2.
Chromatin state and sequence comparison adjacent to the Tbx5 locus. (A) A schematic representation of zebrafish tbx5a locus, ATAC-seq data from 24 hpf zebrafish, and predicted CTCF sites. CNS12 is denoted in yellow. (B) A schematic representation of human TBX5 locus and mVISTA analysis with Tbx5 (Tbx4/5 in Japanese lamprey) coding and the downstream noncoding sequences using the mouse sequence for comparison. CNS12 is labeled in yellow, and peaks at the middle of the label indicate CNS12sh.
Fig. S5.
Chromatin state and alignments of genomic sequence around the Tbx5 locus. (A) A schematic representation of the zebrafish tbx5a locus, ATAC-seq data from 24 hpf zebrafish, and predicted CTCF sites. mVISTA analyses of genomic sequence between Tbx3 and Tbx5 by using the mouse sequence as baseline (B), and gar sequence as baseline (C). (D) Schematic representation of human TBX5 locus. mVISTA analyses of Tbx5 and downstream sequence using the mouse sequence as baseline (E), and gar sequence as baseline (F). Conserved sequences are indicated as peaks. DNA fragments tested in transgenic zebrafish are highlighted in dark gray. DNA fragments marked in light gray were tested in transient injection of reporter construct and did not show GFP expression in the pectoral fin.
Fig. S6.
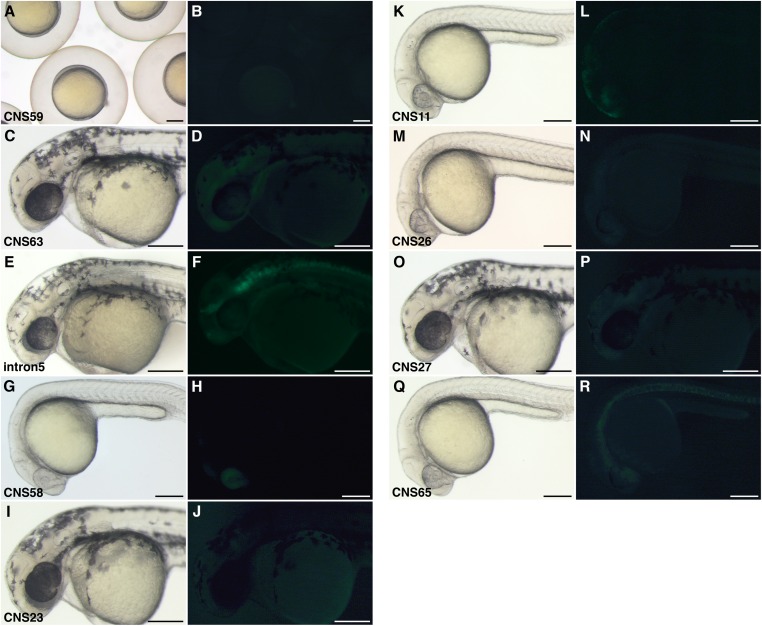
Transgenic zebrafish with DNA fragment around Tbx5 locus. Brightfield (A, C, E, G, I, K, M, O, and Q) and fluorescent (B, D, F, H, J, L, N, P, and R) images. (A and B) CNS59 transgenic zebrafish at 9 hpf. One stable line is screened. (C and D) CNS63 transgenic zebrafish at 36 hpf: one line. (E and F) Tbx5 intron5 transgenic zebrafish at 36 hpf: seven lines. (G and H) CNS58 transgenic zebrafish at 24 hpf: two lines. (I and J) CNS23 transgenic zebrafish at 36 hpf: six lines. (K and L) CNS11 transgenic zebrafish at 24 hpf: one line. (M and N) CNS26 transgenic zebrafish at 36 hpf: one line. (O and P) CNS27 transgenic zebrafish at 36 hpf: one line. (Q and R) CNS65 transgenic zebrafish at 24 hpf: two lines. (Scale bars: 200 μm.)
Fig. 3.
Gar CNS12 drives GFP expression in the pectoral fin of zebrafish. Brightfield (A, D, and G) and fluorescent (B, E, and H) images of gar CNS12 transgenic zebrafish. Lateral views are in A−C and dorsal views are in D−I. GFP expression is found in the dorsal part of the eye and pectoral fin field at 24 hpf (A, B, D, and E), and in the common cardinal vein (arrowhead) and pectoral fin bud at 36 hpf (G and H). Dotted lines outline the pectoral fin. Zebrafish tbx5a in situ hybridization at 24 hpf (C and F) and at 36 hpf (I). (Scale bars: 200 μm.)
Fig. S7.
GFP expression pattern of gar CNS12 transgenic zebrafish. Brightfield (A, C, E, and F) and fluorescent (B, D, and G) images of gar CNS12 transgenic zebrafish. Lateral views in (A, B, and E−G) and dorsal views in (C and D). (A and B) GFP expressions are found in the dorsal eye and pectoral fin bud at 36 hpf. Dotted lines outline the fin bud, in which GFP signals are detected in the mesenchyme. (C and D) GFP are expressed on the posterior edge of the ccv (dotted lines and arrowhead). (E) A 4-wk-old juvenile of the transgenic fish. (F) The juvenile has the pelvic fin bud (black arrowhead). (G) The fin bud does not show GFP signals (white arrowhead). A green emission is detected in the digestive tract. ccv, common cardinal vein; e, eye; pf, pectoral fin. (Scale bars: A−D, 200 μm; E−G, 500 μm.)
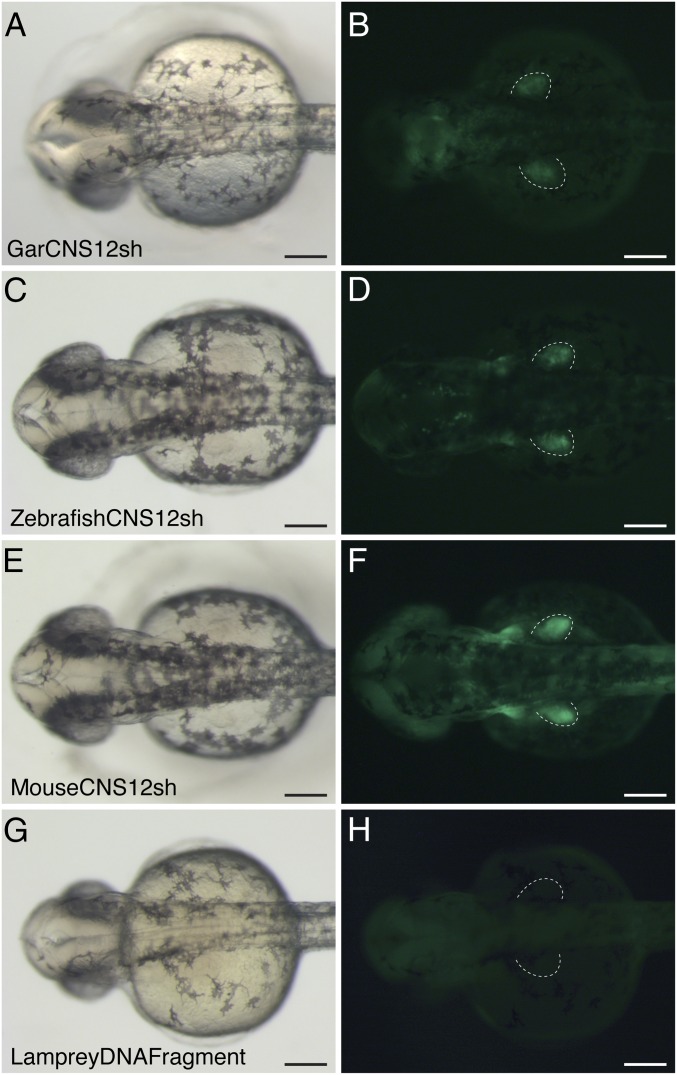
Phylogenetic footprinting revealed a highly conserved domain within CNS12, an ∼200-bp sequence, which was named CNS12sh (CNS12 short) (Fig. 2). To test the evolutionary conservation of CNS12 activity, we isolated orthologous sequences of CNS12sh from gar, zebrafish, and mouse, and assayed their activity in transgenic zebrafish. Analysis of stable lines revealed that all three CNS12sh drove GFP expression in the pectoral fin bud (four lines with gar sequence, two lines with zebrafish sequence, and nine lines with mouse sequence) (Fig. 4). GFP expression in the dorsal part of eye was not detected in these lines. We also cloned a DNA fragment from the Japanese lamprey genome, which was aligned to CNS12 of jawed vertebrates in mVISTA analyses (Fig. 2B and Fig. S8), and tested its activity in transgenic zebrafish. Four transgenic stable lines were established, but GFP expression was undetectable in the pectoral fin (Fig. 4). These results indicate that the function of CNS12 is conserved only among jawed vertebrates.
Fig. 4.
Brightfield (A, C, and E) and fluorescent (B, D, and F) images of CNS12sh transgenic zebrafish from dorsal view at 36 hpf. Gar (A and B), zebrafish (C and D), and mouse (E and F) CNS12sh drive GFP expression in the pectoral fin bud. (G and H) Transgenic zebrafish with Japanese lamprey DNA fragment aligned to jawed vertebrate CNS12 at 36 hpf. Dotted lines outline the pectoral fin. (Scale bars: 200 μm.)
Fig. S8.
Sequence alignment of CNS12 from jawed vertebrates and Japanese lamprey DNA fragment. Gar CNS12sh sequence is highlighted in gray.
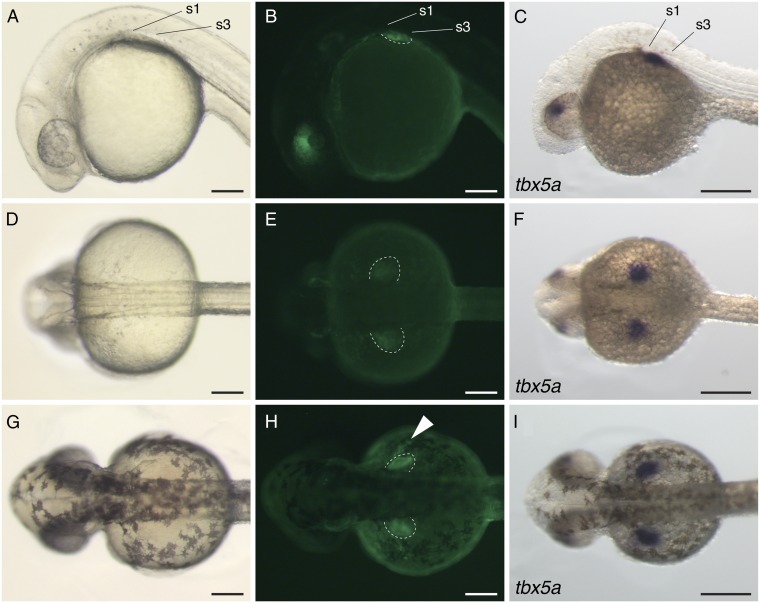
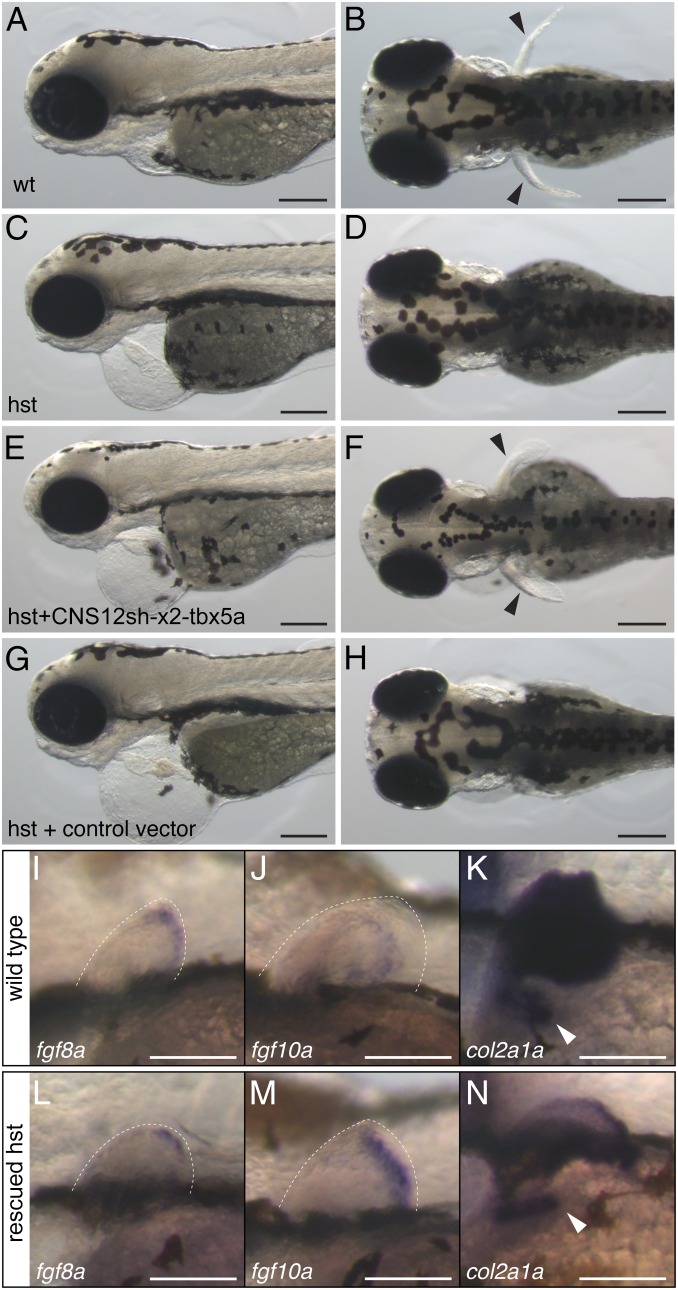
If Tbx5 regulation by CNS12 is associated with pectoral fin initiation, then CNS12 driving Tbx5 expression posterior to the heart and gills should be sufficient to induce pectoral fin development (28). To test this possibility, we used a zebrafish tbx5a mutant, heartstrings (hst), that has an aberrant heart and does not develop a pectoral fin (Fig. 5 A–D) (19). The hst mutation is a premature stop codon in tbx5a exon8, and tbx5a is weakly expressed in the LPM of hst embryos until 32 hpf, but undetectable thereafter (19). We performed injections of a vector carrying two copies of gar CNS12sh and the zebrafish tbx5a coding sequence into hst embryos and assayed development through 3 dpf (Fig. S9). The injected homozygous hst embryos were screened for heart morphology and genotyped by the sequence of tbx5a exon8. Although the heart morphology of hst was not rescued, a number of injected homozygous embryos revealed pectoral fin buds in the region where fin buds appear in the wild-type (n = 22/163) (Fig. 5 E and F). When we injected a control vector, which contains the zebrafish tbx5a coding sequence but lacks the cis-regulatory element (Fig. S9), the injected hst embryos did not develop a pectoral fin (n = 0/52) (Fig. 5 G and H). fgf8a and fgf10a, markers for apical ectodermal ridge and fin mesenchyme, respectively, were expressed in the rescued fin buds in a manner similar to the wild-type (Fig. 5 I, J, L, and M) (37). We further detected col2a1a expression at the base of the rescued fin, indicating that aspects of pectoral girdle development were also restored by the expression construct (Fig. 5 K and N). To test phylogenetic conservation of CNS12sh function, we engineered a construct with two copies of mouse CNS12sh and the zebrafish tbx5a coding sequence, and injected it into hst mutant. We found fin buds in a subset of the injected hst embryos (n = 16/175). In these rescue experiments, fin buds appeared variable in size and laterality, likely due to the mosaicism of injected embryos (Fig. 5 and Fig. S9, seven bilateral and 15 unilateral rescued fins in gar CNS12sh and five bilateral and 11 unilateral rescued fins in mouse CNS12sh). These results indicate that CNS12 driving tbx5a is able to partially rescue the phenotype of hst that lacks a pectoral fin and girdle (Fig. 5 and Fig. S9). Moreover, this ability is conserved between fish and mouse.
Fig. 5.
tbx5a driven by CNS12sh rescues pectoral fin formation in hst. Wild-type (A and B) and hst (C−H) at 3 dpf. Lateral views (A, C, E, and G) and dorsal views (B, D, F, and H). Black arrowheads indicate pectoral fins. fgf8a (I and L), fgf10a (J and M), and col2a1a (K and N) in situ hybridization in the wild-type (I−K) and rescued hst (L−N) at 3 dpf. Dotted lines outline the pectoral fin and white arrowheads indicate the scapulocoracoid. (Scale bars: A−H, 200 μm; I−N, 100 μm.)
Fig. S9.
Various phenotypes of pectoral fins in rescued hst. Schematic of vectors used in the rescue (A) and control (B) experiments. Dorsal views of rescued hst zebrafish at 3 dpf. Arrowheads indicate pectoral fins. (C) The right fin, almost identical to the wild-type fin, is well developed compared with the left one. (D) Likewise in C, the right fin is bigger than the left one, but both right and left fins are less developed compared with the wild-type fin. (E) A small left fin is rescued. (F) A small right fin is rescued. (G and H) tbx5a in situ hybridization in the wild-type (G) and rescued hst (H) at 3 dpf. Dotted lines outline the fin bud. cFos, cFos minimal promoter; CNS12sh, CNS12 short; Drtbx5a, Danio rerio tbx5a coding sequence; Tol2, Tol2 transposon sequence. (Scale bars: C−F, 200 μm; G and H, 100 μm.)
Discussion
Epigenetic, comparative, and functional genomic analyses led to the identification of a Tbx5 fin enhancer, CNS12, in the noncoding region downstream of Tbx5 locus (Figs. 2 and 3). This enhancer drove reporter gene expression in the LPM posterior to the heart, where vertebrates with pectoral appendages exhibit an apomorphic pattern of Tbx5 expression (Figs. 1–3). Importantly, sequence conservation of CNS12 was detected among jawed vertebrates, while remaining undetectable adjacent to the Japanese lamprey Tbx4/5 locus (Fig. 2B and Fig. S8). Indeed, we confirmed the evolutionary conservation of CNS12 activity only among jawed vertebrates (Figs. 2, 4, and 5). Comparative genomic analyses of the noncoding sequence downstream of jawed vertebrate Tbx5 and Japanese lamprey Tbx4/5 repeatedly aligned the same sequence of the Japanese lamprey genome with CNS12, but this lamprey sequence did not show any activity in pectoral fins of transgenic zebrafish (Fig. 4).
In this context, Tbx4/5 expression in amphioxus (Branchiostoma floridae BfTbx4/5 and Branchiostoma lanceolatum BlTbx4/5) becomes phylogenetically relevant. BfTbx4/5 expression was originally described in the caudoventral part of the amphioxus body and was compared with Tbx5 expression in the heart and LPM of jawed vertebrates (14, 15). However, reexamination of these patterns revealed that BlTbx4/5 is expressed in the pharyngeal and posterior mesoderm together with cardiac genes. This broad expression, and its association with cardiac markers, suggests that amphioxus Tbx4/5 is involved in the development of a noncentralized heart (38). Consistent with this observation, marker genes for vertebrate head and trunk mesoderm are expressed in overlapping domains in amphioxus dorsal mesoderm indicating that, in contrast to vertebrates, the mesodermal components of amphioxus are not differentiated along the craniocaudal axis (39). Together with our comparative assays, these findings support the hypothesis that expansion of Tbx5 expression in the LPM of the prospective pectoral domain is an apomorphic feature of jawed vertebrates and is not directly comparable to Tbx4/5 expression of amphioxus.
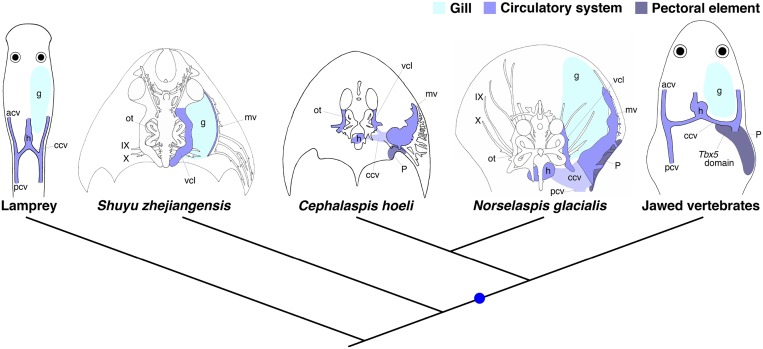
Given the function of Tbx5 and its regulatory element in pectoral fin development (16–22, 28) (Fig. 5), the gain of a Tbx5 expression domain controlled by CNS12 was likely an important step in the acquisition of vertebrate paired appendages. Analysis of Paleozoic osteostracans, armored jawless fish from the Silurian and Devonian, may reveal phylogenetic nodes at which this developmental mechanism arose. Osteostracans have head shields containing a canal for a lateral head vein, which passes along the lateral side of the semicircular canals, and another for a marginal vein, that runs ventrolaterally to the lateral head vein (Fig. S10). Ventromedial to the canals, an ossified pericardial capsule lies posterior to the gill region in osteostracans. Based on the topographic relationships of this capsule and the canals, the common cardinal vein is inferred to reside in the caudal part of the head shield in osteostracans (Fig. S10) (12, 40, 41). Importantly, osteostracans have a pectoral girdle, and portions of the fin skeleton, lying caudally to the heart, gills, and the inferred position of the ccv, a condition much like jawed vertebrates (7–12, 40, 42–45). These observations support the hypothesis that Tbx5 expression and regulation caudal to the heart and gills, characteristic of jawed vertebrates, may have had its origins in Paleozoic jawless taxa.
Fig. S10.
Vertebrate phylogeny and evolution of pectoral elements. Head and anterior trunk regions, together with internal anatomy, are schematized in lamprey, galeaspids (Shuyu zhejiangensis), osteostracans (Cephalaspis hoeli and Norselaspis glacialis), and jawed vertebrates. The heart, lateral head vein, and marginal vein in fossil species are reconstructed based on the ossified capsule and canals. The common cardinal vein and posterior cardinal vein of osteostracans are inferred based on the morphology of the pericardial capsule, lateral head and marginal vein canals, and larval lamprey anatomy (41, 52, 53). Osteostracans possess the pectoral elements that are morphologically comparable to that of placoderms, a paraphyletic assemblage of stem-jawed vertebrates (7–12, 40, 42–45). Those components, the pericardial capsule and pectoral elements, are missing in galeaspids, a sister group of osteostracans and jawed vertebrates (7, 10, 12, 40). Note that the topographical relationship of the pectoral appendage in osteostracans relative to the heart, gills, and ccv is comparable to that of jawed vertebrates. Galeaspids was redrawn from ref. 54, and osteostracans was redrawn from refs. 8, 10, 12, and 41. Blue spot indicates gain of pectoral elements, and hypothesized gain of Tbx5 expression posterior to the heart and CNS12. acv, anterior cardinal vein; ccv, common cardinal vein; g, gills; h, heart; IX, glossopharyngeal nerve (ninth cranial nerve); mv, marginal vein; ot, otic components; p, pectoral element; pcv, posterior cardinal vein; vcl, vena capitis lateralis (lateral head vein); X, vagus nerve (10th cranial nerve).
Materials and Methods
Embryos, Cloning, and in Situ Hybridization.
The protocol for animal experiments was approved by the University of Chicago Institutional Animal Care and Use Committee. Sea lamprey embryos were sampled in the Bronner laboratory at the California Institute of Technology and staged as described (46). Skate embryos were purchased from the Marine Biological Laboratory and staged as described (47). Zebrafish embryos (strain *AB) were obtained from natural mating and staged as described (48). Sea lamprey, skate, and zebrafish embryos were fixed in MEMFA (MOPS, EGTA, magnesium sulfate salts, and formaldehyde) fixative. Total RNAs of these animals were extracted from whole embryos with TRIzol Reagent (Life Technologies) and then used to synthesize cDNAs with SuperScript III system (Thermo Fisher Scientific). DNA fragments were amplified by GoTaq (Promega) and cloned into pCRII-TOPO vector (Invitrogen). RNA probes were synthesized by using SP6 or T7 RNA polymerase (Promega). The accession numbers of cloned genes and all primer sequences used in this study were listed in Tables S1 and S2. In situ hybridization was performed as described (49), with modification of the hybridization solution (5× SSC, pH 4.5, and 50 μg/mL heparin, instead of salt solution and dextran sulfate). The initial stage showing Flt4 (Flt1/4) expression in the common cardinal vein of sea lamprey, skate, and zebrafish embryos was used for the comparison. Embryos were postfixed in MEMFA fixative, washed in TE buffer, and photographed on a Leica M205FA microscope.
Table S1.
List of accession numbers
Table S2.
List of primers
| Gene/position | Forward | Reverse |
| Primers for cloning | ||
| P. marinus Tbx4/5 | TAYAARTTYGCNGAYAAYAA | CCYTTNGCRAANGGRTTRTTYTC |
| AARTTYGCNGAYAAYAARTGG | RAANCCYTTNGCRAANGGRTT | |
| CGCGGCTGGTACTTGTGCATCGAG | ||
| GCGAGTCGGGGTGCACGTAGAG | ||
| Primers for probe synthesis | ||
| L. erinacea Tbx5 | GGGGAGGTTGTACGTTCACCCAGAT | GGGCTGGTGAGAAACTGAGGTCTGA |
| L. erinacea Flt4 | CGGGACTTCTGGACAATCTCACCGA | TACTCCACGATCACCATCAGCGGAC |
| Danio rerio tbx5a (AF179407) | CGGTAGACATCGTACAGGCCTCTCC | ACGGCTTCTTATAGGGGTGTTCTCCTG |
| D. rerio flt4 (NM_130945) | TGCAATGTCAAGCGAGTGAACCCAG | CAATCTGGATGCCGGGGTATGGAGA |
| D. rerio tpma (NM_131105) | CGCCTTGGACAGAGCAGAGCAG | AGTGCGATGGAGAAAAGCGGCA |
| D. rerio fgf8a (NM_131281) | TCCTTCACCTCTTTGCGTTT | CACATCCTGTGCTTCGCTTA |
| D. rerio fgf10a (NM_182870) | ACCAACTCCTCATCGTCTGC | CAATGTCCGATTCCTCTCGT |
| D. rerio col2a1a (NM_131292) | GGTGATCGTGGTGAGATTGGTGCAC | CCTCTGTGTCCCTTCTGGCCTCTTT |
| P. marinus Tbx4/5 | AARTTYGCNGAYAAYAARTGG | CCYTTNGCRAANGGRTTRTTYTC |
| P. marinus Flt1/4 | GACGAGCGAGGATGAAGGGGAGTAC | CCACACCATTATCGGCCAGCAGAAC |
| (ENSPMAT00000007382) | ||
| Primers for L. japonicum Tbx4/5 genome sequence | ||
| Tbx4/5 Gap1 | CGTGTCGCCTTCCTCCGCAGA | TGGATGCGTGGGTGATTTCGTGGAT |
| Tbx4/5 Gap2 | CGCCTCCTCCGCTCTCCTTATCTGA | ACCACGGGAAGAGAGAAAGAGACGC |
| Tbx4/5 Gap3 | TGCGGTCTGTGAACATCGTCACTGG | AACATCTCCTAGCCACCGTGCTTGC |
| Tbx4/5 Gap4 | TGTGGGAGGAGAAATCGGAGGACCC | TGCGGTAATTTCCAGAACTGCGGCT |
| Tbx4/5 Gap5 | GCAACTCACGACTACCCCACTTGCA | GTGGTGACCGTGAGCAGGAATCGAG |
| Tbx4/5 Gap6 | GCAACTCACGACTACCCCACTTGCA | GTGGTGACCGTGAGCAGGAATCGAG |
| Tbx4/5 Gap7 | GAGGGAGGGAGAGAGAGAGGGACAA | CGTCACCGGTGTTGGCGGT |
| Tbx4/5 Gap8 | CTGCTGCCCATCACATCACCCGC | GTGTAGGTGGTCGCGGGCACTTAAG |
| Tbx4/5 Gap9 | CCGTGACCTTTTACATCGTGCACCCAA | CAACCGGGCAATGACACGATTCAGC |
| Tbx4/5 Gap10 | GTGCTCCTCCTGAACATCGCCCATT | ACCTAGTTGCTGTACTGCCGTGCTG |
| Primers for genomic DNA fragment | ||
| Mouse Tbx5 intron2 | CTCCCAGCAAGTCTCCATCATCCCC | AACTTCAGCCACAGTTCACGTTCATGA |
| Mouse CNS12sh | CCTCATCATCATTAGTCATGCTCGCT | GCTGCTTCTGGAGACACTGGTC |
| Gar Tbx5 intron2 | TCTCCATCACCACAGACAACATACACTCA | CCTCCCTGCCTTGGTTATAATCATCTCTGT |
| Gar Tbx5 intron5 | CGCGCGATTGAGGGTTAGAGTTGAA | AGCGTCTGTTAAGTGATTTCAGATGGCAA |
| Gar CNS11 | AGTCGGCAGAGTCAGCTTCCATAGG | GTGCTCACTACGCCTTCCATTTCGG |
| Gar CNS12 | GGCTGCTGTCTTAAATACCTTTCTTGGGC | TGACGGGTGCTATTTGTCCTTTGCC |
| Gar CNS12sh | AAATCATCATTAGTCGTGGCTCATCGTG | GCCTATCTTGATGTTAGTTTGACCCCTG |
| Gar CNS23 | TCTACAGGCACACACAACCCCTCTG | CTGGACTCTGGACTCTGGACTGTGG |
| Gar CNS26 | CAGGGATGGAAGGCTTCAAACACTCA | AGGAATTAAACCCAGGGCCCCAGAG |
| Gar CNS27 | AGGTTACCAGCTACAGTTGTCACACAGATG | GGCGAGTTCATCCAGTTACACTCAGAAGG |
| Gar CNS58 | AGGCATCGATTCCTTCTCCGTGACG | ATGTGGACTGCAGGGGTTGTGATGG |
| Gar CNS59 | GGGGAGGCTGTGTGGAAAACTGAGT | CTTGTAGCACTGGGGCTCTGGGTTC |
| Gar CNS63 | TATAGCGCTCAGTCACGTGGCCTT | TGCAAATCAACACTCAACGGTACAAGATGT |
| Zebrafish tbx5a intron2 | GTAACGTCAACTAGTGCCTCGATGGT | CTTTTTAAAGAAGCATGCAGACAGAGAGTGAG |
| Zebrafish CNS12sh | GTGACCTTCTCGCTGTAAGATGATTGT | ACACATCTTGATTTCATTAACATCTCTTCCTTCT |
| Zebrafish CNS65 | TGTTCCAGGGTCACAATAATGATGCTTTTAAGA | CATGAAGGTTGCTCTGTGTTCTGCATGA |
| SkateTbx5 intron2 | CCAACAAACCACATTCCCCACAGCA | TCCCGCTTTCGTAATGATCATCTCTGTCC |
| Lamprey DNA fragment | TCGATGCGTTCCTCAAAAGTGGCAC | AGGATTAGGGCAGAGGGAGAGGGTG |
| Primers for tbx5a mutant genotype | ||
| Zebrafish tbx5a exon8 | AGTATTCCTGTGAGAACGGCGTTTCC | |
| Zebrafish tbx5a intron8 | AGGTATCAGTGTGTGGTTTAGAGGCACA | |
Phylogenetic Analysis.
Amino acid sequences were collected from the Ensembl (uswest.ensembl.org/index.html), GenBank (www.ncbi.nlm.nih.gov/), and SkateBase (skatebase.org), and then aligned by ClustalW (www.clustal.org/) without gaps. The construction of phylogenetic trees was performed by the maximum-likelihood method the JTT+I+Γ4 model in PhyML (50).
Genomic Analysis.
ATAC-seq data and CTCF sites were reported (34, 35). Open chromatin regions with peak intensity more than 10 (arbitrary units) and without overlapping CTCF sites were regarded as primary candidates for potential enhancers. Genomic sequences of animals used in this study were gathered from Ensembl (www.ensembl.org//uswest.ensembl.org/index.html?redirectsrc=//www.ensembl.org%2Findex.html), University of California, Santa Cruz Genome Browser (genome.ucsc.edu), SkateBase (skatebase.org), Elephant Shark Genome Project (esharkgenome.imcb.a-star.edu.sg), and Japanese Lamprey Genome Project (jlampreygenome.imcb.a-star.edu.sg). Genomic DNA of Japanese lamprey was obtained from the Sugahara laboratory at the Hyogo College of Medicine. The genome assembly gaps adjacent to Japanese lamprey Tbx4/5 locus were cloned and sequenced. The accession number of lamprey genome sequence is listed in Table S2. The comparison of genomic sequences was performed by mVISTA LAGAN program (genome.lbl.gov/vista/mvista/submit.shtml) with the following parameters: 50 bps calc window, 100 bps min cons Width, and 70% cons Identity. We defined CNSs as regions conserved between more than two vertebrate species. Of these regions, CNSs with conservation among more than five species including gar and/or elephant shark were preferentially assessed.
Vector Construction.
Mouse genomic DNA (Swiss–Webster albino mice) was purchased from Promega. Gar, zebrafish, and skate genome DNA were extracted with phenol-chloroform-isoamyl alcohol and chloroform solutions. The DNA fragment of a putative enhancer was amplified by Platinum Taq DNA Polymerase High Fidelity, cloned into pCR8/GW/TOPO vector, and then relocated to pXIG-cfos-EGFP by using LR Clonase II Plus (Life Technologies). The sequence of a putative enhancer was inserted into 5′ of EGFP, and checked by restriction enzyme digestion and sequencing.
Zebrafish Injection.
Zebrafish fertilized eggs (strain *AB) were obtained from natural mating. Male and female heterozygous tbx5a mutant zebrafish were mated to obtain homozygous tbx5a mutant eggs. For transgenic analyses, 25 ng/μL pXIG-cfos-EGFP vector was injected into one- or two-cell stage embryos with 35 ng/μL transposase RNA, 0.2 M KCl, and phenol red. Injected eggs were raised for 3 mo and then outcrossed to *AB fish (34). Zebrafish with GFP expression in any tissue were screened for establishing transgenic stable lines. For the rescue experiment, 25 ng/μL pXIG-cfos-tbx5a vector with two copies of CNS12sh was injected into one-cell stage hst embryos with 50 ng/μL transposase RNA and phenol red. pXIG-cfos-tbx5a vector without the enhancer was used as a control. Genomic DNA was extracted from wild-type, heterozygous, and homozygous hst embryos at 72 hpf, and PCR was performed with zebrafish tbx5a exon8 and intron8 primers (Table S2). PCR products were sequenced to identify homozygous tbx5a mutants. Embryos were photographed on a Leica M205FA microscope.
Acknowledgments
We thank John Westlund for illustrations; Andrew Gehrke, Darcy Ross, Gokhan Dalgin, Igor Schneider, Joyce Pieretti, Julie Szymaszek, Justin Lemberg, Tetsuya Nakamura, Philippe Janvier, and Robert K. Ho for helpful discussions and critical reading of the manuscript; Marianne Bronner and Stephen Green for sea lamprey sampling; Peter Currie, Alysha Heimberg, Catherine Boisvert, and Steve McLeod for elephant shark embryos; Fumiaki Sugahara for Japanese lamprey materials; and José Luis Gómez-Skarmeta for ATAC-seq data. tbx5a mutant zebrafish was kindly provided by Deborah Garrity and Rasha Alnefie, and we are grateful to Marc Ekker, Gary Hatch, Koichi Kawakami, Gembu Abe, and Robert K. Ho for vectors. This work was supported by The Brinson Foundation and the University of Chicago Biological Sciences Division.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the DNA Data Bank of Japan (DDBJ) database (accession nos. LC121589–LC121592).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609997113/-/DCSupplemental.
References
- 1.Thacher JK. Median and paired fins, a contribution to the history of the vertebrate limbs. Trans Conn Acad Arts Sci. 1877;3:281–310. [Google Scholar]
- 2.Mivart S. Notes on the fins of elasmobranchs, with considerations on the nature and homologues of vertebrate limbs. Trans Zool Soc Lond. 1879;10:439–484. [Google Scholar]
- 3.Balfour FM. On the dedopment of the skeleton of the paired fins of Elasmobranchii, considered in relation to its bearings on the nature of the limbs of the vertebrata. Proc Zool Soc Lond. 1881;49(3):656–671. [Google Scholar]
- 4.Gegenbaur C. Elements of Comparative Anatomy. MacMillan and Co.; London: 1878. [Google Scholar]
- 5.Goodrich E. Studies on the Structure and Development of Vertebrates. MacMillan and Co., Limited; London: 1930. [Google Scholar]
- 6.Ruvinsky I, Gibson-Brown JJ. Genetic and developmental bases of serial homology in vertebrate limb evolution. Development. 2000;127(24):5233–5244. doi: 10.1242/dev.127.24.5233. [DOI] [PubMed] [Google Scholar]
- 7.Pieretti J, et al. Organogenesis in deep time: A problem in genomics, development, and paleontology. Proc Natl Acad Sci USA. 2015;112(16):4871–4876. doi: 10.1073/pnas.1403665112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stensiö E. The Downtonian and Devonian vertebrates of Spitsbergen. I, Family Cephalaspidae. Skrifer om Svalbard og Ishavet. 1927;12:1–391. [Google Scholar]
- 9.Coates MI. The origin of vertebrate limbs. Dev Suppl. 1994;(Suppl):169–180. [PubMed] [Google Scholar]
- 10.Janvier P. Early Vertebrates. Oxford Univ Press; Oxford: 1996. [Google Scholar]
- 11.Coates M. The evolution of paired fins. Theory Biosci. 2003;122:266–287. [Google Scholar]
- 12.Janvier P. Homologies and evolutionary transitions in early vertebrate history. In: Anderson JS, Sues H-D, editors. Major transitions Vertebr. Evol. Indiana Univ Press; Indiana: 2007. pp. 57–121. [Google Scholar]
- 13.Ruvinsky I, Oates AC, Silver LM, Ho RK. The evolution of paired appendages in vertebrates: T-box genes in the zebrafish. Dev Genes Evol. 2000;210(2):82–91. doi: 10.1007/s004270050014. [DOI] [PubMed] [Google Scholar]
- 14.Horton AC, et al. Conservation of linkage and evolution of developmental function within the Tbx2/3/4/5 subfamily of T-box genes: Implications for the origin of vertebrate limbs. Dev Genes Evol. 2008;218(11-12):613–628. doi: 10.1007/s00427-008-0249-5. [DOI] [PubMed] [Google Scholar]
- 15.Minguillon C, Gibson-Brown JJ, Logan MP. Tbx4/5 gene duplication and the origin of vertebrate paired appendages. Proc Natl Acad Sci USA. 2009;106(51):21726–21730. doi: 10.1073/pnas.0910153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duboc V, Logan MPO. Regulation of limb bud initiation and limb-type morphology. Dev Dyn. 2011;240(5):1017–1027. doi: 10.1002/dvdy.22582. [DOI] [PubMed] [Google Scholar]
- 17.Minguillon C, Del Buono J, Logan MP. Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev Cell. 2005;8(1):75–84. doi: 10.1016/j.devcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Ahn DG, Kourakis MJ, Rohde LA, Silver LM, Ho RK. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417(6890):754–758. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- 19.Garrity DM, Childs S, Fishman MC. The heartstrings mutation in zebrafish causes heart/fin Tbx5 deficiency syndrome. Development. 2002;129(19):4635–4645. doi: 10.1242/dev.129.19.4635. [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi JK, et al. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development. 2003;130(12):2729–2739. doi: 10.1242/dev.00474. [DOI] [PubMed] [Google Scholar]
- 21.Gros J, Tabin CJ. Vertebrate limb bud formation is initiated by localized epithelial-to-mesenchymal transition. Science. 2014;343(6176):1253–1256. doi: 10.1126/science.1248228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao Q, Stinnett HK, Ho RK. Asymmetric cell convergence-driven zebrafish fin bud initiation and pre-pattern requires Tbx5a control of a mesenchymal Fgf signal. Development. 2015;142(24):4329–4339. doi: 10.1242/dev.124750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka M, et al. Fin development in a cartilaginous fish and the origin of vertebrate limbs. Nature. 2002;416(6880):527–531. doi: 10.1038/416527a. [DOI] [PubMed] [Google Scholar]
- 24.Kokubo N, et al. Mechanisms of heart development in the Japanese lamprey, Lethenteron japonicum. Evol Dev. 2010;12(1):34–44. doi: 10.1111/j.1525-142X.2009.00389.x. [DOI] [PubMed] [Google Scholar]
- 25.Minguillon C, et al. Hox genes regulate the onset of Tbx5 expression in the forelimb. Development. 2012;139(17):3180–3188. doi: 10.1242/dev.084814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smemo S, et al. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet. 2012;21(14):3255–3263. doi: 10.1093/hmg/dds165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domyan ET, et al. Molecular shifts in limb identity underlie development of feathered feet in two domestic avian species. eLife. 2016;5:1–21. doi: 10.7554/eLife.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isogai S, Horiguchi M, Weinstein BM. The vascular anatomy of the developing zebrafish: An atlas of embryonic and early larval development. Dev Biol. 2001;230(2):278–301. doi: 10.1006/dbio.2000.9995. [DOI] [PubMed] [Google Scholar]
- 30.Nishimoto S, Minguillon C, Wood S, Logan MPO. A combination of activation and repression by a colinear Hox code controls forelimb-restricted expression of Tbx5 and reveals Hox protein specificity. PLoS Genet. 2014;10(3):e1004245. doi: 10.1371/journal.pgen.1004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weirauch MT, Hughes TR. Conserved expression without conserved regulatory sequence: The more things change, the more they stay the same. Trends Genet. 2010;26(2):66–74. doi: 10.1016/j.tig.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Komisarczuk AZ, Kawakami K, Becker TS. Cis-regulation and chromosomal rearrangement of the fgf8 locus after the teleost/tetrapod split. Dev Biol. 2009;336(2):301–312. doi: 10.1016/j.ydbio.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 33.Marinić M, Aktas T, Ruf S, Spitz F. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Dev Cell. 2013;24(5):530–542. doi: 10.1016/j.devcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 34.Gehrke AR, et al. Deep conservation of wrist and digit enhancers in fish. Proc Natl Acad Sci USA. 2015;112(3):803–808. doi: 10.1073/pnas.1420208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gómez-Marín C, et al. Evolutionary comparison reveals that diverging CTCF sites are signatures of ancestral topological associating domains borders. Proc Natl Acad Sci USA. 2015;112(24):7542–7547. doi: 10.1073/pnas.1505463112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braasch I, et al. A new model army: Emerging fish models to study the genomics of vertebrate Evo-Devo. J Exp Zoolog B Mol Dev Evol. 2015;324(4):316–341. doi: 10.1002/jez.b.22589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeller R, López-Ríos J, Zuniga A. Vertebrate limb bud development: Moving towards integrative analysis of organogenesis. Nat Rev Genet. 2009;10(12):845–858. doi: 10.1038/nrg2681. [DOI] [PubMed] [Google Scholar]
- 38.Pascual-Anaya J, et al. The evolutionary origins of chordate hematopoiesis and vertebrate endothelia. Dev Biol. 2013;375(2):182–192. doi: 10.1016/j.ydbio.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 39.Onai T, Aramaki T, Inomata H, Hirai T, Kuratani S. Ancestral mesodermal reorganization and evolution of the vertebrate head. Zoological Lett. 2015;1:29. doi: 10.1186/s40851-015-0030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janvier P. Early specializations in the branchial apparatus of jawless vertebrates: A consideration of gill number and size. In: Arratia G, Wilson MVH, Cloutier R, editors. Recent Advances in the Origin and Early Radiation of Vetebrates. Verlag Dr. Friedrich Pfeil; Munich: 2004. pp. 29–52. [Google Scholar]
- 41.Janvier P, Percy L, Potter I. The arrangement of the heart chambers and associated blood vessels in the Devonian osteostracan Norselaspis glacialis. A reinterpretation based on recent studies of the circulatory system in lampreys. J Zool (Lond) 1991;223:567–576. [Google Scholar]
- 42.Janvier P, Arsenault M, Desbiens S. Calcified cartilage in the paired fins of the Osteostracan Escuminaspis laticeps (Traquair 1880), from the Late Devonian of Miguasha (Québec, Canada), with a consideration of the early evolution of the pectoral fin endoskeleton in vertebrates. J Vertebr Paleontol. 2004;24:773–779. [Google Scholar]
- 43.Goujet D, Young G. Placoderm anatomy and phylogeny: New insights. In: Arratia G, Wilson MVH, Cloutier R, editors. Recent Advances in the Origin and Early Radiation of Vetebrates. Verlag Dr. Friedrich Pfeil; Munich: 2004. pp. 109–126. [Google Scholar]
- 44.Carr RK, Lelièvre H, Jackson GL. The ancestral morphotype for the gnathostome pectoral fin revisited and the placoderm condition. In: Elliott DK, Maisey JG, Yu X, Miao D, editors. Morphol. Phylogeny Paleobiogeography Foss. Fishes. Verlag Dr. Friedrich Pfeil; Munich: 2010. pp. 107–122. [Google Scholar]
- 45.Brazeau MD, Friedman M. The origin and early phylogenetic history of jawed vertebrates. Nature. 2015;520(7548):490–497. doi: 10.1038/nature14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tahara Y. Normal stages of development in the lamprey, Lampetra reissneri (Dybowski) Zoolog Sci. 1988;5:109–118. [Google Scholar]
- 47.Ballard WW, Mellinger J, Lechenault H. A series of normal stages for development of Scyliorhinus canicula, the lesser spotted dogfish (Chondrichthyes: Scyliorhinidae) J Exp Zool. 1993;267:318–336. [Google Scholar]
- 48.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 49.O’Neill P, McCole RB, Baker CVH. A molecular analysis of neurogenic placode and cranial sensory ganglion development in the shark, Scyliorhinus canicula. Dev Biol. 2007;304(1):156–181. doi: 10.1016/j.ydbio.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 51.Kuraku S. Impact of asymmetric gene repertoire between cyclostomes and gnathostomes. Semin Cell Dev Biol. 2013;24(2):119–127. doi: 10.1016/j.semcdb.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Percy LR, Potter IC. Description of the heart and associated blood vessels in larval lampreys. J Zool. 1986;208:479–492. [Google Scholar]
- 53.Percy LR, Potter IC. Morphological changes to the heart and associated blood vessels during lamprey metamorphosis. J Zool (Lond) 1988;214:417–432. [Google Scholar]
- 54.Gai Z, Donoghue PC, Zhu M, Janvier P, Stampanoni M. Fossil jawless fish from China foreshadows early jawed vertebrate anatomy. Nature. 2011;476(7360):324–327. doi: 10.1038/nature10276. [DOI] [PubMed] [Google Scholar]