Significance
Klebsiella pneumonia is an important cause of refractory nosocomial infections, the pathogenicity of which is largely a result of the bacteria’s ability to form biofilms on biomedical devices. A 3′,5′-cyclic diguanylic acid (c-di-GMP)–activated transcription activator, MrkH, drives biofilm formation. Here we describe structures of MrkH in its apo- and c-di-GMP–bound states. MrkH consists of two domains, both of which have PilZ-like folds. PilZ domains are known signaling modules, but, to our knowledge, MrkH is the first PilZ-containing protein to function in DNA binding. MrkH shows no homology to any human protein. Hence, our combined data, which uncovered the mechanism of c-di-GMP activation of MrkH, set the stage for the rational development of novel antimicrobial agents that target biofilm formation by K. pneumonia.
Keywords: MrkH, DNA binding motif, PilZ, Klebsiella pneumonia, biofilm
Abstract
The pathogenesis of Klebsiella pneumonia is linked to the bacteria’s ability to form biofilms. Mannose-resistant Klebsiella-like (Mrk) hemagglutinins are critical for K. pneumonia biofilm development, and the expression of the genes encoding these proteins is activated by a 3′,5′-cyclic diguanylic acid (c-di-GMP)–regulated transcription factor, MrkH. To gain insight into MrkH function, we performed structural and biochemical analyses. Data revealed MrkH to be a monomer with a two-domain architecture consisting of a PilZ C-domain connected to an N domain that unexpectedly also harbors a PilZ-like fold. Comparison of apo- and c-di-GMP–bound MrkH structures reveals a large 138° interdomain rotation that is induced by binding an intercalated c-di-GMP dimer. c-di-GMP interacts with PilZ C-domain motifs 1 and 2 (RxxxR and D/NxSxxG) and a newly described c-di-GMP–binding motif in the MrkH N domain. Strikingly, these c-di-GMP–binding motifs also stabilize an open state conformation in apo MrkH via contacts from the PilZ motif 1 to residues in the C-domain motif 2 and the c-di-GMP–binding N-domain motif. Use of the same regions in apo structure stabilization and c-di-GMP interaction allows distinction between the states. Indeed, domain reorientation by c-di-GMP complexation with MrkH, which leads to a highly compacted structure, suggests a mechanism by which the protein is activated to bind DNA. To our knowledge, MrkH represents the first instance of specific DNA binding mediated by PilZ domains. The MrkH structures also pave the way for the rational design of inhibitors that target K. pneumonia biofilm formation.
The opportunistic bacterial pathogen Klebsiella pneumonia is a significant cause of nosocomially acquired infections, particularly among immunocompromised patients. The pathogenicity of K. pneumonia is associated with its ability to form biofilms, which are largely recalcitrant to treatment (1–14). Key to K. pneumonia biofilm development are mannose-resistant Klebsiella-like (Mrk) hemagglutinins or Mrk proteins, which are encoded by the mrkABCDF operon (7–14). These proteins form type 3 fimbriae, which are extended filamentous structures. The MrkA protein is the pilin fibrial subunit, MrkB and MrkC function as the periplasmic chaperone and usher translocase for MrkA, and MrkD forms the tip adhesion subunit (10–13). Studies showed that type 3 fimbriae can adhere to a number of inert surfaces that are present on biomedical devices such as polystyrene, polypropylene, glass, and stainless steel (6–14). The expression of type 3 fimbriae components was recently shown to be activated by increasing intracellular concentrations of the second messenger, 3′,5′-cyclic diguanylic acid (c-di-GMP) (7). c-di-GMP sets off this cascade by interacting with the 26-kDa MrkH protein, which functions as a c-di-GMP–regulated activator of the type 3 fimbriae genes (7).
MrkH was originally identified in a screen looking for genes essential for K. pneumonia type 3 fimbriae and thus biofilm formation (7). KO of the mrkH gene essentially abrogated biofilm formation by K. pneumonia on a variety of surfaces (7). Isolation and cloning of mrkH revealed that it encoded a protein with a putative c-di-GMP–binding PilZ domain within its C-terminal region, and subsequent studies demonstrated that it bound c-di-GMP (5). c-di-GMP was first discovered as an allosteric effector of cellulose synthase in Gluconacetoobacter xylinus and has since been shown to be one of the most important and widespread second messengers in bacteria (15–17). Studies have established that c-di-GMP synthesis is carried out by GGDEF-containing diguanylate cyclases, whereas the degradation of the second messenger is mediated by EAL and HD-GYP motif-containing phosphodiesterases (17). In addition to biofilm formation, processes regulated by c-di-GMP include motility, virulence, and the cell cycle (15–17). Moreover, recent work has revealed that c-di-GMP serves as the switch that controls Streptomyces development and secondary metabolite production (18). Our understanding of the proteins and networks that drive c-di-GMP–dependent processes are, however, still in its infancy, largely because of the difficulty in identifying c-di-GMP–binding motifs within effector proteins. Currently, the best-characterized c-di-GMP–binding motifs include GGDEF, EAL, HD-GYP motifs, and PilZ motifs (15–17). PilZ motifs were first revealed by bioinformatics approaches and are ubiquitous in bacteria. The biological outputs of most PilZ proteins are currently unknown, but these proteins have been implicated in a wide range of signaling processes (19). The PilZ motif was named after the single-domain c-di-GMP–binding pilus protein in Pseudomonas aeruginosa and is also present in the first identified c-di-GMP–binding protein, cellulose synthase (20, 21). Although PilZ proteins share little to no overall sequence homology, they are recognizable by the presence of two key motifs that mediate c-di-GMP binding, an arginine-rich motif 1, also called the c-di-GMP switch, located near the N-terminal region of the domain with the consensus RxxxR, and a centrally located motif, motif 2, with the consensus D/NxSxxG (19–21). To date, structures of PilZ proteins solved in complex with c-di-GMP include the single PilZ domains of Alg44, CeSA, and PA4608 and the PilZ proteins PA0042 and PP4397, which contain a second domain (22–26). Of these proteins, only Alg44 and CeSA have clear functions, which are the regulation of alginate and cellulose synthesis, respectively (24, 26).
Although a diverse set of DNA-binding proteins have been experimentally identified as c-di-GMP–binding proteins, MrkH is thus far the only DNA-binding protein predicted to harbor a PilZ domain (18, 27–31). Data indicate that c-di-GMP binding to MrkH stimulates its ability to bind a consensus DNA sequence called the MrkH box, TATCAA, located upstream of the −35 site in the mrkABCDF promoter (6, 8). Binding to this site by MrkH activates the transcription of the mrkABCDF genes. MrkH was also shown to activate transcription from the mrkHI promoter, thereby revealing a positive autoregulatory loop in K. pneumonia biofilm development (6). MrkH functions as an activator by recruiting RNA polymerase to nonoptimal promoters via an interaction with the α subunit of RNA polymerase (6, 8). MrkH is predicted to contain two domains, an N-terminal domain of 106 residues, which shows no homology to any known protein, and a 115-residue C-terminal PilZ domain. Secondary structure predictions indicating the presence of five β-strands and two α-helices in the N domain lead to the suggestion that it harbors a LytTR DNA-binding domain (7). However, there are currently no structures available for MrkH, and hence its fold and the molecular mechanism by which it is regulated by c-di-GMP remains unknown. To gain insight into these questions, we undertook a structural and functional dissection of the MrkH protein. Here we show that MrkH is a monomeric protein with a two-domain architecture composed of a C-terminal PilZ domain and an N-terminal domain that also contains a PilZ-like fold. The MrkH–c-di-GMP structure reveals a c-di-GMP–binding motif present in the N domain that collaborates with C-domain PilZ motifs 1 and 2 to mediate binding of an intercalated c-di-GMP dimer. In the apo state, the c-di-GMP binding motifs make contacts with each other, leading to the stabilization of a distinct elongated conformation. As MrkH is key for biofilm formation, these structures set the stage for the development of unique inhibitors that specifically target this process in K. pneumonia.
Results and Discussion
Crystal Structure of K. pneumonia MrkH–c-di-GMP Complex.
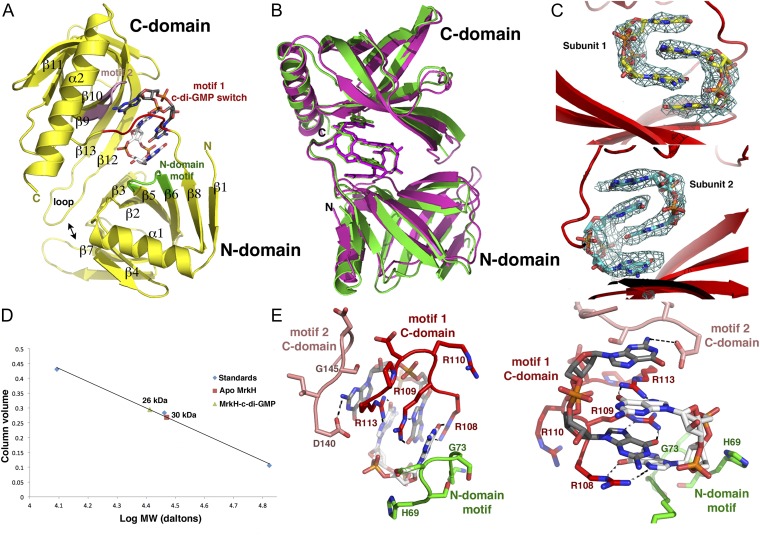
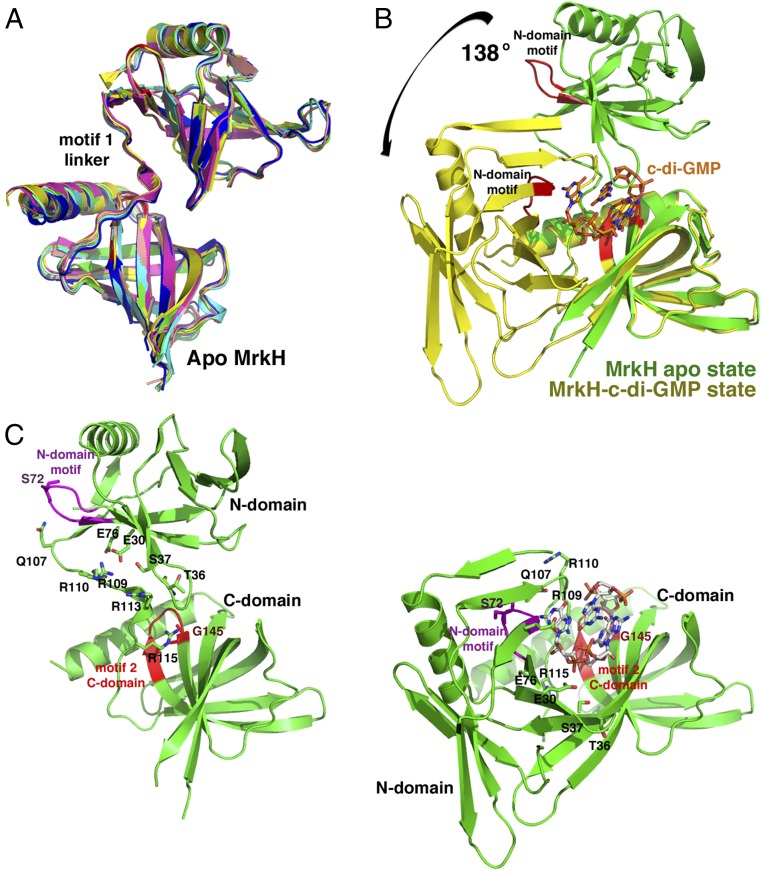
To gain insight into the molecular mechanism by which MrkH is regulated by c-di-GMP, we first determined the structure of the MrkH–c-di-GMP complex to 2.90-Å resolution. The structure was solved by mercury single-wavelength anomalous diffraction (SAD), and contained two MrkH molecules in the crystallographic asymmetric unit (ASU; Table S1 and SI Materials and Methods). Clear electron density was observed in the experimental SAD map for c-di-GMP molecules, which bind each MrkH subunit as intercalated dimers (Fig. 1 A–C). Following model construction, the structure was refined to a final Rwork/Rfree of 22.1%/28.0% (Table S1). The structure reveals that MrkH consists of two domains, an N-terminal domain (i.e., N domain) from residues 1 to 106 linked to a C-terminal domain (i.e., C domain) from residues 112 to 234 (Fig. 1A). The domains of the two MrkH subunits in the ASU can be superimposed with rmsd values of 0.6–0.7 Å, and overlays of the complete subunits results in an rmsd of 1.8 Å (Fig. 1B). The higher rmsd obtained for the latter superimposition reflects a slight shift in the domains of the two subunits relative to each other. Nonetheless, the two MrkH subunits in the structure use the same mode of binding to c-di-GMP (Fig. 1B).
Table S1.
Data collection and refinement statistics for MrkH and MrkH–c-di-GMP
| MrkH structure | MrkH–c-di-GMP | apo MrkH (1) | apo MrkH (2) |
| PDB ID code | 5KGO | 5KEC | 5KED |
| Space group | P212121 | C2221 | P21 |
| Cell dimensions | |||
| a, b, c, Å | 65.6,74.6,135.5 | 44.8,93.4,206.4 | 53.5,207.8,53.9 |
| α, β, γ, ° | 90.0,90.0, 90.0 | 90.0,90.0,90.0 | 90.0,120.2,90.0 |
| Resolution, Å | 74.6–2.90 | 45.6–1.95 | 103.9–2.65 |
| Rsym | 0.127 (1.03) | 0.073 (0.511) | 0.050 (0.349) |
| I/σI | 5.2 (1.2) | 7.2 (1.5) | 12.8 (2.6) |
| Completeness, % | 94.1 (96.5) | 91.2 (79.6) | 92.3 (68.5) |
| Redundancy | 3.2 (3.2) | 2.7 (2.1) | 3.5 (3.1) |
| Refinement | |||
| Resolution, Å | 74.6–2.90 | 45.6–1.95 | 103.9–2.65 |
| Rwork/Rfree, % | 22.1/28.0 | 20.2/23.6 | 23.0/27.6 |
| rmsds | |||
| Bond lengths, Å | 0.006 | 0.004 | 0.003 |
| Bond angles, ° | 1.224 | 0.790 | 0.626 |
Values in parentheses are for highest-resolution shell.
Fig. 1.
Structure of the MrkH–c-di-GMP complex. (A) Ribbon diagram of MrkH–c-di-GMP structure showing secondary structural elements and c-di-GMP–binding motifs. The C-domain PilZ motif 1, C-domain PilZ motif 2, and the N-domain c-di-GMP motif are colored red, pink, and green, respectively. (B) Overlay of the two independent MrkH–c-di-GMP complexes in the crystal. (C) Fo-Fc omit maps (contoured at 2.7 σ) calculated after removal of the c-di-GMP molecules and subjecting the structure to multiple cycles of refinement. Clear density (blue mesh) is observed for the c-di-GMP intercalated dimers bound to each MrkH monomer. (D) SEC analyses of apo- and c-di-GMP–bound MrkH indicating that both are monomers. (E) Two close-up views of the MrkH–c-di-GMP interactions with the residues (residue numbering according to ref. 7) from C-domain PilZ motifs 1 and 2 and the N-domain motif (colored as in Fig. 1A).
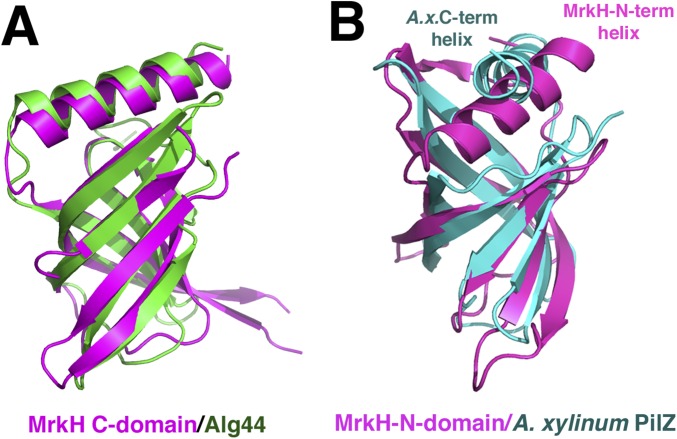
As predicted, the MrkH C-domain possesses a PilZ fold, which is characterized by the presence of two antiparallel β-sheets (Fig. 1A). The MrkH C domain also contains an elongated helix, α2, that is attached C-terminal to its β-barrel. DALI (distance matrix alignment) searches revealed that the C domain shows the strongest structural similarity to the Alg44 PilZ domain, with which it can be superimposed with an rmsd of 1.9 Å for 103 corresponding Cα atoms (Fig. S1A) (26). The MrkH N domain was predicted to harbor a DNA-binding, LytTR-like fold (7). However, the structure shows that, instead, it consists of a β-barrel arrangement similar to PilZ domains. This was confirmed by DALI searches, which revealed strong structural homology to PilZ domains, in particular the Acetobacter xylinum cellulose synthase, with which 66 of its residues superimpose with an rmsd of 2.5 Å (Fig. S1B). In addition to the PilZ-like β-barrel, the MrkH N domain contains an extra N-terminal β-strand–α-helix motif as well as a β-strand at its C terminus (Fig. 1A). Interestingly, although the only protein showing significant sequence homology to MrkH is its homolog from Citrobacter koseri (33% identity to the Klebsiella protein), there is a large group of so-called YcgR-like proteins that are predicted to contain two PilZ domains. However, the functions of most of these proteins are currently unknown (7, 22).
Fig. S1.
MrkH is a two-domain protein with N and C domains containing PilZ-like folds. (A) The MrkH C domain (magenta) shows strong structural homology to the Alg44 PilZ domain. (B) DALI search reveals that MrkH N domain (magenta) has the strongest similarity to the Acetobacter xylinum PilZ domain (cyan). Note that the MrkH N domain has an N-terminal helix rather than the C-terminal helix found in typical PilZ domains.
Most bacterial transcription factors are dimeric, and it was initially proposed that MrkH must function as a dimer (6). However, to our knowledge, no studies have been carried out to assess the oligomeric state of MrkH. Examination of the crystal packing and PISA (proteins, interfaces, structures and assemblies) analyses of the MrkH–c-di-GMP structure failed to reveal a potential oligomerization interface (32). Thus, these findings suggested that MrkH, at least in its c-di-GMP–bound form, is monomeric. To assess the MrkH oligomeric state in solution, we therefore carried out size exclusion chromatography (SEC) analyses of the apo- and c-di-GMP–bound states. The molecular weights (MWs) of monomeric and dimer MrkH would be 26 kDa and 52 kDa, respectively. The SEC analyses of apo MrkH resulted in a calculated MW of 30 kDa, whereas the MrkH–c-di-GMP complex produced an MW of 26 kDa. Thus, these data indicate that both states are monomeric, with the apo form perhaps adopting a more extended or irregular shape compared with the c-di-GMP–bound state (Fig. 1D).
MrkH–c-di-GMP Interactions.
Similar to other PilZ proteins, the MrkH PilZ C domain has two c-di-GMP–binding motifs, an arginine-rich motif, called motif 1, located at the N terminus of the domain, and a centrally localized motif, called motif 2 (Fig. 1 A and E). PilZ arginine-rich motifs have the consensus RxxxR, and, in MrkH, this motif is encompassed within residues 109–113 and has the sequence RRDPR. The second PilZ c-di-GMP–binding motif has the consensus D/NxSxxG, and, in MrkH, corresponds to residues 140–145, with the sequence DISDGG. Previous structures of proteins that contain a C-terminal PilZ domain attached to an N-terminal domain in complex with c-di-GMP revealed that only the C-terminal PilZ domain made contact to the cyclic nucleotide; the VCA0042 structure revealed one contact (4.0 Å) from an N-terminal isoleucine to the c-di-GMP (22, 23). In contrast, the MrkH N domain forms part of the c-di-GMP interaction network, making a number of contacts to one face of the bound c-di-GMP intercalated dimer. These contacts are provided by a motif from the N domain composed of residues 69–74, HSDSGK, which is somewhat similar to a PilZ motif 2 in sequence and in the fact that it harbors a β–turn–β structure (Fig. 1 A and E).
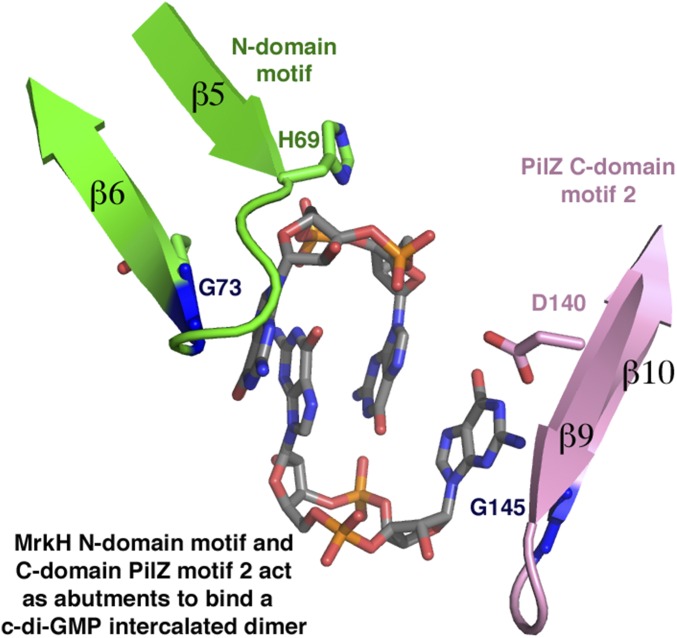
The unique MrkH N-terminal c-di-GMP–binding motif and motif 2 from its C domain act as abutments at each end of the intercalated c-di-GMP dimer, whereas motif 1 encircles the center of the c-di-GMP molecule (Fig. 1 A and E and Fig. S2). Residues from motif 1 contribute most of the contacts to the guanine nucleobases. Specifically, the side chains of Arg108, Arg109, and Arg113 “read” three of the four guanines via contacts to the O6 and N7 atoms of the nucleobases (Fig. 1E). These arginines also provide cation-π stacking interactions with the bases (Fig. 1E). The fourth guanine is recognized by contacts from the side chain of motif 2 residue Asp140 and the carbonyl oxygen of Gly145. The close approach of the guanine to residue 145 in the C-domain motif 2 indicates that this residue must be a glycine. Similarly, Gly73 of the MrkH N-domain motif packs tightly against the guanine at the opposite end of the c-di-GMP dimer (Fig. 1E). In addition to base interactions, residues His69 and Lys74 from the N-domain c-di-GMP–binding motif and C-domain residues Asn185, Gln203, Ser205, and Gln207 make contacts to the ribose and phosphate moieties of the intercalated c-di-GMP dimer.
Fig. S2.
The MrkHPilZ C-domain motif 2 and N-domain motif act as abutments for specifically binding a c-di-GMP intercalated dimer. Key glycines (blue) in each motif, G145 from motif 2 and G73 from the N-domain motif, allow tight packing of the outward-facing guanine bases of the intercalated c-di-GMP dimer. Motif 2 is colored pink and the N-domain motif is colored green.
Previous structural and functional analyses on PilZ domains showed that the identity of the residue N-terminal to motif 1, called the X position (XRxxxR), helps determine whether the given protein binds a monomer or dimer of c-di-GMP (22–26). Proteins with an arginine or lysine in the X position bound an intercalated dimer, whereas those with a leucine at this location, such as VC0042, bound a c-di-GMP monomer (22). Although there is an arginine in the X position of MrkH that contributes to its interactions with a c-di-GMP dimer, a key determinant in specifying the c-di-GMP dimer in MrkH appears to be the coordinated binding by the PilZ motifs and the N-domain motif (Fig. 1A). Indeed, binding of a c-di-GMP dimer by MrkH allows optimal contacts from its C-domain PilZ motifs 1 and 2 as well as the N-domain c-di-GMP binding motif. This complexation also leads to the formation of a highly compact MrkH conformation in which its N domain and C domain are closely apposed, allowing interdomain interactions. In particular, the extended C-domain β12–loop–β13 region of MrkH, which is not present in other PilZ domains solved thus far, docks over the β2–β3 and β7 region of the N domain allowing hydrophobic contacts between residues in these regions and hydrogen bonds from C-domain residues Asp192 and Ser198 to N-domain residue His55. Further, residues in the motif 1 linker, Arg110 and Arg115, make salt bridges to N-domain residues Glu5 and Glu28, respectively. These contacts are clearly observed in one MrkH–c-di-GMP complex, whereas, in the other subunit, the electron density for the loop between β12 and β13 is less well defined.
Analyses of Cyclic Nucleotide Binding by MrkH: Testing Structure-Based Predictions.
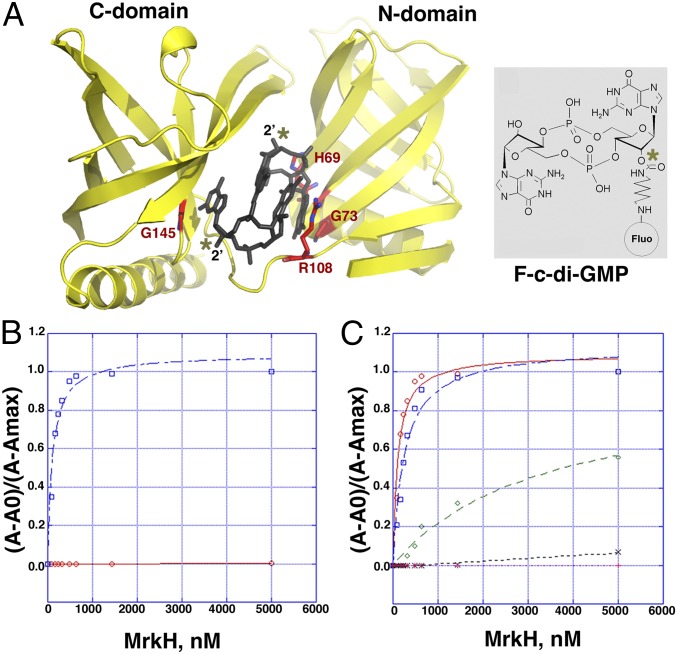
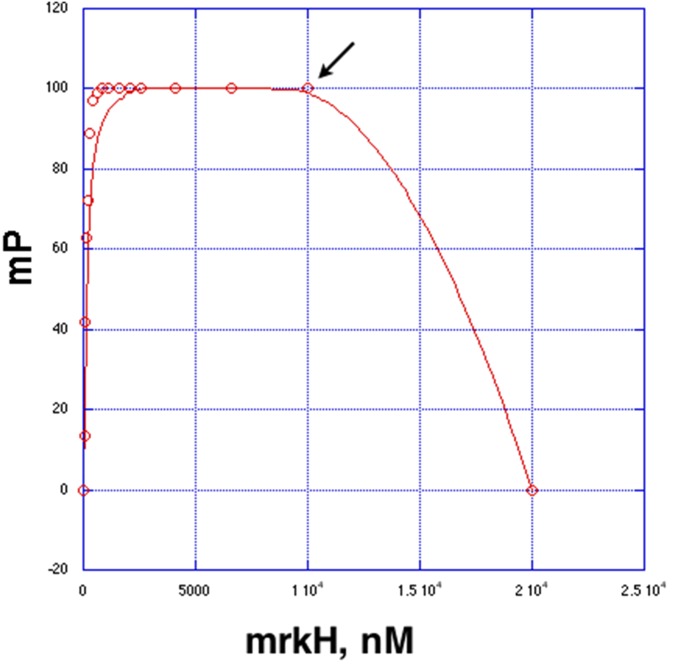
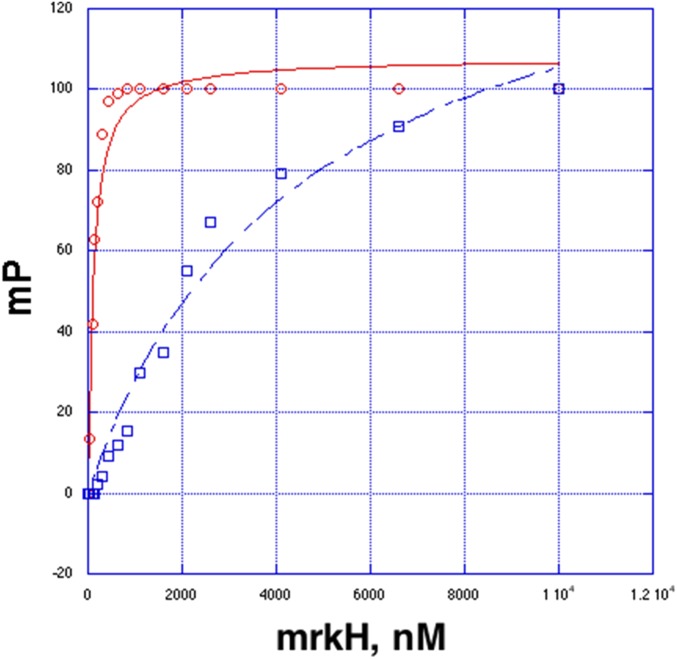
Previous filter-binding studies demonstrated that MrkH bound c-di-GMP but did not provide a quantification of this interaction (5). We sought to obtain a quantitative method that could also be used to assess binding by mutant proteins. Our structure shows that the 2′ hydroxyl groups on each of the c-di-GMP molecules in the intercalated dimer bound to MrkH are not involved in interactions with MrkH and point into the solvent (Fig. 2A). This structural feature allowed us to use the fluoresceinated c-di-GMP analog 2′-O-[6-(fluoresceinyl)aminohexylcarbamoyl]-cyclic diguanosine monophosphate (2′-Fluo-AHC-c-di-GMP; henceforth referred to as F-c-di-GMP), which contains a fluorescein moiety attached to one of its 2′ ribose hydroxyls in fluorescence polarization (FP)-based binding assays. By using this method, we first showed that WT MrkH bound F-c-di-GMP saturably, with a Kd of 107 ± 13 nM (Fig. 2B). Notably, the F-c-di-GMP could be competed specifically with nonfluoresceinated c-di-GMP (Fig. S3). Finally, a stoichiometry experiment was performed with WT MrkH and revealed a binding ratio of 2:1.2 c-d-GMP:MrkH subunit, consistent with the structural data (Fig. S4). As expected from the structure, assays that used the similar fluoresceinated c-di-AMP analog (2′-fluo-AHC-c-di-GMP) revealed no binding by MrkH (Fig. 2B).
Fig. 2.
FP analyses of c-di-GMP and c-di-AMP binding to WT MrkH and c-di-GMP binding to mutant MrkH proteins. (A) Ribbon diagram of MrkH with bound c-di-GMP (dark gray sticks) showing the residues mutated to assess the affects on c-di-GMP binding as red sticks. The location of the 2´ hydroxyls in each of the c-di-GMP molecules (asterisks) reveals that they make no interactions with the protein and face the solvent, thus representing ideal locations for attachment of the fluoresceinated label. (Right) Chemical structure of the F-c-di-GMP, which was used to obtain binding affinities of WT and mutant MrkH proteins. The asterisk shows the 2´ location of the attached label. (B) Isotherms of WT MrkH binding to F-c-di-GMP (blue) and 2′-O-(6-[fluoresceinyl]aminohexylcarbamoyl)-cAMP (F-c-di-AMP; red). (C) Binding by MrkH mutants to c-di-GMP. Binding isotherms for WT MrkH, MrkH(H69A), MrkH(G145E), MrkH(R108A), and MrkH(G73E) are colored red, blue, black, pink, and green, respectively.
Fig. S3.
Fluorescence polarization competition experiment. WT MrkH was added to the reaction tube containing 1 nM F-c-di-GMP, resulting in a binding curve (MrkH was added to a final concentration of 10 μM). The Kd for binding was determined as 113 ± 19 nM. Then, 10 μM nonfluoresceinated c-di-GMP was added (arrow), which resulted in apparent competition with the fluoresceinated c-di-GMP as revealed by the decrease in millipolarization units to 0.
Fig. S4.
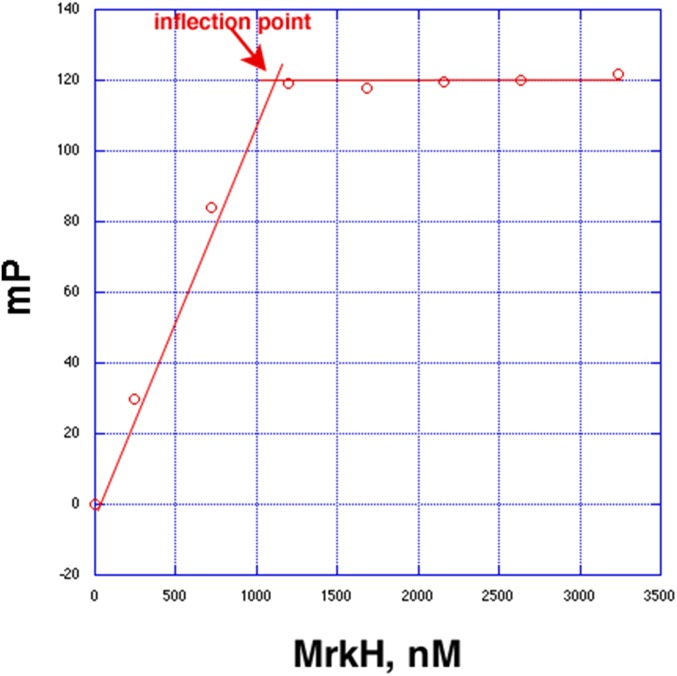
Determination of the stoichiometry of MrkH binding to c-di-GMP. The inflection point occurs at an MrkH monomer concentration of ∼1.2 μM (red arrow), indicating the shift from high-affinity binding to no binding.
To test the structure-based hypothesis that MrkH employs residues from its C-domain PilZ motifs 1 and 2 and its unique N-domain c-di-GMP binding motif for c-di-GMP complexation, we mutated residues in these regions and tested the effects of these mutations on c-di-GMP binding. Specifically, MrkH(R108A), MrkH(G145E), MrkH(G73E), and MrkH(H67A) were generated, and FP binding assays were performed (Fig. 2 A and C). MrkH(R108A) and MrkH(G145E), which contained mutations in key C-domain c-di-GMP–binding residues, R108 from motif 1 and G145 from motif 2, were completely defective in binding (Fig. 2C). Substitution of His67 from the N-domain motif to an alanine resulted in a twofold reduction in binding, whereas MrkH(G73E), which contains a mutation in the central glycine in the N-domain motif, was highly defective in binding, revealing a Kd of ∼5 μM (Fig. 2C and Fig. S5). These combined data, which are consistent with the structure, demonstrate that MrkH is exquisitely selective for binding cyclic guanine nucleotides compared with cyclic adenine nucleotides. The analyses also show that the residues that contact the guanine bases from C-domain motifs 1 and 2 are essential for c-di-GMP binding and likely serve as the initial c-di-GMP docking site but that the N-domain c-di-GMP binding motif is also required for high-affinity binding.
Fig. S5.
Fluorescence polarization analyses of MrkH(G73E) and WT proteins. These experiments were carried out to high MrkH concentrations to obtain near-saturable binding of the MrkH(G73E) mutant. The x and y axes are MrkH concentration (in nanomoles) and millipolarization units, respectively. Calculated Kd values are 113 ± 19 nM and 4,500 ± 900 nM for the WT and MrkH(G73E) mutant, respectively.
Structures of apo MrkH Reveal Specific State Stabilized by c-di-GMP–Binding Motifs; c-di-GMP Binding as Conformational Toggle.
Motif 1 of PilZ proteins has been called the c-di-GMP switch because it is typically flexible and disordered in the absence of c-di-GMP and folds upon binding the second messenger. The resultant structural changes are posited to be key to the functions of the proteins (22). In the case of MrkH, binding c-di-GMP has been shown to activate DNA binding by the protein. To glean insight into this stimulatory mechanism, we determined structures of apo MrkH. Two crystal forms of apo MrkH were obtained under different conditions, and their structures solved to resolutions of 2.65 Å and 1.95 Å (SI Materials and Methods). One crystal form contained four MrkH subunits in the ASU and the other contained two MrkH subunits. Thus, the combined structures provided six crystallographically independent views of the MrkH apo conformation. Detailed crystal packing analyses of the apo forms revealed a possible dimer; however, no similar dimer was found in the c-di-GMP bound structure and, as noted, SEC indicated that apo MrkH is monomeric (Fig. 1D). Overlays of the six apo MrkH structures reveal that they all adopt the same elongated conformation (Fig. 3A). This state is noticeably distinct from the compact c-di-GMP–bound form of MrkH. Indeed, even though the individual MrkH domains are essentially unchanged between the apo and c-di-GMP–bound states (rmsds of ∼0.6 for the C-domain and N-domain overlays), analyses reveal that there is a large 138° rotation and as much as 60 Å translation between domains in going from one state to the other (Fig. 3 B and C).
Fig. 3.
Structures of apo MrkH reveal the use of c-di-GMP–binding residues in stabilization of its extended state. (A) Superimpositions of the six apo subunits captured in two different apo MrkH crystal forms. The figure highlights the fact that apo MrkH adopts an elongated conformation (note the well-ordered linker between domains). (B) Overlay of the C-domains of the apo (green) and c-di-GMP–bound MrkH (yellow) showing the dramatic conformational change that is induced upon c-di-GMP binding. (C) Side-by-side comparison of apo- and c-di-GMP–bound MrkH. The apo form is largely stabilized by contacts between motif 1 linker residues and C-domain motif 2 (red) and N-domain motif (magenta) residues, which are the same regions and residues that mediate c-di-GMP binding. The figure shows the apo- and c-di-GMP–bound state with the C domains in the identical orientations to underscore the dramatic movement of the domains from one state to the next.
Strikingly, analysis of the apo MrkH structure reveals that it is stabilized in its elongated conformation by contacts from residues located in motif 1 of the c-di-GMP binding linker to residues in C-domain PilZ motif 2 and the N-domain c-di-GMP–binding motif (Fig. 3C). As a result of these interactions, the PilZ motif 1 linker in apo MrkH adopts a well-ordered structure that includes a small central helix (Fig. 3A). Specific contacts include hydrophobic packing and hydrogen bonds from motif 1 linker residue Arg115 to motif 2 residue Gly145. The small size of Gly145 is crucial in enabling this interaction, just as it is to allow binding of c-di-GMP (Fig. 3C). Motif 1 residues Gln107, Arg109, and Arg110 hydrogen bond to N-domain c-di-GMP–binding motif residues Ser72 (its carbonyl oxygen), Glu30, and Glu76, respectively (Fig. 3C). Arg113, which, in the c-di-GMP–bound MrkH conformation, specifically contacts a guanine base as well as forming stacking interactions with guanine bases, makes hydrogen bonds to N-domain residue Ser37 and the carbonyl oxygen of Thr36 in the apo MrkH structure. The fact that the apo MrkH structure employs some of the same sets of residues to stabilize an open conformation as it does to interact with c-di-GMP provides a mechanism by which c-di-GMP binding can act as conformational toggle between two stable states as binding of the c-di-GMP second messenger stabilizes the closed state. As c-di-GMP binding has been shown to be required for high-affinity DNA binding by MrkH, these structural findings suggest these conformational differences as the key to this function.
MrkH: A Transcription Regulator with PilZ Domains as DNA Binding Motifs.
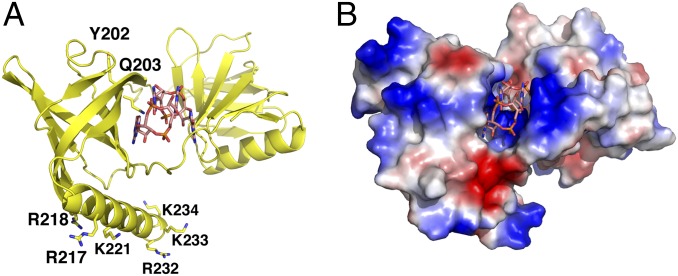
Previous studies showed that complexation of MrkH with c-di-GMP is required for high affinity DNA binding by the protein (6, 8). The structural data on apo and c-di-GMP–bound MrkH suggests a mechanism by which c-di-GMP activates DNA binding, which is through stabilization of a compact form very distinct from the extended apo state. To date, the known DNA binding motifs that have been identified and well characterized from bacterial proteins include the helix–turn–helix (HTH), winged helix–turn–helix, ribbon–helix–helix, and LytTR motifs. Initial analyses of the MrkH sequence lead to the suggestion that it contained a LytTR motif within its N domain, which was the domain hypothesized to mediate DNA binding (7). However, the MrkH structures reveal that its N domain harbors a PilZ-like fold. To our knowledge, PilZ domains themselves have never been implicated in DNA binding, and therefore the finding that the MrkH structure contains only PilZ or PilZ-like domains reveals the first such case. Moreover, although very few biochemical analyses have been carried out on MrkH, a recent study in which serine/alanine insertions were generated in various regions of the protein implicated the MrkH C domain in binding. Specifically, serine/alanine insertions between residues DNA Tyr202–Gln203 and Arg217–Arg218 abrogated DNA binding (8). The MrkH–c-di-GMP structure shows that Gln203 makes contacts to c-di-GMP, and hence mutagenesis at this site would be predicted to impact the interaction of MrkH with c-di-GMP, thus indirectly affecting DNA binding. Insertion mutagenesis at this position, which is part of the protein core, might also impair folding. Residues Arg217 and Arg218, by contrast, are located on the surface of the C-terminal helix of the PilZ C domain. This helix is notably basic, containing six lysines and arginines on its exposed face (Fig. 4A). Electrostatic surface representation of the protein shows that this helix is adjacent to other basic regions of the protein that may be involved in DNA binding (Fig. 4B). However, although our data indicate that the apo- and c-di-GMP–bound forms of MrkH are monomeric, gel shifts carried out with MrkH-box–containing DNA fragments that were several hundred base pairs in length showed an initial shift followed by a large super shift that suggests the possibility that multiple MrkH proteins may load onto the DNA and be required for high-affinity binding (6, 8). Clearly, more structural and functional data will be required to deduce the specific mechanism of this interaction.
Fig. 4.
Mapping DNA binding residues on the MrkH–c-di-GMP structure. (A) Ribbon diagram of MrkH–c-di-GMP showing the location of residues or regions implicated in DNA binding, which are Tyr202, Gln203, R217, and R218. Also shown are additional basic residues that cluster on the MrkH C-terminal helix (8). (B) Electrostatic surface representation of the MrkH–c-di-GMP complex shown in the same orientation as in A.
In summary, we showed here that the key regulator of biofilm formation in K. pneumonia, MrkH, binds c-di-GMP by using a binding site comprised of three motifs. Of particular note, the MrkH C-domain PilZ motif 2 and the newly described c-di-GMP–binding motif located in its N domain serve as abutments that specify binding of an intercalated c-di-GMP dimer (Fig. 1A). As a result of these interactions, MrkH adopts a highly compact structure. The motif 1 linker region of most PilZ domains is disordered in the absence of the second messenger. By sharp contrast, in apo MrkH, this region is well ordered large part because of its contacts to residues within motif 2 and the N-domain motif. Hence, MrkH employs the same motifs to not only bind c-di-GMP but stabilize a specific apo state. This allows c-di-GMP binding to facilitate toggling between the elongated apo and compact c-di-GMP states. Interestingly, as noted, the functions of most PilZ proteins are currently unknown. The finding that PilZ domains can be used in DNA binding leads to the intriguing possibility that some of these orphan proteins may be DNA-binding proteins. Future studies will be needed to address this question and also determine the precise molecular mechanism used by the MrkH PilZ domains in DNA binding.
SI Materials and Methods
Expression and Purification of Klebsiella pneumonia MrkH.
An artificial gene encoding K. pneumonia mrkH, codon-optimized for expression in Escherichia coli was subcloned into the pET15b vector such that it expressed an N-terminal his-tag for purification (Genscript). The resultant vector was transformed into E. coli C41(DE3) cells. For protein expression, the mrkH expressing cells were grown to an OD600 of 0.6–0.8 and induced with 1 mM isopropyl β-d-1-thio-galactopyranoside for 3 h at 37 °C. Cells were lysed in buffer A [25 mM Tris, pH 7.5, 300 mM NaCl, 5% (wt/vol) glycerol, 1 mM β-mercaptoethanol] using a microfluidizer, and cell debris was removed by centrifugation at 35,000 × g. The MrkH-containing lysate was loaded onto a Ni-NTA column, and the column was extensively washed with increasing concentrations of imidazole in buffer A. The protein was eluted with 0.1–1.0 M imidazole and was >95% pure at this step. The his-tag was cleaved by using a thrombin capture cleavage kit, and the tag was removed by buffer exchanging the protein into buffer A using a 10 kDa MW Centricon concentrator. The MrkH mutants [numbering according to Wilksch et al. (7)], MrkH(H69A), MrkH(G73E), MrkH(R108A), and MrkH(G154E), were expressed and purified as per the WT protein.
Crystallization and Structure Determination of MrkH–c-di-GMP.
The his-tag–free MrkH was used for crystallization trials with c-di-GMP. The protein was concentrated just before crystallization setups as it precipitated over time at concentrations of 5 mg/mL or higher. To obtain MrkH–c-di-GMP crystals suitable for data collection, the protein (in buffer A) was concentrated to 10 mg/mL and c-di-GMP was added to a final concentration of 1 mM, and this mixture was then combined 1:1 with 20% (wt/vol) PEG 3350, 0.1 M Bis-Tris propane/HCl, pH 6.5, 0.2 M sodium fluoride. Crystals were obtained at room temperature and took several days to multiple weeks to grow. Notably, small crystals could be obtained with protein concentrations in the 3–5-mg/mL range (and 1 mM c-di-GMP), but, to obtain crystals large enough for data collection, higher concentrations (10 mg/mL) were needed. These crystals were cryopreserved by first equilibrating the crystal drops against a reservoir with increasing concentrations of PEG 3350 to a final concentration of 45%. The equilibrated crystals were then looped and placed directly in the cryostream. Crystals take the space group P212121. A native X-ray intensity data set was collected to 2.90-Å resolution at the ALS beamline 8.3.1 and processed with MOSFLM. The structure could not be solved by molecular replacement by using the PilZ structures in the protein database as search models. Thus, a mercury derivative was generated, and SAD data were collected for phasing. The data were collected to 2.95 Å at ALS beamline 8.3.1 and processed with MOSFLM. Eight mercury sites were located in Phenix Autsol (33) and used to aid in tracing via the positions of the cysteines. There are two MrkH–c-di-GMP complexes per crystallographic ASU. The model generated from SAD phasing was refined to an Rfree of 32%, and the structure was then used in molecular replacement against the native data set. Multiple rounds of refitting and refinement resulted in Rwork/Rfree values of 22.1%/28.0%. The final model includes two MrkH subunits, with residues 5–192 and 198–234 of one subunit and 5–192 and 196–234 of the other subunit, and two intercalated c-di-GMP dimers. MolProbity revealed 89% of residues in the Ramachandran favored regions and 2% in outlier regions (34). The latter residues are in poorly ordered regions or loops. Final model refinement statistics are presented in Table S1.
Crystallization and Structure Determination of Two apo MrkH Crystal Forms.
To obtain apo MrkH crystals, the his-tag–free MrkH (in buffer A) was concentrated to 20 mg/mL before setups. Crystals were obtained at room temperature by mixing the protein solution at a 1:1 ratio with a crystallization reagent composed of 2 mM MgCl2, 2 mM CaCl2, 0.1 M sodium Hepes, pH 7.5, 20% (wt/vol) ethylene glycol, and 10% (wt/vol) PEG 8000. The crystals were cryopreserved by dipping them for 1 s in the crystallization solution supplemented with ethylene glycol to a final concentration of 30%. The crystals take the monoclinic space group P21. Crystals took one to several days to grow. Data were collected at ALS beamline 5.0.2 and processed with MOSFLM. The structure contains four MrkH molecules in the ASU. The c-di-GMP–bound conformation of the full-length MrkH was not successful in molecular replacement. Hence, the structure was solved by using individual MrkH domains in multiple steps. The domains were first pruned to remove the c-di-GMP linker region. Then, residues 5–101 of the N domain and residues 116–220 of the C domain were used in molecular replacement searches. In the first step of molecular replacement, the MrkH N domain was used as a search model in Phaser, which successfully placed four N domains. The resultant partial N-domain model was then input as a static structure and four C domains were located sequentially by using Molrep. After initial rigid body and xyz refinement, clear density was observed for the linker region in each of the four subunits. After multiple rounds of rebuilding and refinement in Phenix (33), the Rwork/Rfree ratio converged to 23.0%/27.6% (Table S1). The model includes residues 3–192 and 197–234 of two subunits and 5–192 and 197–234 of the other two subunits and 37 water molecules. MolProbity analyses revealed 91% of residues in favored regions of the Ramachandran plot with 0 outliers (34).
A second apo MrkH crystal form was obtained by concentrating the protein to 10 mg/mL and mixing it at a 1:1 ratio with a reagent consisting of 30% (wt/vol) PEG 600, 0.1 M Hepes, pH 7.5, 50 mM lithium sulfate, and 10% (wt/vol) glycerol. The crystals, which were obtained at room temperature, took 2 d to grow and were cryopreserved from the drop. The crystals diffracted to 5 Å, but the diffraction was significantly improved by a combination of dehydration and annealing, which ultimately allowed data to 1.95-Å resolution to be collected at 100 K at ALS beamline 8.3.1. The crystals take the orthorhombic space group C2221. There are two MrkH subunits in the ASU, and the structure was solved in Molrep by using one of the apo P21 subunits as a search model. The structure was refined in Phenix, and the final model contains two subunits with residues 5–86, 91–192, and 197–234 of one subunit and 3–192 and 197–234 of the other subunit and 151 water molecules. The final Rwork/Rfree values are 20.2%/23.6% (33). MolProbity analyses revealed 97% of residues in the favored region of the Ramachandran plot with 0 outliers (34). Final refinement statistics are provided in Table S1.
SEC.
SEC was used to probe the MWs of MrkH in solution by using a HiLoad 26/600 Superdex 75 prep-grade column. Experiments were performed in a buffer containing 200 mM NaCl, 5% (wt/vol) glycerol, 20 mM Tris⋅HCl, pH 7.5, and 1 mM β-mercaptoethanol. For SEC analysis of the MrkH–c-di-GMP complex, c-di-GMP was included in the buffer at a concentration of 10 μM.
Analysis of MrkH and MrkH Mutant Binding to F-c-di-GMP and F-c-di-AMP.
To measure c-di-GMP and c-di-AMP binding by MrkH, F-c-di-GMP or F-c-di-AMP were used as the fluoresceinated ligands. These molecules are conjugated via a 9-atom spacer to one of the 2′ hydroxyl groups of the c-di-GMP or c-di-AMP. Binding was carried out at 25 °C in a buffer of 150 mM NaCl and 25 mM Tris⋅HCl, pH 7.5, which contained 1 nM F-c-di-GMP or F-c-di-AMP. Increasing concentrations of MrkH or MrkH mutant were titrated into the reaction mixture to obtain the binding isotherms. The resulting data were plotted using KaleidaGraph, and curves were fit to deduce binding affinities.
To determine the binding stoichiometry of c-di-GMP to MrkH, an FP assay was used. Here, the binding buffer and conditions were the same as those used in the binding affinity determination experiments except that the concentration of the c-di-GMP was increased to 2 μM, which is 20-fold greater than the Kd (by using a solution containing 1 nM F-c-di-GMP and 1.999 μM c-di-GMP), thereby ensuring stoichiometric binding. MrkH was titrated into the binding solution, and the graph of the resulting data shows a linear increase in the observed millipolarization until saturation of the c-di-GMP by MrkH, after which the millipolarization values showed no increase. The inflection point occurs at an MrkH monomer concentration of ∼1.2 μM, which, when divided by the concentration of c-di-GMP (2 μM), indicates a stoichiometry of one MrkH subunit to two c-di-GMP molecules.
Materials and Methods
The gene encoding the K. pneumonia MrkH protein was purchased from Genscript and subcloned into pET15b such that a cleavable hexahistidine-tag (his-tag) was expressed for purification. The expressed protein was recovered from lysates and purified via Ni-NTA chromatography. Crystal structures were solved and refined by using Phenix and analyzed with MolProbity (33, 34). Structure factor amplitudes and coordinates have been deposited in the Protein Data Bank under the ID codes 5KEC and 5KED for the apo MrkH structures and 5KGO for the MrkH–c-di-GMP complex. Details on purification, crystallization, structure determination, and biochemical assay are provided in SI Materials and Methods.
Acknowledgments
The authors acknowledge Advanced Light Source (ALS) beamlines 8.3.1 and 5.0.2 for data collection with a special thanks to Jane Tanamachi, Stacey Ortega, and Corie Ralston. This work was supported by National Institutes of Health Grant GM115547 (to M.A.S.). The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231. Beamline 8.3.1 at the ALS is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives Grant MR-15-328599 and Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.K. is a Guest Editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5KEC, 5KGO, and 5KED).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607503113/-/DCSupplemental.
References
- 1.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandes A, Dias M. The microbiological profiles of infected prosthetic implants with an emphasis on the organisms which form biofilms. J Clin Diagn Res. 2013;7(2):219–223. doi: 10.7860/JCDR/2013/4533.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy CN, Mortensen MS, Krogfelt KA, Clegg S. Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect Immun. 2013;81(8):3009–3017. doi: 10.1128/IAI.00348-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahlhut SG, Struve C, Krogfelt KA, Reisner A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunol Med Microbiol. 2012;65(2):350–359. doi: 10.1111/j.1574-695X.2012.00965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson JG, Murphy CN, Sippy J, Johnson TJ, Clegg S. Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J Bacteriol. 2011;193(14):3453–3460. doi: 10.1128/JB.00286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan JW, et al. Positive autoregulation of mrkHI by the cyclic di-GMP-dependent MrkH protein in the biofilm regulatory circuit of Klebsiella pneumoniae. J Bacteriol. 2015;197(9):1659–1667. doi: 10.1128/JB.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilksch JJ, et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7(8):e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang J, et al. Transcriptional activation of the mrkA promoter of the Klebsiella pneumoniae type 3 fimbrial operon by the c-di-GMP-dependent MrkH protein. PLoS One. 2013;8(11):e79038. doi: 10.1371/journal.pone.0079038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu CC, et al. Fur-dependent MrkHI regulation of type 3 fimbriae in Klebsiella pneumoniae CG43. Microbiology. 2012;158(Pt 4):1045–1056. doi: 10.1099/mic.0.053801-0. [DOI] [PubMed] [Google Scholar]
- 10.Di Martino P, Cafferini N, Joly B, Darfeuille-Michaud A. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res Microbiol. 2003;154(1):9–16. doi: 10.1016/s0923-2508(02)00004-9. [DOI] [PubMed] [Google Scholar]
- 11.Sebghati TA, Korhonen TK, Hornick DB, Clegg S. Characterization of the type 3 fimbrial adhesins of Klebsiella strains. Infect Immun. 1998;66(6):2887–2894. doi: 10.1128/iai.66.6.2887-2894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tarkkanen AM, Virkola R, Clegg S, Korhonen TK. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect Immun. 1997;65(4):1546–1549. doi: 10.1128/iai.65.4.1546-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murphy CN, Clegg S. Klebsiella pneumoniae and type 3 fimbriae: Nosocomial infection, regulation and biofilm formation. Future Microbiol. 2012;7(8):991–1002. doi: 10.2217/fmb.12.74. [DOI] [PubMed] [Google Scholar]
- 14.Jagnow J, Clegg S. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology. 2003;149(pt 9):2397–2405. doi: 10.1099/mic.0.26434-0. [DOI] [PubMed] [Google Scholar]
- 15.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sondermann H, Shikuma NJ, Yildiz FH. You’ve come a long way: c-di-GMP signaling. Curr Opin Microbiol. 2012;15(2):140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 18.Tschowri N, et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014;158(5):1136–1147. doi: 10.1016/j.cell.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22(1):3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 20.Ryan RP, Tolker-Nielsen T, Dow JM. When the PilZ don’t work: Effectors for cyclic di-GMP action in bacteria. Trends Microbiol. 2012;20(5):235–242. doi: 10.1016/j.tim.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Alm RA, Bodero AJ, Free PD, Mattick JS. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J Bacteriol. 1996;178(1):46–53. doi: 10.1128/jb.178.1.46-53.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benach J, et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26(24):5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko J, et al. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J Mol Biol. 2010;398(1):97–110. doi: 10.1016/j.jmb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Fujiwara T, et al. The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A. Biochem Biophys Res Commun. 2013;431(4):802–807. doi: 10.1016/j.bbrc.2012.12.103. [DOI] [PubMed] [Google Scholar]
- 25.Habazettl J, Allan MG, Jenal U, Grzesiek S. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J Biol Chem. 2011;286(16):14304–14314. doi: 10.1074/jbc.M110.209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitney JC, et al. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem. 2015;290(20):12451–12462. doi: 10.1074/jbc.M115.645051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krasteva PV, et al. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science. 2010;327(5967):866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W, He ZG. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012;40(22):11292–11307. doi: 10.1093/nar/gks923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fazli M, et al. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol Microbiol. 2011;82(2):327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 30.Leduc JL, Roberts GP. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J Bacteriol. 2009;191(22):7121–7122. doi: 10.1128/JB.00845-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin KH, et al. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol. 2010;396(3):646–662. doi: 10.1016/j.jmb.2009.11.076. [DOI] [PubMed] [Google Scholar]
- 32.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]