Significance
Maternal inheritance of mitochondria and mitochondrial genes is a major developmental paradigm in mammals. Propagation of paternal, sperm-contributed mitochondrial genes, resulting in heteroplasmy, is seldom observed in mammals, due to postfertilization targeting and degradation of sperm mitochondria, referred to as “sperm mitophagy.” Our and others’ recent results suggest that postfertilization sperm mitophagy is mediated by the ubiquitin–proteasome system, the major protein-turnover pathway that degrades proteins and the autophagic pathway. Here we demonstrate that the co-inhibition of the ubiquitin-binding autophagy receptors, sequestosome 1 (SQSTM1) and gamma-aminobutyric acid receptor-associated protein (GABARAP), and the ubiquitinated protein dislocase valosin-containing protein (VCP)-dependent pathways delayed postfertilization sperm mitophagy. Our findings provide the mechanisms guiding sperm mitochondrion recognition and disposal during preimplantation embryo development, which prevents a potentially detrimental effect of heteroplasmy.
Keywords: mitochondria, mtDNA, ubiquitin, autophagy, mitophagy
Abstract
Maternal inheritance of mitochondria and mtDNA is a universal principle in human and animal development, guided by selective ubiquitin-dependent degradation of the sperm-borne mitochondria after fertilization. However, it is not clear how the 26S proteasome, the ubiquitin-dependent protease that is only capable of degrading one protein molecule at a time, can dispose of a whole sperm mitochondrial sheath. We hypothesized that the canonical ubiquitin-like autophagy receptors [sequestosome 1 (SQSTM1), microtubule-associated protein 1 light chain 3 (LC3), gamma-aminobutyric acid receptor-associated protein (GABARAP)] and the nontraditional mitophagy pathways involving ubiquitin-proteasome system and the ubiquitin-binding protein dislocase, valosin-containing protein (VCP), may act in concert during mammalian sperm mitophagy. We found that the SQSTM1, but not GABARAP or LC3, associated with sperm mitochondria after fertilization in pig and rhesus monkey zygotes. Three sperm mitochondrial proteins copurified with the recombinant, ubiquitin-associated domain of SQSTM1. The accumulation of GABARAP-containing protein aggregates was observed in the vicinity of sperm mitochondrial sheaths in the zygotes and increased in the embryos treated with proteasomal inhibitor MG132, in which intact sperm mitochondrial sheaths were observed. Pharmacological inhibition of VCP significantly delayed the process of sperm mitophagy and completely prevented it when combined with microinjection of autophagy-targeting antibodies specific to SQSTM1 and/or GABARAP. Sperm mitophagy in higher mammals thus relies on a combined action of SQSTM1-dependent autophagy and VCP-mediated dislocation and presentation of ubiquitinated sperm mitochondrial proteins to the 26S proteasome, explaining how the whole sperm mitochondria are degraded inside the fertilized mammalian oocytes by a protein recycling system involved in degradation of single protein molecules.
The inheritance pattern of the mitochondrial genome does not follow Mendelian rules as mtDNA is predominantly or exclusively inherited from the mother in almost all eukaryotic species studied, which is referred to as maternal inheritance of mtDNA (1, 2). The proteolytic ubiquitin–proteasome system (UPS) for substrate-specific, regulated protein recycling has been implicated in the targeted degradation of paternal mitochondria (sperm mitophagy) in mammals and other taxa. Mammalian sperm mitochondria are already modified with ubiquitin during spermatogenesis and ultimately processed by proteasome-mediated proteolysis (3). Specific cell-permeant inhibitors of proteasomal chymotrypsin-like activity, MG132 and lactacystin, blocked the progression of sperm mitophagy after porcine fertilization, and the resumption of sperm mitochondrion degradation was observed once the MG132 (a reversible inhibitor) was removed from the zygotes (4). Our early study reported that sperm tails in zygotes were surrounded by lysosome-like structures, which suggested that the end point mechanism of ubiquitinated sperm mitochondrion degradation might involve lysosomes (3). This early evidence showed the possibility for the involvement of the lysosomal pathway before it became clear at a molecular level that cellular mitophagy requires a compartmentalized organelle/protein aggregate degradation process termed autophagy, the endpoint of which is the fusion of an autophagosome vesicle that contains sequestered organelle remnants with a lysosome (5).
Three recent studies in Caenorhabditis elegans reported that the lysosome-dependent autophagic pathway is associated with the degradation of sperm mitochondria by the embryo (6–8). Sperm mitochondria in C. elegans are engulfed by autophagosomes and gradually degraded by the 16-cell stage of embryonic development. Similarly, a very recent study in Drosophila implicates the autophagic pathway in postfertilization degradation of the sperm mitochondrial derivative (9). In a mammalian model, the autophagy-related proteins microtubule-associated protein 1 light chain 3 (LC3), sequestosome 1 (SQSTM1), and gamma-aminobutyric acid receptor-associated protein (GABARAP) were detected in the mitochondrial region of mouse spermatozoa and found to dissociate from sperm mitochondria after fertilization, possibly supplanted by ubiquitin (6, 7). Such observations suggested that the mechanism involving both UPS and the autophagy cascade might regulate the elimination of sperm mitochondria in mammals. However, a recent study of mouse embryos once again blurred the role of autophagy in sperm mitochondrial degradation. Spermatozoa from a transgenic mouse bearing red fluorescent protein labeled mitochondria were used to fertilize oocytes expressing GFP-tagged autophagosome protein LC3. However, no association was found between GFP-autophagosomes and red fluorescent sperm mitochondria in the zygotes (10). Although no studies of species other than mouse were conducted, the authors concluded that sperm mitophagy was not involved in maternal inheritance of mitochondria in mammals. However, other mammalian models such as the aforementioned porcine zygote, or branches of the autophagic pathway other than the LC3-dependent one, were not taken into consideration.
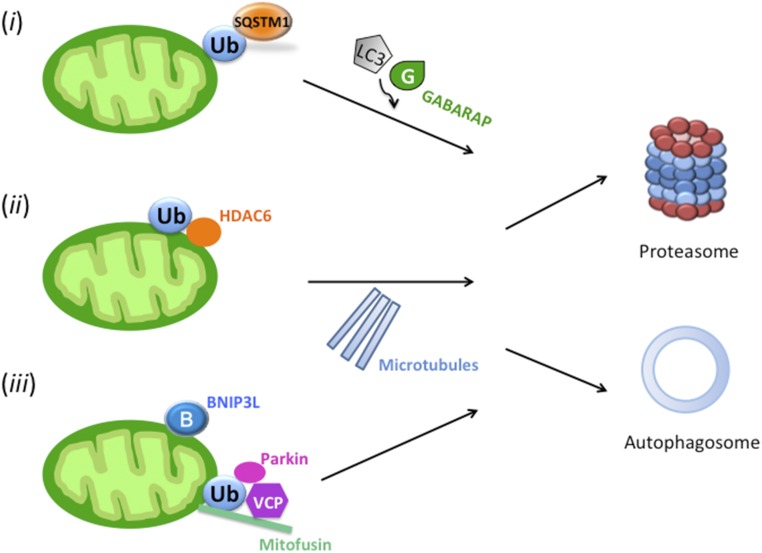
Here we consider that at least three well-characterized pathways involving both autophagy and UPS may act during sperm mitochondrial degradation in mammals (Fig. 1): (i) ubiquitin-binding autophagy receptor SQSTM1 could recognize ubiquitinated mitochondrial protein cargo and interact with autophagosome-binding ubiquitin-like modifiers, such as LC3 and/or GABARAP; (ii) ubiquitinated proteins could be extracted from mitochondria and form aggresomes, the protein aggregates induced by ubiquitin-binding adaptor protein HDAC6 that transports them along the microtubules to the autophagophore; and (iii) protein dislocase, the valosin-containing protein (VCP), could extract and present the ubiquitinated mitochondrial membrane proteins to the 26S proteasome. Data presented here indicate that instead of the canonical LC3-mediated pathway, sperm mitophagy in higher mammals relies on a combined action of SQSTM1-dependent autophagy and VCP-mediated presentation of ubiquitinated sperm mitochondrial proteins to the 26S proteasome.
Fig. 1.
Putative pathways participating in the elimination of sperm mitochondria by coordinated activities of autophagy and the ubiquitin–proteasome system. (i) The ubiquitin-binding autophagy receptor SQSTM1 could recognize ubiquitinated mitochondrial protein cargo and interact with autophagosome-binding ubiquitin-like modifiers, such as LC3 and/or GABARAP to transport them toward autophagosome. (ii) Ubiquitinated proteins could be extracted from mitochondria and form aggresomes, the protein aggregates induced by ubiquitin-binding adaptor protein HDAC6 that transports them along the microtubules to the autophagophore. (iii) Protein dislocase VCP could extract and present the ubiquitinated mitochondrial membrane proteins to the 26S proteasome.
Results
Ubiquitin-Binding Autophagy Receptor SQSTM1 Associates with Sperm Mitochondria After Fertilization.
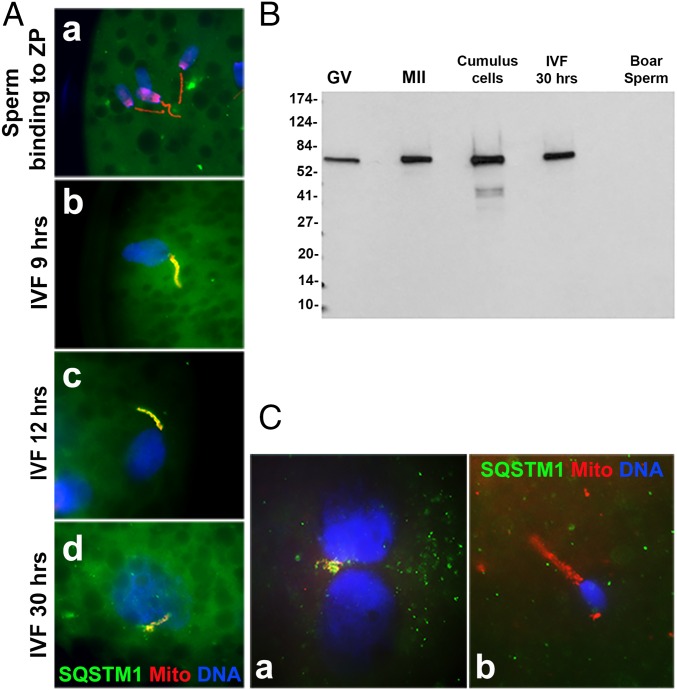
To detect the presence of SQSTM1 in porcine zygotes and its association with sperm mitochondria after fertilization, oocytes were inseminated with fresh boar spermatozoa prelabeled with red fluorescent MitoTracker, then cultured and sampled at 9, 12, and 30 h after fertilization. The zygotes were labeled with anti-SQSTM1 antibody and the DNA stain DAPI (Fig. 2 and SI Appendix, Fig. S1). The SQSTM1 was detected in the midpiece/mitochondrial sheath of sperm tail in fertilized oocytes (Fig. 2 A, b–d). Some sperm mitochondria labeled with SQSTM1 were still detectable in zygotes cultured for 30 h after in vitro fertilization (IVF) (Fig. 2 A, d). However, there was no SQSTM1 signal in spermatozoa bound to zona pellucida (ZP) of oocytes (Fig. 2 A, a). As anticipated, we detected the 75-kDa protein band corresponding to SQSTM1 in germinal vesicle (GV) stage and metaphase II (MII) oocytes and zygotes by Western blotting; however, SQSTM1 protein was not detectable in sperm protein extracts (Fig. 2B). Therefore, the result of Western blotting was consistent with the immunofluorescence staining of SQSTM1, which suggested that SQSTM1 was absent from boar spermatozoa until fertilization, and the SQSTM1 protein associated with sperm mitochondria inside zygotes originating instead from oocyte cytoplasm.
Fig. 2.
The ubiquitin-binding autophagy receptor SQSTM1 associated with sperm mitochondria after porcine (A and B) and rhesus monkey (C) fertilization. (A) The SQSTM1 protein (green) was not detectable in the zona-bound spermatozoa before sperm incorporation into oocyte cytoplasm (a); it became associated with sperm mitochondria by 9 h postfertilization (b; very early stage of paternal pronucleus formation was shown) and remained detectable at 12 (c) and 30 h postfertilization (d). (Original magnification, 1,000×.) (B) Western blotting detected the predicted 75-kDa SQSTM1 protein band in porcine GV and MII oocytes, cumulus cells and zygotes (IVF, 30 h), but not in boar spermatozoa (boar sperm). (C) The SQSTM1 protein was detected in the sperm mitochondria at the pronuclear stages of zygotic development in rhesus monkey ICSI embryos (a), but not yet associated with the sperm mitochondrial sheath early after ICSI (b).
In addition to porcine zygotes, we have examined the presence of SQSTM1 in early-stage rhesus monkey zygotes generated by intracytoplasmic sperm injection (ICSI). Consistent with observations in porcine gametes, SQSTM1 labeling was present on the sperm tail midpiece in rhesus pronuclear-stage zygotes (Fig. 2 C a), but there was no detectable signal of SQSTM1 in spermatozoa early after ICSI (Fig. 2 C, b).
Identification of Candidate SQSTM1-Substrate Proteins in Boar Sperm Tail Extracts.
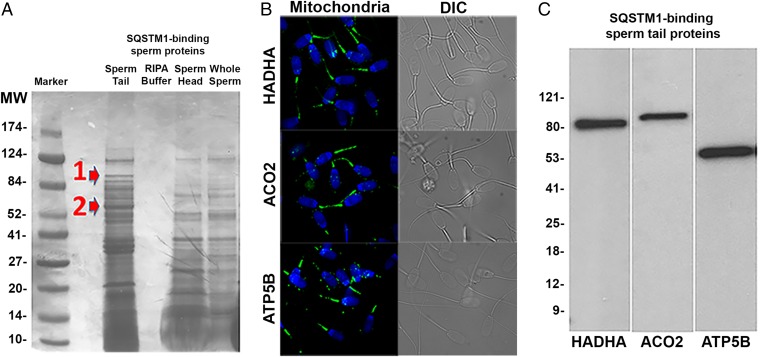
To support the involvement of ooplasmic SQSTM1 in the recognition of sperm mitochondrial proteins, we used a synthetic SQSTM1-derived ubiquitin-associated domain (UBA) to purify solubilized whole boar sperm tail proteins. Boar sperm head extracts were processed as control. The eluted UBA domain-binding proteins were separated on PAGE gels (Fig. 3A), excised, and subjected to Nano LC-Nanospray MS/MS. Such a proteomic analysis identified four proteins with high ion scores for known amino acid sequences, three of which were in fact known mitochondrial proteins; they were the mitochondrial trifunctional enzyme subunit alpha (HADHA), mitochondrial aconitase 2 (ACO2), and mitochondrial ATP synthase H+ transporting F1 complex β-subunit (ATP5B). β-Tubulin was the fourth identified protein. Significance scores and sequence coverage data are shown in SI Appendix, Fig. S2. We confirmed immunolocalization of HADHA, ACO2, and ATP5B to the sperm tail midpiece by immunofluorescence (Fig. 3B), whereas these mitochondrial proteins were not detectable in sperm mitochondria after fertilization (SI Appendix, Fig. S3). Western blotting confirmed target specificity of HADHA, ACO2, and ATP5B antibodies in boar sperm tail extracts (Fig. 3C). Copurification of ubiquitinated sperm proteins with the UBA domain was confirmed by Western blotting with anti-ubiquitin antibody FK1, specific to polyubiquitinated protein conjugates (SI Appendix, Fig. S4).
Fig. 3.
Affinity purification and identification of the ubiquitinated mitochondrial proteins from boar sperm tails. (A) Boar sperm heads and tails were solubilized with RIPA buffer and the resultant extracts were incubated with a synthetic SQSTM1-derived UBA domain, immobilized on agarose beads. The eluted sperm proteins were separated on the gel, then stained with Coomassie blue. Arrows mark protein bands that are conspicuously different between sperm head and tail fractions and were excised for proteomic identification. (B) Immunofluorescence localization of HADHA, ACO2, and ATP5B (all in green), three proteins prevalent in the ubiquitinated sperm tail protein fraction, with high ion scores. (C) Western blotting confirmed the presence of HADHA, ACO2, and ATP5B in the ubiquitinated sperm tail protein fraction. Bands of anticipated size were observed for each of the identified proteins.
The Cross-Talk Between UPS and Autophagy During Sperm Mitochondrion Degradation in Porcine Zygote.
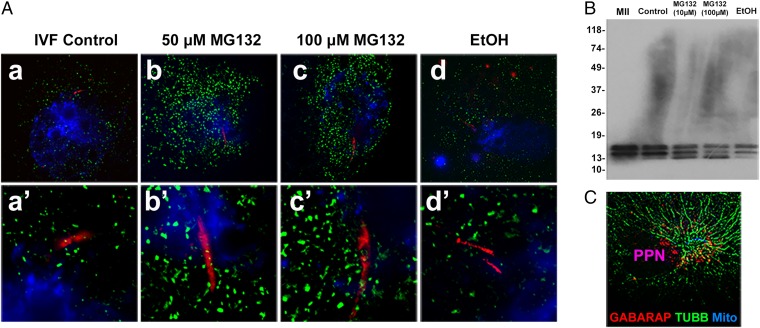
We further investigated mitophagy-associated ubiquitin receptors that could act downstream of, or in parallel with, ooplasmic SQSTM1 and examined the cross-talk between UPS and autophagy in porcine zygotes treated with the proteasomal inhibitor MG132, previously shown to inhibit porcine sperm mitophagy. Fertilized oocytes were cultured for up to 36 h in the presence of MG132 (50 and 100 μM) and immunostained with an antibody specific to ubiquitin-like protein modifier GABARAP, serving as a marker for autophagophore/autophagosome formation. The GABARAP-containing structures surrounded, but seldom overlapped, the sperm mitochondria (Fig. 4A and SI Appendix, Fig. S5A), reflecting the autophagophore/autophagosome formation around the sperm mitochondria and paternal pronucleus. Another significant finding was that the accumulation of GABARAP-containing aggregates was significantly increased in the presence of MG132, (Fig. 4A and SI Appendix, Fig. S5 A and B). The corresponding intensity profiles of GABARAP labeling are shown in SI Appendix, Fig. S5C. Intact sperm mitochondrial sheaths were present in the embryos treated with MG132, suggesting that autophagy may compensate for impaired proteasomal proteolysis, but this compensation alone is not sufficient to achieve sperm mitophagy under proteasome-blocking conditions (Fig. 4A). Altogether, the presence of intact sperm mitochondrial sheaths and increased accumulation of GABARAP around paternal pronuclei of zygotes incubated with MG132 are consistent with the primary role of proteasomal proteolysis in sperm mitophagy as well as with the compensatory action of the GABARAP-associated autophagic pathway under proteasome-inhibiting conditions. Western blotting confirmed the presence of appropriate size GABARAP protein bands in control and MG132-treated zygotes. The GABARAP protein migrated as a triplet of bands in which the third, lowest mass band was strongly induced in the presence of MG132 (Fig. 4B).
Fig. 4.
Detection of ubiquitin-like protein modifier GABARAP, a mitophagy receptor, in control and MG132-treated porcine zygotes. (A) The accumulation of GABARAP-containing aggregates/autophagophores (green) was detected around the sperm tail (red; MitoTracker) and paternal pronuclei (blue; DAPI) at 30 h postinsemination in untreated control (a and a′), at 50 μM MG132 (b and b′), 100 μM MG132 (c and c′), and in vehicle control (d and d′; EtOH). Note the increased GABARAP particle content in the MG132-treated zygotes. Note that very few of the GABARAP+ green particles were colocalized with the red fluorescent sperm mitochondria. (Original magnification, 1,000×.) (B) Western blotting of GABARAP in porcine zygotes cultured for 30 h after fertilization (i.e., 36 h after sperm addition), with or without MG132 (10/100 μM). The GABARAP protein migrated as a triplet of bands. The lowest mass band was amplified in the presence of MG132. Last lane is a vehicle control (EtOH). (C) Ooplasmic, autophagophore-associated GABARAP (red) recruited along sperm aster microtubules (green) in a fertilized oocyte cultured for 30 h after IVF. Well-developed sperm aster microtubules were formed in the vicinity of paternal pronucleus, showing that GABARAP recruitment to the sperm aster was concomitant with the pronuclear development and degradation of the sperm mitochondria. Image was processed by 2D deconvolution. PPN, paternal pronucleus; TUBB, β-tubulin.
Because the transport of protein aggregates toward autophagophore/autophagosome may occur along microtubule tracks, we examined the enrichment of GABARAP within the microtubule-based sperm aster organized after fertilization by the sperm centriole/zygotic centrosome. Fertilized oocytes were collected at 30 h after IVF and double immunolabeled with anti–β-tubulin and anti-GABARAP antibodies. The GABARAP+ autophagophores were recruited along and sometimes, but not always, colocalized with sperm aster microtubules, which emanated from the connecting piece of the sperm tail associated with the paternal pronucleus (Fig. 4C and SI Appendix, Fig. S6 A and B). It is thus plausible that sperm aster microtubules help in transporting and recruiting GABARAP and autophagophores to the vicinity of sperm tail mitochondria after fertilization.
Immunolocalization of Autophagy-Related Proteins in the Boar Spermatozoa.
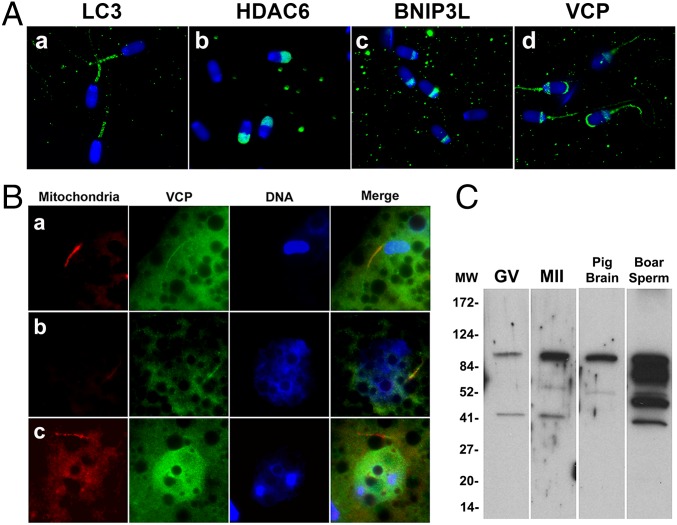
To identify sperm mitophagy determinants that may already be present in the sperm mitochondria before fertilization, boar spermatozoa were immunolabeled with antibodies against autophagy associating proteins LC3, HDAC6, BNIP3L, and VCP. Various autophagy determinants (Fig. 1, diagram) were detected in the sperm mitochondria and other sperm accessory structures before fertilization (Fig. 5A and SI Appendix, Fig. S7A). LC3, which is known to interact with autophagosome/phagophore, was present in the sperm tail midpiece (Fig. 5 A, a). HDAC6, responsible for the microtubule-dependent transport of ubiquitinated protein aggregates toward the autophagophores, was prominent in the acrosomal region (Fig. 5 A, b). Mitophagy receptor BNIP3L, an outer mitochondrial membrane component (11) that induces autophagy in response to hypoxia (12), was abundant in the proximal section of the postacrosomal sheath of the sperm head, but not associated with the mitochondrial sheath of the sperm tail (Fig. 5 A, c). On the contrary, VCP, which is implicated in the displacement of ubiquitinated proteins from mitochondrial membranes (for presentation to 26S proteasome), was most prominent in the mitochondrial sheath, although also detected in the postacrosomal sheath and in the subacrosomal layer of the sperm head perinuclear theca (Fig. 5 A, d). Whereas the LC3, HDAC6, and BNIP3L were detected in the sperm mitochondria and/or other sperm accessory structures, they seemed to dissociate from the sperm head/tail after fertilization (SI Appendix, Fig. S7B), which was not the case for VCP (Fig. 5B). Therefore, further investigation focused on the association of VCP with sperm mitochondria after fertilization.
Fig. 5.
Immunolocalization and inhibition of autophagy determinants implicated in sperm mitophagy. (A) Immunolocalization of candidate mitophagy determinants in the boar spermatozoa before fertilization: LC3 (a) was present in the midpiece/mitochondrial sheath of the sperm tail. HDAC6 (b) was present in the acrosomal region of the sperm head. BNIP3L (c) was also detectable in the sperm head, particularly in the postacrosomal sheath. VCP (d) was detected in the apical ridge of the acrosome, in the sperm head postacrosomal sheath and in the sperm tail midpiece. Nuclear DNA was counterstained with DAPI (blue). (B) Colocalization of VCP (green) with the sperm mitochondria (red) in porcine zygotes at the early stage of sperm-nuclear decondensation (a), at 30 h after IVF (b), and at 30 h after IVF in the presence of MDBN, a specific inhibitor of VCP (c). Intact sperm mitochondria were present in embryos cultured for 30 h in the presence of MDBN. (C) Western blotting of VCP in the porcine oocytes and sperm extracts. The anticipated band of 97 kDa, corresponding to VCP protein, was detected in GV oocytes, MII-oocytes, pig brain (third lane), and boar spermatozoa (fourth lane).
Ubiquitinated Protein Dislocase VCP Associates with Sperm Mitochondria After Fertilization.
Based on the above observations, we hypothesized that the sperm mitochondrial membrane proteins in porcine zygotes could be extracted and delivered to ooplasmic proteasomes as the individual ubiquitinated protein molecules via the VCP-dependent pathway. Western blotting with anti-VCP antibody detected the anticipated 97-kDa VCP protein in boar spermatozoa, and GV stage and MII oocytes (Fig. 5C and SI Appendix, Fig. S8).
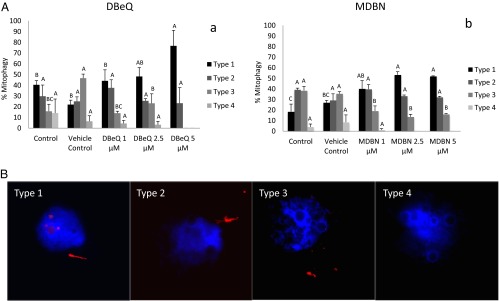
To examine the role of VCP in sperm mitophagy, we performed a controlled trial with two selective small molecule inhibitors of VCP, such as 3,4-methylenedioxy-β-nitrostyrene (MDBN) and N,N′-dibenzylquinazoline-2, 4-diamine (DBeQ), anticipating that both DBeQ (reversible) and MDBN (irreversible) should inhibit sperm mitophagy. Under control conditions, the colocalization of VCP with the sperm mitochondria was readily observed at the early stage of sperm nuclear decondensation (Fig. 5 B, a) and still seen as late as 30 h after IVF (Fig. 5 B, b; only some of the 30-h zygotes still had detectable sperm mitochondria). Similarly, VCP remained detectable in sperm mitochondria of zygotes treated transiently with 5 μM MBDN (Fig. 5 B, c). Sperm mitochondria in the fertilized oocytes were partially or completely degraded at 30 h after IVF, whereas intact sperm mitochondria were observed with a significantly higher frequency in zygotes transiently treated with MDBN for the first 6 h after IVF. To categorize the degradation patterns of the sperm mitochondria in porcine oocytes harvested 30 h after IVF with or without DBeQ and MDBN, the distribution of four different types of sperm mitochondrial sheaths (Fig. 6B) was assessed, reflective of the progress of sperm mitophagy. The incidence of type 1, intact sperm mitochondrial sheaths gradually increased with increasing dose of both VCP inhibitors (Fig. 6A); this change was statistically significant at 2.5- and 5-μM concentrations of both inhibitors, DBeQ and MDBN, compared with a lower dose of inhibitors and vehicle control (Fig. 6A). Altogether, transient inhibition of VCP activity during the first 6 h of IVF delayed the progress of sperm mitophagy in the porcine zygote.
Fig. 6.
(A) The effect of specific VCP inhibitors: DBeQ (reversible inhibitor) (a) and MDBN (irreversible) (b) on sperm mitophagy in porcine zygotes examined at 30 h after IVF. (A) Porcine oocytes were fertilized for 6 h with or without VCP inhibitors and then washed and cultured in an inhibitor-free embryo culture medium for an additional 24 h. The incidence of type 1, intact sperm mitochondrial sheaths gradually increased with increasing dose of both VCP inhibitors. Experiments were repeated three times for each treatment group (10–20 oocytes per group). Values are shown as the mean percentages of sperm mitophagy ± SEM. Different superscripts A, B, and C in each diagram present a significant difference at P < 0.05. (B) Four different phenotypes of the sperm mitochondrial sheath, reflective of the progression of sperm mitochondrion degradation after fertilization. Type 1 displays a straight, intact mitochondrial sheath. Type 2 represents a distorted, corkscrew-shaped mitochondrial sheath. Type 3 shows scattered or clumped, nearly completely degraded sperm mitochondria. Type 4 represents the absence of sperm mitochondria; only the remains of outer dense fibers that take up the red MitoTracker stain nonspecifically are present.
Combined Treatment with a VCP Inhibitor and Autophagy-Targeting Antibodies Prevents Postfertilization Sperm Mitophagy.
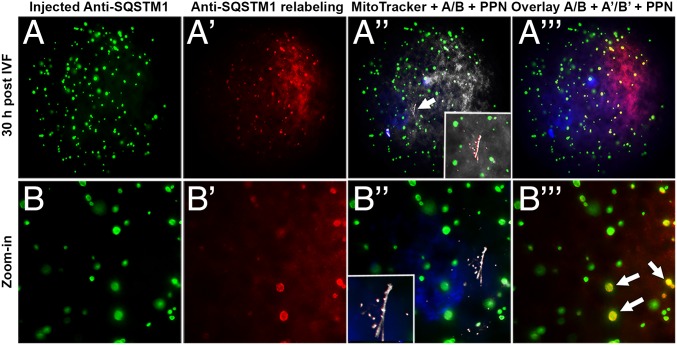
We next examined the combined postfertilization effect of VCP inhibitor treatment and the microinjection of antibodies specific to SQSTM1 and/or GABARAP into MII oocytes. Preinjection of MII oocytes with anti-SQSTM1 antibody alone or mixed with anti-GABARAP antibody caused the formation of SQSTM1+ aggregates throughout the ooplasm (Fig. 7 A and B), not seen in sham-injected zygotes (SI Appendix, Fig. S9A). Preinjection of MII oocytes with anti-SQSTM1 antibody alone prevented the postfertilization binding of ooplasmic SQSTM1 to sperm mitochondria (Fig. 7 A′ and B′), whereas the translocation of SQSTM1 to sperm mitochondria was detectable in noninjected and sham-injected zygotes (SI Appendix, Fig. S9 A′ and B′). Also, the formation of GABARAP-containing aggregates/autophagosomes increased around the paternal pronucleus. The GABARAP-containing aggregates were not detected in noninjected and sham-injected zygotes (SI Appendix, Fig. S10 A′ and B′).
Fig. 7.
Preinjection of MII oocytes with anti-SQSTM1antibody prevented the postfertilization binding of ooplasmic SQSTM1 to sperm mitochondria. Oocytes preinjected with mouse anti-SQSTM1 antibody (green) were cultured for 30 h after fertilization, fixed, labeled with green fluorescent anti-mouse secondary antibody, then washed, and relabeled with anti-SQSTM1 antibody followed by red fluorescent anti-mouse IgG. In oocytes preinjected with anti-SQSTM1 antibody, SQSTM1+ aggregates (A and B, green) were formed throughout ooplasm. Neither the injected anti-SQSTM1 antibody (A′′ and B′′) nor the ooplasmic SQSTM1 (A′′′ and B′′′) associated with sperm mitochondria (MitoTracker; arrow in A′′) in the oocytes preinjected with anti-SQSTM1 antibody. Some of the aggregates triggered by antibody injection appeared to attract ooplasmic SQSTM1, as shown by red–green overlap (arrows in B′′′). PPN, paternal pronucleus.
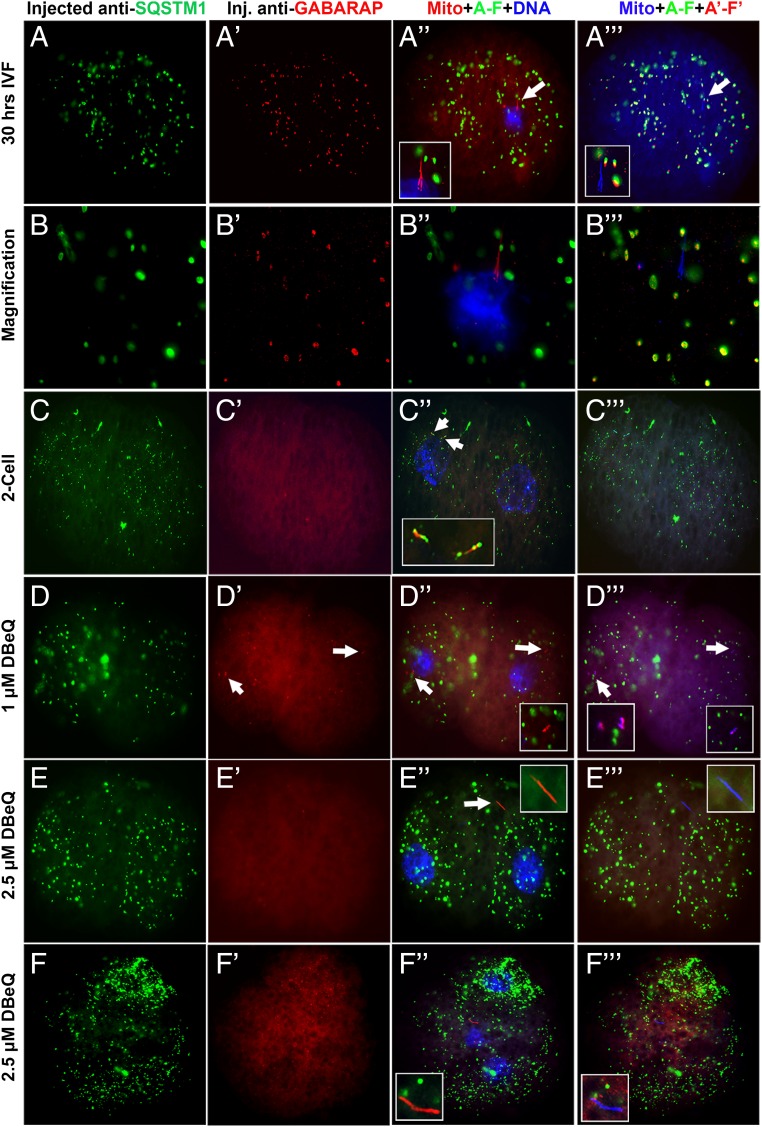
Co-injection of MII oocytes with anti-SQSTM1 and anti-GABARAP antibodies, followed by IVF resulted in the formation of large ooplasmic aggregates in which SQSTM1 and GABARAP colocalized throughout zygotic cytoplasm without detectable association of either protein with the mitochondrial sheath (Fig. 8 A′′′ and B′′′). Whereas the sperm mitophagy was completed before first embryo cleavage under control conditions, large remnants of mitochondrial sheaths were observed in some two-cell embryos after anti-SQSTM1 and anti-GABARAP antibody co-injection (Fig. 8 C′′ and C′′′), but not after the injection of anti-SQSTM1 antibody alone (SI Appendix, Fig. S11). Strikingly, intact sperm mitochondria were observed in two-cell embryos co-injected with anti-SQSTM1 and anti-GABARAP antibodies, and incubated to two-cell stage in the presence of the VCP inhibitor DBeQ (Fig. 8 E′′ and E′′′). The association of GABARAP with sperm mitochondria was observed in two-cell embryos treated with 1 µM DBeQ (Fig. 8D′′′) but not in those treated with 2.5 µM DBeQ (Fig. 8E′′′). Intact sperm mitochondria were clearly observed in some embryos during the transition from two- to four-cell stage (Fig. 8 F′′ and F′′′).
Fig. 8.
Concomitant inhibition of autophagy and substrate–proteasome presentation prevented postfertilization sperm mitophagy without affecting embryo cleavage. (A–B′′′) The formation of large ooplasmic aggregates in which SQSTM1 and GABARAP colocalized throughout zygotic cytoplasm without obvious association of either protein with the mitochondrial sheath was observed in one-cell zygotes obtained by IVF of the oocytes preinjected with a combination of mouse anti-SQSTM1 and rabbit anti-GABARAP antibodies. (C–C′′′) The aforementioned co-injection delayed sperm mitophagy until the two-cell stage, at which the association of SQSTM1 with sperm mitochondria reappeared. (D–F′′′) Sperm mitophagy was blocked in the early embryos grown from the in vitro fertilized oocytes preinjected with anti-SQSTM1 and anti-GABARAP antibodies and cultured in the presence of the specific VCP inhibitor DBeQ (D–D′′′, 1 µM DBeQ; E–F′′′, 2.5 µM DBeQ) until the blastocyst stage. In some cases, intact sperm mitochondria were clearly observed in the resultant two-cell embryos (arrows in D′′, D′′′, and E′′ and E′′′) and occasionally displayed the binding of injected anti-GABARAP antibody (Insets in D′′′). Intact mitochondrial sheaths were still present in embryos transitioning from two- to four-cell stages (F–F′′′).
Discussion
Uniquely, the outer mitochondrial membrane of sperm mitochondria is covered by a keratinous structure composed of cysteine-rich proteins, known as the mitochondrial capsule. The cysteine-rich proteins could stabilize the mitochondrial capsule, and cement mitochondria with each other in the sheath, resulting in a protein-rich aggregate structure (13). Such structure may be similar to aggresomes, the aggregates of ubiquitinated proteins subject to autophagy, which would make the mitochondrial sheath susceptible to autophagy after fertilization. Accordingly, boar sperm mitochondrial sheaths are recognized by the aggresome-detecting probes both before and after fertilization (SI Appendix, Fig. S12). Mitochondrial membrane and matrix proteins such as prohibitin and TFAM, as well as those identified in the present study by copurification with the UBA domain of SQSTM1 are ubiquitinated (14, 15). Thus, it is possible that the whole sperm mitochondrial sheath is recognized by the oocyte autophagy machinery as one large aggresome. Extraction of ubiquitinated mitochondrial proteins by VCP could initially destabilize the sperm mitochondrial membrane, leading to breakup of the mitochondrial sheath into fragments and even individual mitochondria. Sperm mitophagy by zygote could then be completed by autophagosome. Accordingly, our imaging studies show that except when inhibited by DBeQ or MDBN, the VCP dissociates from sperm mitochondria early after fertilization and appears to be supplanted by ooplasmic SQSTM1, which functions as a ubiquitin receptor in mitophagy.
Mitochondrial degradation by autophagy (mitophagy), mediates maternal inheritance of mitochondrial DNA in mammals (16). The elimination of sperm-borne mitochondria upon fertilization assures normal preimplantation development. Our early studies determined that UPS activity is essential for the degradation of paternal mitochondria in fertilized mammalian oocytes (3, 4, 17–19). Attempts have been made to identify the mechanism of maternal inheritance of mitochondria and their mtDNA in various organisms. A mechanism combining multiple mitophagy/autophagy-related pathways, including SQSTM1, has recently been proposed in Drosophila melanogaster (9). Likewise, studies of the nematode worm C. elegans reported that sperm mitochondria inside the embryo were surrounded by autophagosomes and subsequently degraded by autophagic pathway (6, 7). The same studies also provided comparative data suggesting that sperm mitophagy might be conserved between nematode and mouse models, indicating the autophagosomal markers, such as SQSTM1, LC3, and GABARAP were recruited to mouse sperm tail structures after fertilization. However, to truly extend such a notion to mammals as taxon, the involvement of sperm mitophagy in postfertilization should be investigated in other, higher mammalian models, such as porcine and nonhuman primate models used in the present study to examine the relationship between UPS and autophagy during sperm mitophagy.
Contradicting the above studies, some findings in the mouse challenged the role of the autophagic pathway, suggesting that sperm mitochondria did not associate with GFP-tagged LC3 protein in the four-cell embryos and never associated with lysosomes (10). Lack of LC3 function in sperm mitophagy may be in agreement with our finding that LC3 protein did not congregate to sperm mitochondria after porcine fertilization (SI Appendix, Fig. S7B), even though LC3 protein was detectable in sperm mitochondria before fertilization. However, the conclusion that autophagy is not involved in murine sperm mitochondrion degradation is not sustainable in light of our and others’ data indicating alternative routes for autophagy involvement, including the ooplasmic SQSTM1-dependent autophagic pathway and the VCP-mediated proteasomal degradation of mitochondrial proteins. Very recently, a paternal mitochondrial derivative in Drosophila was shown to be degraded after fertilization by concerted synergy of endocytotic and autophagy pathways (9). Similar to our observations in mammals, the fly paternal mitochondrial derivative became ubiquitinated and attracted SQSTM1 right after fertilization (9). Next, the ubiquitinated mitochondrial derivative was separated from the axoneme and sequestered into autophagosome. These observations in Drosophila agree with our finding of postfertilization association of SQSTM1 with boar and primate sperm mitochondria (Fig. 2 A and C). Such an alternative route to sperm mitophagy may thus be conserved in higher mammals, including primates. Ubiquitination of paternal mitochondrial proteins could be strictly required as a determinant of sperm mitophagy, whereas the prefertilization ubiquitination of sperm mitochondria in C. elegans was not required for their degradation (6). These interspecies differences may shed light on the mechanisms that assure species specificity of sperm mitophagy, because mitophagy does not occur in the interspecific crosses, resulting in heteroplasmy (20, 21).
The C-terminal amino acids of SQSTM1 bind noncovalently to ubiquitin. In vitro experiments using ubiquitin-conjugated Sepharose beads support the interaction of SQSTM1 with ubiquitin (22). Consistent with the proposed role of SQSTM1 in sperm mitophagy, three of four major SQSTM1-binding sperm tail proteins were of mitochondrial origin (SI Appendix, Fig. S2). The fourth major SQSTM1/UBA domain binding protein was a species of tubulin, some isoforms of which may also be ubiquitinated in the sperm tail (14). Ubiquitination of sperm mitochondria was observed at the secondary spermatocyte stage of spermatogenesis (3). In mammals, ubiquitination of sperm mitochondrial proteins has been implicated in their proteasomal degradation after fertilization (4, 17).
The identities of SQSTM1 copurifying sperm mitochondrial proteins, including HADHA, ACO2, and ATP5B, were confirmed by Western blotting and their presence in the sperm mitochondria was detected by immunofluorescence. The mitochondrial trifunctional protein encoded by the Hadha gene is a heterooctamer composed of four α- and four β-subunits, catalyzing consecutive steps in mitochondrial β-oxidation of long chain fatty acids, which is an important energy generation system (23). This protein is located in the inner mitochondrial membrane (24) and its deficiency may cause defects of respiratory chain function (25). Another protein of interest that copurified with SQSTM1 is mitochondrial aconitase, which catalyzes the interconversion of citrate to isocitrate in the tricarboxylic acid cycle. There are two isozymes of aconitase in eukaryotes: one is in the matrix of mitochondria, and the other is localized in the cytosol (26). The third SQSTM1-copurifying mitochondrial protein, the mitochondrial ATP synthase ATP5B, catalyzes ATP synthesis by using electrochemical proton gradient across the inner membrane of mitochondria (27).
The SQSTM1 protein is able to engage ubiquitinated proteins by binding to multiubiquitin chains via the UBA domain (28–30). Furthermore, the LIR (LC3-interacting region) or LRS (LC3 recognition sequence) of SQSTM1 recognizes LC3, an autophagosomal protein (30). The LC3-dependent autophagy has been implicated in the degradation of sperm mitochondria and sperm-mitochondrion–associated membranous organelles in C. elegans (6). The SQSTM1 directly interacts with LC3 to degrade ubiquitinated proteins by autophagy in HeLa cells (28, 30). The siRNA-mediated deletion of Sqstm1 function prevents recruitment of LC3 to autophagosome, suggesting a close relation between SQSTM1 bodies and LC3 (29). However, it is not established whether this interaction would operate during sperm mitophagy in mammalian zygotes. Our results indicate that SQSTM1, but not LC3, participates in sperm mitophagy after porcine fertilization, although LC3 is detectable in the sperm mitochondria before fertilization. Such a conclusion is consistent with recently published studies of Drosophila sperm mitophagy (9) and suggests that sperm mitophagy in mammals and other taxa involves a noncanonical ubiquitin-recognizing autophagic pathway independent of LC3.
The GABARAP protein was found to colocalize with the sperm tail midpiece in fertilized mouse oocytes (6). The Gabarap-null cells failed to form autophagic membranes, essential for autophagosome formation (31). Our findings in fertilized porcine oocytes indicate that GABARAP could connect UPS and autophagy pathways during postfertilization sperm mitophagy. We also found that the inhibition of proteasomal proteolysis caused an accumulation of GABARAP+ autophagophores, a process accompanied by the delayed sperm mitophagy, supporting the role for the GABARAP-related autophagosome formation in porcine zygotic development.
In fertilized porcine oocytes, microtubules emanating from the sperm aster near the decondensed sperm head participated in pronuclear movement and apposition to the maternal pronucleus (32, 33). In the present study, the GABARAP/autophagophore recruitment to the sperm aster was centered on the sperm mitochondrial sheath and was concomitant with pronuclear development. Sperm aster microtubules may thus be instrumental in the recruitment of GABARAP and other autophagy receptors to the vicinity of the sperm mitochondrial sheath at an early stage of zygotic development. On the basis of our results, we conclude that GABARAP is involved in the process of zygotic autophagosome formation and directly links both UPS and autophagy; however, GABARAP seldom associated with the sperm mitochondrial sheath in the porcine zygote, suggesting that similar to LC3, GABARP may not be directly required for mammalian zygotic sperm mitophagy. Recruitment of GABARAP to sperm aster, on the other hand, could facilitate the degradation of other insoluble sperm accessory structures such as sperm head perinuclear theca and redundant nuclear envelopes, and the striated columns of the sperm tail connecting piece, somewhat similar to the autophagy of the sperm membranous organelle in the C. elegans embryo (6, 7). Association of the GABARAP+ structures with sperm aster microtubules agrees with studies showing that microtubule disruption with nocodazole interferes with the formation of aggresomes (34).
Protein dislocase VCP is an ATPase involved in the extraction of ubiquitinated proteins from the outer mitochondrial membrane and their subsequent presentation to the 26S proteasome for proteasomal degradation of the mitochondrial proteins in somatic cells (35). Parkin, an E3 ubiquitin ligase, initiates the ubiquitination of outer mitochondrial membrane proteins. VCP has therefore been implicated in parkin-dependent degradation of the mitochondrial proteins by proteasomal proteolysis, to prime redundant, dysfunctional, or damaged mitochondria for mitophagy (36). The Vcp gene mutation caused defective autophagosome maturation, resulting in dysfunctional autophagy (37). Therefore, some mitochondrial proteins in the oocyte-incorporated sperm mitochondria could be degraded by VCP-dependent proteasomal degradation, which would prime the sperm mitochondria for SQSTM1-mediated mitophagy. We used two specific small molecule VCP inhibitors, MDBN and DBeQ, to delay sperm mitophagy postfertilization. We detected VCP protein in the mitochondrial sheaths of boar spermatozoa both before fertilization and after the incorporation into porcine oocytes; VCP remained associated with sperm mitochondria after fertilization, whereas we cannot rule out that VCP molecules from ooplasm were also recruited to sperm mitochondria postfertilization. Such late recruitment would explain the increased intensity of sperm mitochondrion-associated VCP immunolabeling in zygotes transiently treated with the irreversible VCP inhibitor MDBN. Because both MDBN and DBeQ delayed the progression of sperm mitochondrion degradation after porcine fertilization, it is plausible that the early stages of postfertilization sperm mitophagy depend upon VCP activity.
Antibody microinjection was used to test the effect of SQSTM1 and GABARAP interference on mammalian sperm mitophagy. Others have shown that microinjection of anti-LC3 antibody blocked the movement of autophagosomes, which is required for their docking to the lysosome (38). In our study, oocyte preinjection with anti-SQSTM1 antibody prevented translocation of ooplasmic SQSTM1 to the sperm mitochondria. Furthermore, the GABARAP+ autophagosomes accumulated around male pronuclei and sperm mitochondria in oocytes preinjected with anti-SQSTM1 antibody, which suggests that GABARAP was involved in zygotic autophagosome formation. However, preinjection with anti-SQSTM1 antibody alone was not sufficient to block sperm mitophagy, suggesting that SQSTM1 could cooperate with GABARAP to facilitate sperm mitophagy. Accordingly, sperm mitophagy was completely blocked in zygotes co-injected with anti-SQSTM1 and anti-GABARAP antibodies and subsequently treated with the specific VCP inhibitor DBeQ. Thus, sperm mitophagy may be mediated by UPS via combined action of SQSTM1, GABARAP, and VCP.
In conclusion, the present study indicates that postfertilization sperm mitophagy employs an unconventional, ubiquitin-recognizing autophagic pathway independent of canonical autophagy receptors such as LC3 or GABARAP. Ooplasmic SQSTM1 was identified as one of the ubiquitin-binding autophagy receptors that exclusively associate with boar and rhesus monkey sperm mitochondria after fertilization. The GABARAP could still be involved in zygotic autophagosome formation, albeit nonessentially. These results suggest that sperm mitophagy in higher mammals may rely on a combined action of SQSTM1-dependent autophagy and VCP-mediated presentation of ubiquitinated sperm mitochondrial proteins to the 26S proteasome. Altogether, our observations explain how a large organelle assembly, such as the sperm mitochondrial sheath, could be degraded with the help of the 26S proteasome holoenzyme capable of degrading only one protein molecule at a time.
Materials and Methods
A detailed description of procedures used in the study is provided in SI Appendix, SI Materials and Methods.
Isolation of Boar Sperm Heads and Tails.
A total of 1 × 109 fresh spermatozoa per milliliter were washed by using PBS (pH 7.4) and collected by centrifugation at 800 × g for 5 min. Sperm pellets were sonicated (digital sonifier, Branson) at 30% intensity for 1 min in 30% (wt/vol) sucrose in PBS. The sonicated sperm pellets of 30% (wt/vol) sucrose in PBS were loaded on the top of 70% (wt/vol) sucrose in PBS. The sample was isolated by centrifugation at 100,000 × g for 1 h in a Beckman ultracentrifuge using an SW 28 rotor (Beckman Coulter). Sperm heads and tails were separated by different concentration of sucrose in the gradient column. The sperm heads were collected from the pellet at the bottom of the centrifuge tube, while the sperm tails accumulated on top of 70% sucrose solution. Isolated sperm heads and tails were examined under a light microscope to confirm the purity of the head and tail fractions.
Purification of Ubiquitinated Proteins from Boar Sperm Extracts.
To obtain sperm extracts, the sperm fractions were resuspended in RIPA buffer (200 mM Tris⋅HCl, pH 8; 150 mM NaCl; 1% Triton X-100; 0.5% sodium deoxycholate; 0.1% SDS; and 1 mM PMSF). Resuspended fractions were sonicated at 30% intensity for 1 min in RIPA buffer.
An agarose-immobilized p62/SQSTM1-derived ubiquitin pathway-associated domain was washed with Tris-buffered solution (TBS; 50 mM Tris, 150 mM NaCl, pH 7.4) and centrifuged at 800 × g for 1 min at 4 °C in a Sorvall Biofuge Fresco (Kendro Laboratory Products). The supernatant from SQSTM1-agarose was removed. The sperm extracts from heads and tails were centrifuged at 800 × g for 10 min, and 50 μL of the collected supernatant was mixed with 30 μL of SQSTM1-agarose and incubated for 24 h at 4 °C on a rocker set to low speed. After 24 h of incubation, the sperm sample was centrifuged at 4,000 × g for 1 min at 4 °C. The supernatant was collected for control, and SQSTM1-agarose with sperm proteins was washed with TBS three times. To elute ubiquitinated proteins, SQSTM1-agarose binding sperm proteins were boiled with loading buffer [50 mM Tris, pH 6.8, 150 mM NaCI, 2% (wt/vol) SDS, 20% (vol/vol) glycerol, 5% (vol/vol) β-mercaptoethanol, and 0.02% bromophenol blue] for 5 min and then centrifuged at 4,000 × g for 5 min at 20 °C. The supernatants were separated on 4–20% gradient polyacrylamide gels (PAGEr Gold Precast gels, Lonza Rockland) and stained with Coomassie blue.
Inhibitor Treatments, Mitophagy Assessment, and Statistical Analysis.
To examine the interplay between the autophagic pathway and UPS during sperm mitochondrion degradation in fertilized oocytes, a reversible inhibitor of the proteasomal chymotrypsin-like and caspase-like proteolytic activities, MG132 was used. Porcine oocytes were inseminated and co-incubated for 6 h with fresh boar spermatozoa prelabeled with MitoTracker. The oocytes were then cultured with/without an addition of MG132 (10–100 μL) for an additional 30 h, at which time they were fixed and processed for immunofluorescence with anti-GABARAP and/or anti-SQSTM1 antibodies to evaluate the degradation of sperm mitochondria. Some such zygotes were harvested for Western blotting. For vehicle controls, ethanol (100%) was used at a volume equivalent to MG132 volume added to oocytes.
Two inhibitors of the ubiquitin-binding protein dislocase VCP, including DBeQ (reversible inhibitor) and MDBN (irreversible inhibitor) were used to inhibit VCP activity after fertilization. Porcine oocytes were incubated with fresh boar spermatozoa prelabeled with MitoTracker for 6 h in addition to the VCP inhibitors (final concentration: 0, 1, 2.5, and 5 μM DBeQ/MDBN) at the time of fertilization. The oocytes were washed and further cultured for 24 h postinsemination, at which time they were fixed and processed for immunofluorescence with anti-VCP antibody to evaluate the degradation patterns of sperm mitochondria. The DMSO was used as vehicle control without inhibitors. Four distinct phenotypes of the sperm mitochondrial sheath, reflective of the progression of sperm mitochondrion degradation, were established (4) and the degradation rate of sperm mitochondria was calculated in zygotes (10–20 oocytes per treatment per replicate). Experiments were replicated three times for each treatment group. Analyses of variance were carried out using the Statistical Analysis Software package in a completely randomized design. Duncan’s multiple range test was used to compare values of individual treatment when the F-value was significant (P < 0.05).
Antibody Microinjection.
Microinjection of antibodies was performed by the Eppendorf Microinjection System. A 100-μL drop of TL-Hepes-PVA was placed on a Petri dish and covered with mineral oil. Injection needles were loaded with 2 μL of anti-SQSTM1 and/or anti-GABARAP antibodies, or control mouse/rabbit IgG. The microinjection was performed under the following conditions: antibody concentration of 0.5 mg/mL, an injection pressure of 1.0–2.0 psi, and an injection time between 0.1–1 s. Porcine MII oocytes were co-injected with equal Ig concentrations of antibodies specific for SQSTM1 and/or GABARAP, followed by IVF, and cultured with/without the VCP inhibitor DBeQ (1 or 2.5 μM) until the blastocyst stage of development. Controls included sham injection of nonimmune mouse/rabbit serum as negative control and no injection as IVF control.
Supplementary Material
Acknowledgments
We thank Beverly DaGue (C. W. Gehrke Proteomics Center, University of Missouri) for protein sample analysis. Editorial and clerical assistance by Ms. Kathy Craighead is appreciated. This project was a supported by Agriculture and Food Research Initiative Competitive Grant 2013-67015-20961 from the US Department of Agriculture National Institute of Food and Agriculture (to P.S.) and by seed funding from the Food for the 21st Century Program of the University of Missouri (to P.S.). Work in the laboratory of S.M. was supported by the National Institutes of Health, Office of Research Infrastructure Programs (formerly National Center for Research Resources), Division of Comparative Medicine Grant R01-RR016581.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605844113/-/DCSupplemental.
References
- 1.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA. 1980;77(11):6715–6719. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basse CW. Mitochondrial inheritance in fungi. Curr Opin Microbiol. 2010;13(6):712–719. doi: 10.1016/j.mib.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Sutovsky P, et al. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol Reprod. 2000;63(2):582–590. doi: 10.1095/biolreprod63.2.582. [DOI] [PubMed] [Google Scholar]
- 4.Sutovsky P, McCauley TC, Sutovsky M, Day BN. Early degradation of paternal mitochondria in domestic pig (Sus scrofa) is prevented by selective proteasomal inhibitors lactacystin and MG132. Biol Reprod. 2003;68(5):1793–1800. doi: 10.1095/biolreprod.102.012799. [DOI] [PubMed] [Google Scholar]
- 5.Seglen PO, Gordon PB, Holen I. Non-selective autophagy. Semin Cell Biol. 1990;1(6):441–448. [PubMed] [Google Scholar]
- 6.Al Rawi S, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334(6059):1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 7.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334(6059):1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Q, Li H, Xue D. Elimination of paternal mitochondria through the lysosomal degradation pathway in C. elegans. Cell Res. 2011;21(12):1662–1669. doi: 10.1038/cr.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Politi Y, et al. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev Cell. 2014;29(3):305–320. doi: 10.1016/j.devcel.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Luo SG, et al. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci USA. 2013;110(16):6334–6339. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kraft C, Peter M, Hofmann K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12(9):836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 12.Novak I, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11(1):45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cataldo L, Baig K, Oko R, Mastrangelo MA, Kleene KC. Developmental expression, intracellular localization, and selenium content of the cysteine-rich protein associated with the mitochondrial capsules of mouse sperm. Mol Reprod Dev. 1996;45(3):320–331. doi: 10.1002/(SICI)1098-2795(199611)45:3<320::AID-MRD9>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Thompson WE, Ramalho-Santos J, Sutovsky P. Ubiquitination of prohibitin in mammalian sperm mitochondria: Possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol Reprod. 2003;69(1):254–260. doi: 10.1095/biolreprod.102.010975. [DOI] [PubMed] [Google Scholar]
- 15.Antelman J, et al. Expression of mitochondrial transcription factor A (TFAM) during porcine gametogenesis and preimplantation embryo development. J Cell Physiol. 2008;217(2):529–543. doi: 10.1002/jcp.21528. [DOI] [PubMed] [Google Scholar]
- 16.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20(1):31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutovsky P, et al. Ubiquitin tag for sperm mitochondria. Nature. 1999;402(6760):371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- 18.Sutovsky P, Navara CS, Schatten G. Fate of the sperm mitochondria, and the incorporation, conversion, and disassembly of the sperm tail structures during bovine fertilization. Biol Reprod. 1996;55(6):1195–1205. doi: 10.1095/biolreprod55.6.1195. [DOI] [PubMed] [Google Scholar]
- 19.Sutovsky P, Van Leyen K, McCauley T, Day BN, Sutovsky M. Degradation of paternal mitochondria after fertilization: Implications for heteroplasmy, assisted reproductive technologies and mtDNA inheritance. Reprod Biomed Online. 2004;8(1):24–33. doi: 10.1016/s1472-6483(10)60495-6. [DOI] [PubMed] [Google Scholar]
- 20.Kaneda H, et al. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92(10):4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shitara H, et al. Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics. 2000;156(3):1277–1284. doi: 10.1093/genetics/156.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vadlamudi RK, Joung I, Strominger JL, Shin J. p62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, belongs to a new class of ubiquitin-binding proteins. J Biol Chem. 1996;271(34):20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 23.Gannavaram S, Sharma P, Duncan RC, Salotra P, Nakhasi HL. Mitochondrial associated ubiquitin fold modifier-1 mediated protein conjugation in Leishmania donovani. PLoS One. 2011;6(1):e16156. doi: 10.1371/journal.pone.0016156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibdah JA, et al. Lack of mitochondrial trifunctional protein in mice causes neonatal hypoglycemia and sudden death. J Clin Invest. 2001;107(11):1403–1409. doi: 10.1172/JCI12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson S, et al. Combined enzyme defect of mitochondrial fatty acid oxidation. J Clin Invest. 1992;90(4):1219–1225. doi: 10.1172/JCI115983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantu D, Schaack J, Patel M. Oxidative inactivation of mitochondrial aconitase results in iron and H2O2-mediated neurotoxicity in rat primary mesencephalic cultures. PLoS One. 2009;4(9):e7095. doi: 10.1371/journal.pone.0007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geyik E, et al. Investigation of the association between ATP2B4 and ATP5B genes with colorectal cancer. Gene. 2014;540(2):178–182. doi: 10.1016/j.gene.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 28.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282(33):24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 29.Bjørkøy G, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584(7):1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Weidberg H, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29(11):1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura S, et al. Human sperm aster formation and pronuclear decondensation in bovine eggs following intracytoplasmic sperm injection using a Piezo-driven pipette: A novel assay for human sperm centrosomal function. Biol Reprod. 2001;65(5):1359–1363. doi: 10.1095/biolreprod65.5.1359. [DOI] [PubMed] [Google Scholar]
- 33.Terada Y, Simerly CR, Hewitson L, Schatten G. Sperm aster formation and pronuclear decondensation during rabbit fertilization and development of a functional assay for human sperm. Biol Reprod. 2000;62(3):557–563. doi: 10.1095/biolreprod62.3.557. [DOI] [PubMed] [Google Scholar]
- 34.Seibenhener ML, et al. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24(18):8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Mol Biol Cell. 2011;22(3):291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka A, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010;191(7):1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ju JS, et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009;187(6):875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33(1):109–122. doi: 10.1247/csf.08005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.