Significance
Many phages use a contractile tail to inject their genomes into the host bacterial cell. These complex assemblages are capable of specifically recognizing the cell surface, contracting upon cell binding, penetrating the cell wall, and creating a channel in the cell membrane through which the phage genome can pass. Remarkably, bacteria have co-opted phage tails and adapted them into protein-injecting nanomachines directed at other bacteria and even eukaryotic cells. Here, we elucidate the composition and assembly pathway of the baseplate of the contractile-tailed phage Mu. Our work demonstrates that the “simple” Mu-like baseplates are the only baseplate type encoded in phage genomes integrated within bacteria, and these baseplates are thus the most likely evolutionary precursors of the tail-derived bacterial nanomachines.
Keywords: baseplate, assembly, contractile tail, phage Mu
Abstract
Contractile phage tails are powerful cell puncturing nanomachines that have been co-opted by bacteria for self-defense against both bacteria and eukaryotic cells. The tail of phage T4 has long served as the paradigm for understanding contractile tail-like systems despite its greater complexity compared with other contractile-tailed phages. Here, we present a detailed investigation of the assembly of a “simple” contractile-tailed phage baseplate, that of Escherichia coli phage Mu. By coexpressing various combinations of putative Mu baseplate proteins, we defined the required components of this baseplate and delineated its assembly pathway. We show that the Mu baseplate is constructed through the independent assembly of wedges that are organized around a central hub complex. The Mu wedges are comprised of only three protein subunits rather than the seven found in the equivalent structure in T4. Through extensive bioinformatic analyses, we found that homologs of the essential components of the Mu baseplate can be identified in the majority of contractile-tailed phages and prophages. No T4-like prophages were identified. The conserved simple baseplate components were also found in contractile tail-derived bacterial apparatuses, such as type VI secretion systems, Photorhabdus virulence cassettes, and R-type tailocins. Our work highlights the evolutionary connections and similarities in the biochemical behavior of phage Mu wedge components and the TssF and TssG proteins of the type VI secretion system. In addition, we demonstrate the importance of the Mu baseplate as a model system for understanding bacterial phage tail-derived systems.
Bacteriophages (phages) are the dominant predators of bacteria and the most numerous biological entities on earth. An overwhelming majority of phages rely on a tail, which is responsible for specific binding to the bacterial host and injection of the phage genome into the cell. Contractile phage (myophage) tails, one of the three classes of tails, possess an outer sheath that surrounds a central tube. Upon contact with a host cell, the sheath contracts and drives the tail tube through the cell envelope and the genome is subsequently injected. Contractile tails are the most complex type of tail and are associated with phages bearing the largest genomes (1), suggesting that they may be the most powerful phage-genome delivery system. The remarkable capabilities of contractile phage tails are highlighted by the prevalence in bacteria of structures evolved from them, such as the type VI secretion system (T6SS) (2) and Photorhabdus virulence cassettes (PVCs) (3). Thus, characterization of contractile phage tails is vital both to increase our understanding of these complex molecular machines and to provide insight into mechanisms of bacterial pathogenesis.
The tail tube is attached at one end to the phage head and at the other end to the baseplate. The baseplate is a key component of the contractile tail because it mediates both host cell binding and genome injection. The baseplate of Escherichia coli phage T4, which has been intensively investigated, is comprised of six wedges assembled radially around a ring-shaped central hub (4). Attached to the wedges at the periphery of the baseplate are the tail fibers required for host cell binding; a centrally positioned spike protein is involved in membrane penetration (5). Although the T4 baseplate has long served as the paradigm for contractile phage tails, it is composed of more proteins and thus is much more complex than many other contractile tail baseplates. For this reason, we have undertaken studies on E. coli phage Mu, a contractile-tailed phage with a much simpler baseplate.
Sequence comparisons indicate that the baseplate of phage Mu is composed of only six proteins (5). Among these, the baseplate hub component Mup44 (6), the baseplate spike protein Mup45 (7), and the baseplate wedge subunit Mup46 are clearly related to the T4 baseplate proteins, gp27, gp5, and gp25, respectively. Mup47 is postulated to be located in the baseplate wedge because of its sequence similarity to gpJ, a proven wedge protein of E. coli phage P2 (8). The location and roles of the other putative baseplate proteins, Mup43 and Mup48, have not been experimentally addressed, but several lines of evidence point to Mup43 forming the upper ring of the hub that is attached to the tail tube (9). The Mu baseplate is an attractive object for investigation because it has been little studied, yet is representative of the simplest type of contractile tail baseplate (5). This “simple” type of baseplate is encoded frequently in both phages and prophages (phage genomes integrated into bacterial genomes) and we hypothesize that it is a more relevant model for comparison with the T6SS and other bacterial phage tail-derived apparatuses. In this study, we have undertaken biochemical and bioinformatic studies to characterize the composition and assembly of the phage Mu baseplate and determine the relationship of this baseplate to the diverse baseplates encoded in phages and prophages, as well as bacterial virulence operons.
Results
Protein Composition of the Phage Mu Baseplate.
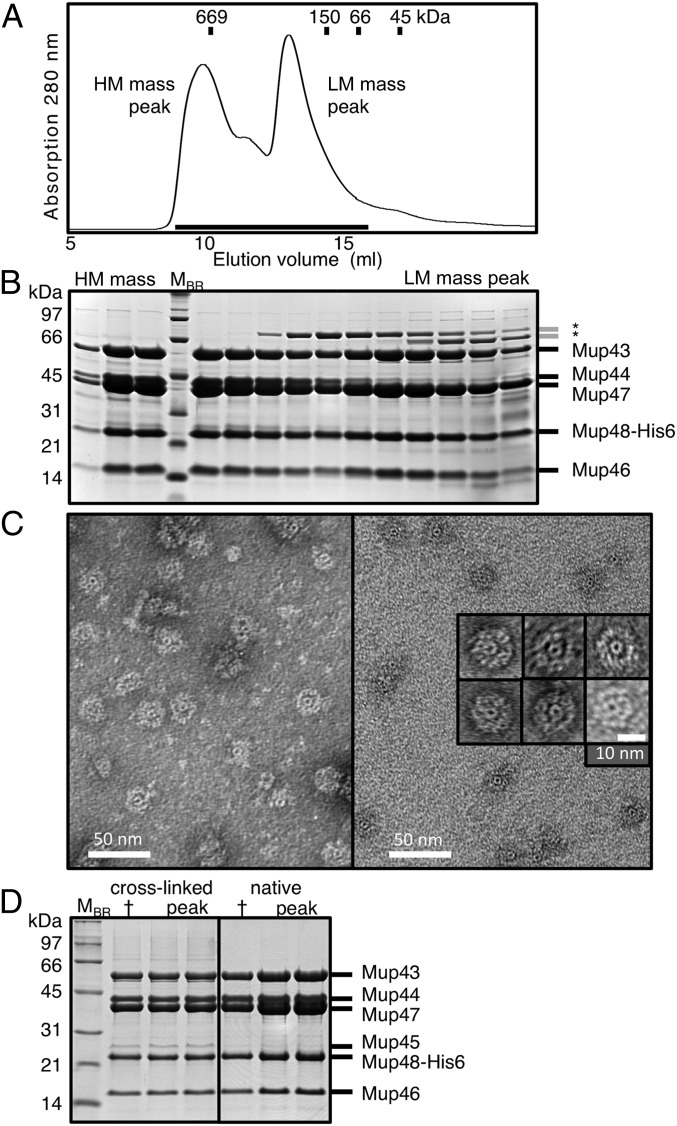
Previous sequence-based analyses suggested that Mu genes 43 to 48 encode the essential simple myophage baseplate components (5, 9). To prove that these components are necessary and sufficient for the formation of the Mu baseplate, we expressed these genes from a plasmid in their natural configuration with a hexahistidine (His6) tag encoded at the C terminus of Mup48. Protein purification using Ni-affinity chromatography followed by size-exclusion chromatography (SEC) yielded a high molecular (HM) mass complex of ∼900 kDa and a lower molecular (LM) mass complex of ∼200 kDa (Fig. 1A). The HM mass complex contained Mup43, Mup44, Mup46, Mup47, and Mup48 as determined by SDS/PAGE (Fig. 1B) and mass spectrometry (Fig. S1A). The isolation of these five proteins in the same elution fractions, even though only Mup48 was His6-tagged, demonstrated that these proteins were interacting within a large complex. Examination of the HM mass complex by electron microscopy (EM) using negative staining revealed disk-shaped objects (∼23 nm in diameter) with an outer ring surrounding a smaller doughnut-shaped central ring (∼6 nm diameter) (Fig. 1C). These structures are likely intact Mu baseplates, and their size is consistent with that estimated from examination of whole phage particles (10). We also observed structures that appeared to be partially disassembled baseplates. These structures had intact inner rings, but parts of the outer rings were missing (Fig. S2A).
Fig. 1.
Purification of Mu baseplate complexes. (A) A Mup43–48 complex containing a Mup48–His6 fusion was purified by Ni-affinity chromatography, and then fractionated by SEC. Eluted proteins were found in an HM mass and a LM mass peak. (B) Samples from each fraction collected across the two peaks, indicated by the line under the peaks in A, were analyzed on Coomassie-stained SDS/PAGE gels. MBR, protein molecular mass markers. Asterisks indicate contaminating bacterial proteins. (C) Negatively stained electron micrographs of phage Mu baseplate rings isolated from the HM mass fraction. (D) Mup43–48 complexes from normal cultures or from cells treated with formaldehyde to covalently cross-link the proteins were processed side-by-side as in A and their HM mass peak fractions analyzed on Coomassie-stained SDS/PAGE gels. Tryptic digest of the extra gel band coupled with mass spectrometry identified with greater than 95% probability Mup45 as the main protein component (79% sequence coverage, 16 detected unique peptides). The cross (†) indicates a fraction sample stored at −20 °C to serve as negative control for protein degradation over time.
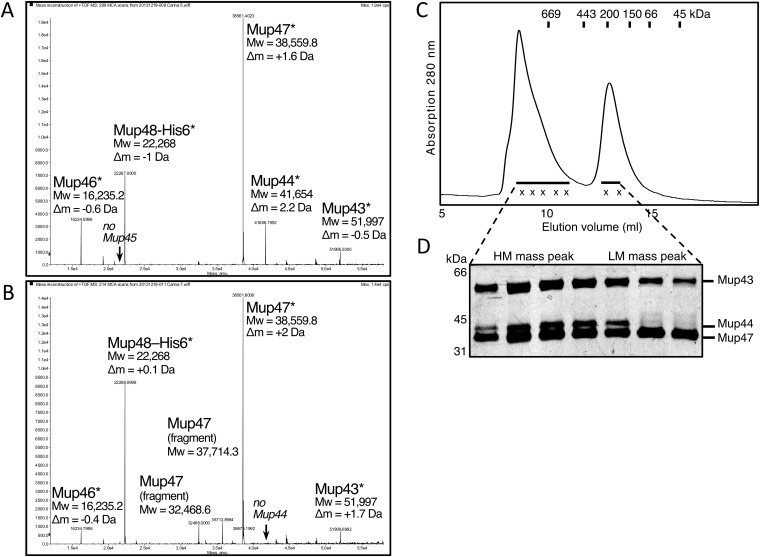
Fig. S1.
Mass spectrometry of Mup43–48 SEC peaks. Deconvoluted m/z mass spectra of the (A) baseplate containing HM mass peak and (B) LM mass peak. The protein name, its calculated molecular mass for the (modified) molecule, and the mass difference are listed next to the corresponding MS peak. Mup44, Mup46, Mup47, and Mup48 show loss of the N-terminal start–methionine (indicated by asterisk) because of E. coli aminopeptidase activity. In the LM mass peak spectrum, we also identified degradation fragments corresponding to N-terminally truncated Mup47 that are commonly observed on SDS/PAGE for constructs containing Mup47. (C) Mup43–48 HM mass peak, refractionated by SEC in running buffer lacking magnesium and calcium, disintegrates, thereby replicating the original two-peak SEC profile with the (D) loss of hub protein Mup44 in the LM mass peak (silver-stained SDS/PAGE, detail view).
Fig. S2.
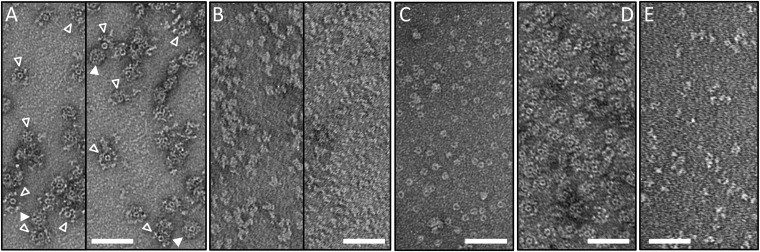
Instability of Mu baseplates. (A) Negatively stained electron micrographs of the Mup43–48 baseplate HM mass peak after SEC purification reveals intact baseplate rings (solid triangles) and to varying extent baseplates exhibiting various states of disintegration (open triangles) showing wedge protein subcomplexes that are detached from the central hub ring or missing. (B) Mup43–48 SEC LM mass peak contains amorphous structures likely being baseplate wedge proteins but lacks the central hub rings. (C) Mup44 hub control. (D) Using the same purification protocol as for the tail spike Mup45-containing construct in A, baseplate-like rings still assemble for a gene 45 deletion construct (Mup43–44–46–47–48His6, pCB100). (E) Negative-stain electron micrograph of wedge Mup46/47/48His6 (SEC peak fraction). The amorphous structures resemble those of the LM mass peak in B. (Scale bars, 50 nm.)
The LM mass complex isolated by SEC contained all of the proteins found in the HM mass complex with the exception of Mup44 (Fig. 1B and Fig. S1B). This complex likely forms by dissociation of Mup44 from the HM mass complex because refractionation of the purified HM mass complex by SEC resulted in reappearance of the LM mass complex lacking Mup44 (Fig. S1 C and D). Distinctive ring-containing complexes were not discernible in electron micrographs of negatively stained samples of the LM mass complex, implying that Mup44 is required to maintain the ring structure (Fig. S2B). To confirm that the central doughnut-shaped rings visible in the electron micrographs of the HM mass complex were composed of Mup44, we purified this protein on its own. The size and shape of the Mup44 complex was identical to the central ring in the HM mass complex (Fig. S2C). Taken together, these results show that intact Mu baseplates can be produced and purified using our plasmid expression construct.
A surprising finding in the experiments described above was the absence of the central spike protein, Mup45, in the purified baseplate complexes, even though its encoding gene was present in the expression plasmid used. To determine whether Mup45 was unstably bound to the baseplate and lost during purification, we added formaldehyde cross-linker to the cell culture and repeated the purification. In this case, we did observe Mup45 coeluting with the other baseplate components in the HM mass peak (Fig. 1D), implying that this protein binds weakly to the baseplate complex. To demonstrate that Mup45 was not required for the formation of the baseplate, we showed that the same HM mass baseplate complex as observed above could be purified from cells bearing a plasmid construct lacking gene 45 (Fig. S2D).
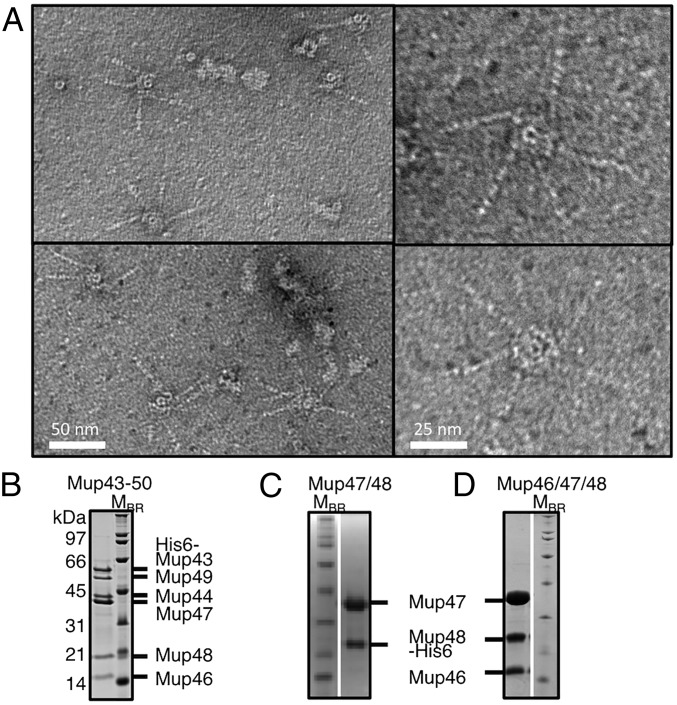
To determine whether our observed baseplate structures were able to bind tail fibers, we created a construct expressing genes 43 to 50, where genes 49 and 50 encode the tail fiber and tail-fiber assembly protein, respectively. Using this construct, with an N-terminal His6-tag encoded on Mup43, we isolated a complex that contained baseplate structures with tail fibers attached as observed by EM (Fig. 2A). The presence of these structures indicated that the plasmid-produced baseplate complexes are genuine intermediates in tail assembly capable of incorporating fibers (Fig. 2B). The number of attached fibers per baseplate ranged from one to five, which deviates from the six fibers per baseplate expected, given the known sixfold symmetry of all characterized baseplates. It is possible that fibers dissociated from the baseplates during EM grid preparation because detached fibers were also frequently observed on these grids.
Fig. 2.
Purification of baseplates with attached fibers and assembly intermediates. (A) Representative electron micrographs of purified Mup43–50 complexes, which include tail fibers. (B) The peak fraction of the HM mass complex of Mup43–50 isolated by SEC was analyzed by SDS/PAGE. The Mup43–50 complex was purified using Ni-affinity chromatography by using an N-terminally His6-tagged Mup43 then fractionated by SEC (Fig. S3G). SDS/PAGE gels are shown of the Mup47/Mup48His6 complex (C) and the Mup46/Mup47/Mup48His6 complex (D), after Ni-affinity purification and SEC fractionation (see Fig. S5 for extended SEC data).
The Assembly Pathway and Stoichiometry of the Mu Baseplate.
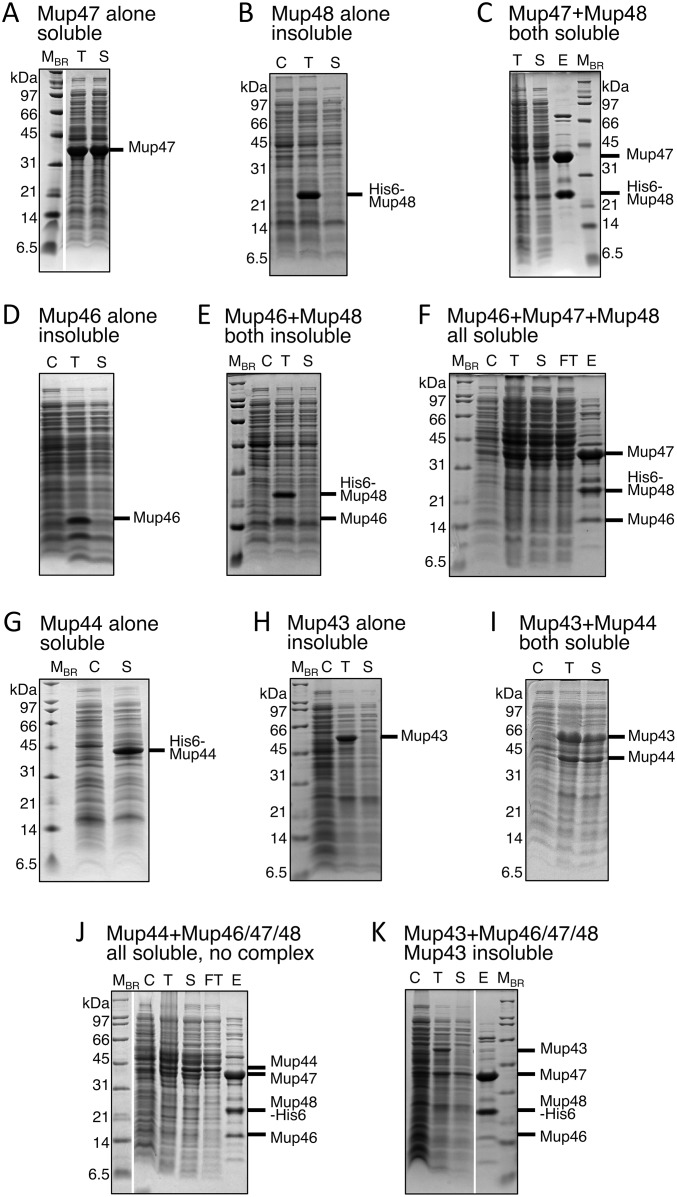
To elucidate the pathway of baseplate assembly we constructed plasmids that produced various combinations of baseplate proteins (Table S1). We discovered that among Mup46, Mup47, and Mup48, only Mup47 could be produced on its own in soluble form (Figs. S3A and S4A). Mup46 and Mup48 were insoluble when produced individually or together (Fig. S4 B, D, and E). However, overproduction of His6-tagged Mup48 together with Mup47 yielded a stable soluble complex (Fig. S4C) that could be copurified by Ni-affinity chromatography followed by SEC (Fig. 2C and Fig. S3B). In addition, the coexpression of genes encoding Mup46, Mup47, and Mup48 resulted in the formation of a complex containing all three proteins (Fig. 2D and Fig. S3C). Because the homologs of Mup46 and Mup47 are baseplate wedge proteins in phage P2 (8), we concluded that the Mup46/47/48 complex constitutes the baseplate wedge of phage Mu. Examination of this complex by EM revealed no distinctive structures resembling baseplates (Fig. S2E). Coproduction of Mup46/47/48 with Mup44, which forms a soluble trimeric ring on its own (6, 11) (Figs. S3E and S4G), did not result in a complex containing all four proteins (Fig. S4J). Putative baseplate hub component Mup43 was insoluble when produced on its own (Fig. S4H) or with the Mup46/47/48 wedge (Fig. S4K), but formed a soluble complex with Mup44 (Fig. S4I). We conclude that Mup43 and Mup44 comprise a baseplate hub that must form before interaction with the baseplate wedges comprised of Mup46/47/48 (Fig. 3).
Table S1.
List of expression constructs with test expression results
| Construct | Gene(s) | Vector | Affinity tag | Tag position | Expression |
| pCB98 | 43 | pCDF1b | — | — | Insoluble |
| pCB40 | 44 | pCDF1b | His6 | N-terminal | Soluble |
| pCB94 | 46 | pAD100 | — | — | No |
| pCB101 | 46 | pCDF1b | — | — | Insoluble |
| pCB6 | 47 | pET32b* | Trx-His6-TEV | N-terminal | Soluble |
| pCB68 | 47 | pCDF1b | — | — | Soluble |
| pCB15 | 48 | pAD100 | FLAG-His6 | C-terminal | No |
| pCB44 | 48 | p15TV-L | His6-TEV | N-terminal | Insoluble |
| pCB108 | 48 | pCDF1b | — | — | No, reduced culture growth |
| pCB119 | 43–44 | pCDF1b | — | — | Soluble, Mup43, -44 |
| pCB82 | 43–44–45–46–47–48 | pAD100 | FLAG-His6 | C-terminal | Soluble, Mup43, -44, -46, -47, -48; 45† |
| pCB83 | 44–45–46–47–48 | pAD100 | FLAG-His6 | C-terminal | Soluble, Mup44, -46, -47, -48 |
| pCB85 | 46–47–48 | pAD100 | FLAG-His6 | C-terminal | Soluble, Mup46, -47, -48 |
| pCB111 | 46–47–48 | p15TV-L | His6-TEV | N-terminal | Soluble, Mup46, -47, -48 |
| pCB14 | 47–48 | pAD100 | FLAG-His6 | C-terminal | Soluble, Mup47, -48 |
| pCB63 | 47–48 | pET32b* | Trx-His6-TEV | N-terminal | Soluble, Mup47, -48 |
| pCB100 | 43–44–46–47–48 | pCB85‡ | FLAG-His6 | C-terminal | Soluble, Mup43, -44, -46, -47, -48 |
| pCB105 | 43–44–45–46–47–48–49–50 | p15TV-L | His6-TEV | N-terminal | Soluble, Mup43, -44, -46, -47, -48, -49 ,-50 |
| pCB127 | 43–44–45–46–47–48–49–50 | pAD100 | His6 | N-terminal | Soluble, Mup43, -44, -46, -47, -48, -49, -50 |
Vector backbones pCDF1b and pET21d (EMD Millipore), pAD100 (33), p15TV-L (Clinical Genomics Centre, University Health Network, Toronto, ON, Canada). Tagged ORF in multicistronic constructs are underlined. His6, hexahistidine tag; Trx, thioredoxin tag.
Modified expression vector pET32b (Novagen), enterokinase protease cleavage site replaced with tobacco virus etch (TEV) protease site.
Detected upon cross-linking.
Cloned into 5′ NcoI site, introducing a 3′ ribosome binding site.
Fig. S3.
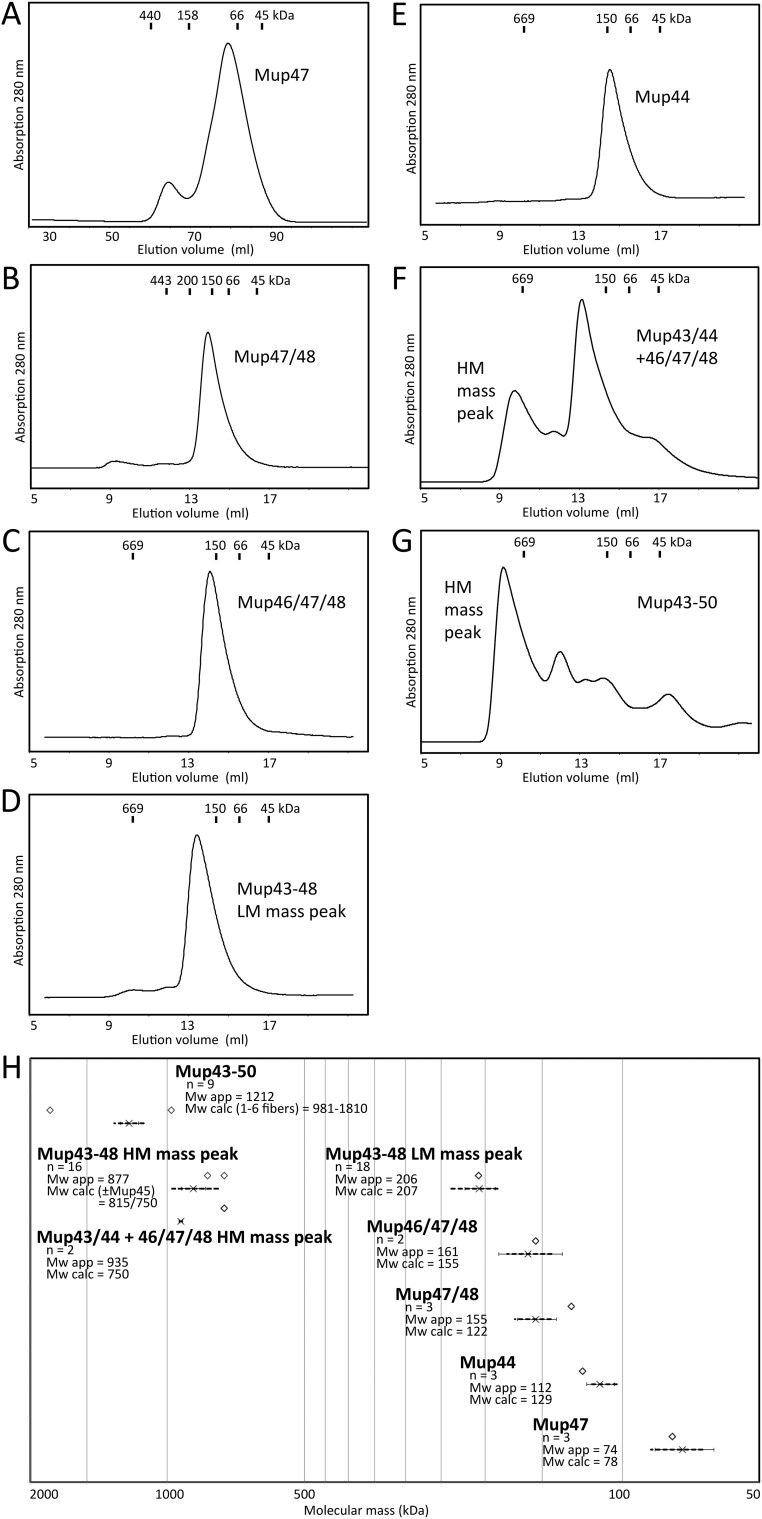
Average molecular mass of Mu baseplate complexes/components determined by SEC. Representative size-exclusion chromatograms of phage Mu baseplate components and subcomplexes. (A) Mup47 (construct pCB6), after tag proteolysis. Note: Mup47 tends to form higher molecular aggregates dependent on concentration; therefore, the larger Superdex XK 16/60 column was used. (B) Mup47/48His6 (pCB14). (C) Mup46/47/48His6 (pCB85), wedge. (D) Mup43–48His6 (pCB82), LM mass peak. (E) His6–Mup44 (pCB40). (F) Mup43/44 + 46/47/48His6 (pCB119 + pCB85). (G) His6–Mup43–50 (pCB127). (H) Average apparent molecular mass (Mw app) is shown with error bars (SD) and data range (dashed lines). Predicted molecular mass (Mw calc), or range, of complexes are indicated above by open diamonds. n, number of runs/ protein batches.
Fig. S4.
Recombinant expression of putative Mu baseplate genes, and protein purification by Ni-affinity chromatography, where applicable. (A) Mup47 (construct pCB68). (B) His6–Mup48 (pCB44). (C) Mup47 (pCB68) with His6–Mup48 (pCB44). (D) Mup46 (pCB101). (E) Mup46 (pCB101) with His6–Mup48 (pCB44). (F) Mup46/47/48His6 (pCB85). (G) His6–Mup44 (pCB40). (H) Mup43 (pCB98). (I) Mup43/44 (pCB119). (J) Mup44 (pCB40) with Mup46/47/48His6 (pCB85). (K) Mup43 (pCB98) with Mup46/47/48His6 (pCB85) on Coomassie-stained SDS/PAGES. C, noninduced cells; E, Ni-NTA agarose eluate; FT, Ni-NTA agarose flowthrough; His6, hexahistidine tag; MBR, protein molecular mass markers; S, soluble lysate fraction; T, total cell lysate.
Fig. 3.
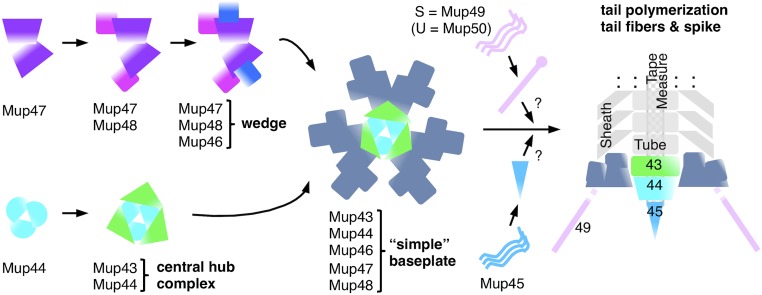
Model of the simple baseplate assembly pathway in phage Mu. Soluble protein Mup47 and hub protein Mup44 initiate the formation of two subcomplexes: wedge (Mup46/47/48) and central hub complex (Mup43/44). A Mup47 dimer interacts first with Mup48 and then this complex interacts with Mup46. The complete wedge contains two copies of each subunit. Three of these wedge dimers assemble around the trimeric central hub complex Mup43/44, forming the baseplate. Mup45 and the tail fibers may assemble onto the baseplate after joining of the wedges and hub, but this timing is not established.
Fig. S5.
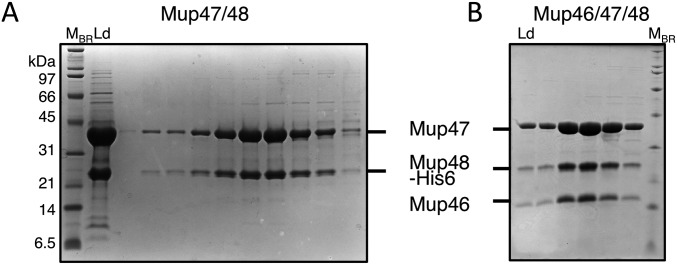
Purification of baseplate wedge complexes using SEC. Samples from fractions across the peak obtained for (A) Mup47/48His6 (pCB14) and (B) Mup46/47/48His6 (pCB85) were analyzed on Coomassie-stained SDS/PAGE gels. His6, hexahistidine tag; Ld, input; MBR, protein molecular mass markers.
Densitometry of SDS/PAGE gels revealed that the subunits of the Mup47/48 and Mup46/47/48 complexes were present in equal relative amounts (Tables S2 and S3). The SEC elution volumes of Mup47, Mup47/48, and Mup46/47/48 were consistent with each of these complexes containing two molecules of each subunit. For example, the Mup46/47/48 complex eluted at an apparent molecular mass of ∼160 kDa (Fig. S3 C and H). A 2:2:2 complex of these proteins accounts for this size (predicted molecular mass = 155 kDa). Densitometry of complete baseplate complexes showed that there were twice as many molecules of Mup46/47/48 compared with the number of Mup43 and Mup44 molecules. Because Mup44 forms a trimer and three copies of its homolog, gp27, are present in the phage T4 baseplate (12), we conclude that the Mu baseplate is composed of Mup43/44/46/47/48 at a 3:3:6:6:6 ratio. Thus, a trimeric hub composed of Mup43 and Mup44 assembles with three dimeric Mup46/47/48 baseplate wedges to form the baseplate (Fig. 3). The dimeric nature of the Mu baseplate wedge contrasts with the T4 wedge, which contains subunits ranging in number from one to three and does not form a dimer before interaction with the hub complex (13).
Table S2.
Densitometric volumes
 |
Representative Coomassie-stained SDS/PAGE gels of peak fractions isolated using SEC.
Table S3.
Densitometric volume-derived molar ratios
| Protein | Mup46-48His6 peak fraction | Mup43-48His6 HM mass peak | Mup43-48His6 LM mass peak | ||||||
| Normalized to | Mup47 | Mup44 | Mup43 | ||||||
| Sample/-fold | 1x | 2.5x | 5x | 1x | 2.5x | 5x | 1x | 2.5x | LM mass Pool |
| Mup43 | 0.98 | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | |||
| Mup44 | 1.00 | 1.00 | 1.00 | ||||||
| Mup47 | 1.00 | 1.00 | 1.00 | 1.75 | 1.68 | 1.57 | 2.33 | 2.29 | 2.18 |
| Mup48His6 | 0.98 | 0.90 | 0.92 | 1.81 | 1.87 | 1.97 | 2.23 | 2.63 | 2.20 |
| Mup46 | 1.02 | 1.11 | 1.10 | 1.30 | 1.78 | 2.05 | 1.94 | 2.40 | 2.04 |
Molar ratios derived from densitometric volumes in Table S2.
Defining the Broadly Conserved Components of the Simple Contractile Tail Baseplate.
After defining the proteins comprising the Mu baseplate, we investigated their conservation among contractile-tailed phages. To circumvent the failure of sequence similarity searches (PSI-BLAST) to identify all homologs of a given baseplate protein because of the large diversity of phage proteins, we used the canonical arrangement of phage genes to aid the annotation of additional baseplate proteins (Fig. 4). Searches with these proteins identified homologs and Pfam and TIGRfam protein families that corresponded to phage baseplate functions. We found 26 protein families (Dataset S1) that contained homologs of the Mu baseplate proteins. We identified additional homologs using the protein family-associated Hidden Markov Models (HMMs) and confirmed the relatedness between protein families assigned to the same baseplate function with the HMM-based profile-profile comparison tool, HHpred. In the process of annotating baseplate proteins we discovered 25 new families of baseplate proteins (Dataset S1).
Fig. 4.
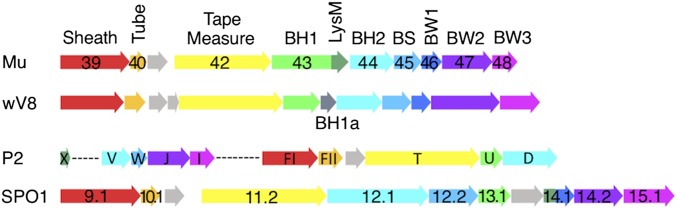
Synteny of the baseplate gene region of phage genomes. A schematic view of the tail-encoding regions of diverse contractile-tailed phages is shown. Arrows represent ORFs and are drawn to scale. ORFs are colored according to function. Names of genes are included for characterized phages. Three of the most common gene arrangements are shown here. Phage wV8 is a genome encoding at BH1a protein. Dashed lines in the P2 genome represent regions of nontail genes that are not shown. Phage genomes represented are E. coli phages Mu, P2, and wV8, and Bacillus subtilis phage SPO1.
Using our collection of protein families and associated HMMs, we analyzed 207 contractile-tailed phage genomes. We found that 103 encoded “simple” baseplates comprised of homologs of each of the six phage Mu baseplate proteins (Dataset S2), and 76 phages encoded baseplates similar to E. coli phage T4 (Dataset S3). Of the 28 remaining phage genomes, 14 encoded baseplates weakly related to the Mu baseplate (i.e., homologs to some of the Mu baseplate proteins could be identified), and 14 possessed few or no recognizable baseplate proteins (Dataset S3). The prevalence of phages bearing Mu-like baseplates led us to introduce a standard nomenclature for their baseplate proteins. The families including the baseplate hub proteins, Mup43 and Mup44, are referred to as BH1 and BH2, respectively. The baseplate spike protein family, which includes Mup45, is called BS, and the three baseplate wedge protein families of Mup46, Mup47, and Mup48, are referred to as BW1, BW2, and BW3, respectively (Table S4). Some phages that encode all of the simple baseplate proteins possess larger baseplates than phage Mu (Dataset S2). For example, the SPO1-like family of phages (e.g., Listeria phage A511) has very large BW3 and BH2 proteins (on average 860 and 820 residues vs. 240 and 340 residues for other simple baseplates) (Dataset S2). Larger baseplate proteins like these often comprise one or more functional domains in addition to the conserved family-defining domain. The SPO1-like phages and others also possess additional baseplate proteins that are not broadly conserved. In addition to the six absolutely conserved protein families, 51 of 103 simple baseplates include a LysM domain. These ∼60-residue domains may be encoded separately or fused to BH1 or BW1 family proteins (9). Strikingly, of the remaining 52 simple baseplate phages lacking LysM domains, we discovered that 46 encode a small protein close to the BH1-encoding gene (60% are immediately downstream), which we refer to as BH1a. Only one baseplate region encodes both a LysM domain and a BH1a protein. The previously unrecognized BH1a family of proteins may play a similar role as the LysM domains.
Table S4.
Nomenclature of simple baseplate components and their homologs in phages Mu, P2, T4, and the T6SS
| Phage proteins | T6SS | Component | Functional annotation | ||
| Mu | P2 | T4 | |||
| Mup43 | P2 gpU | —* | —* | BH1 | Baseplate hub protein 1 |
| Mup44 | P2 gpD | T4 gp27 | VgrG† | BH2 | Baseplate hub protein 2 |
| Mup45 | P2 gpV | T4 gp5 | VgrG† | BS | Baseplate spike protein |
| Mup46 | P2 gpW | T4 gp25 | TssE | BW1 | Baseplate wedge protein 1 |
| Mup47 | P2 gpJ | T4 gp6‡ | TssF | BW2 | Baseplate wedge protein 2 |
| Mup48 | P2 gpI | —* | TssG | BW3 | Baseplate wedge protein 3 |
No homologs could be identified.
BH2-BS fusion protein.
N-terminal region.
To extend our bioinformatic analysis to phage genomes present as prophages within bacterial genomes, we surveyed 2,119 bacterial genomes. We identified 560 putative contractile-tailed prophages where a phage major head protein was encoded within 30 ORFs of both a tail sheath protein and BW1 protein, which are conserved in all types of contractile tails. Among these 560 putative prophages, 418 (75%) encoded all six conserved components of the simple baseplate and another 65 (11%) encoded close to complete simple baseplates with only one or two conserved components that could not be annotated (Dataset S4). No putative prophage contained a cluster of genes (i.e., more than two within 20 genes) encoding phage T4-specific baseplate proteins as detected by the T4 baseplate-specific HMMs listed in Dataset S1. These results imply that prophages harbor only simple contractile tail baseplates.
In surveying bacterial genomes for prophages, we also identified 112 operons encoding putative contractile tail-like bacteriocins, also known as R-tailocins (14). These operons displayed genes required for production of a contractile tail, but lacked any genes encoding a head protein. These putative R-tailocin operons were indistinguishable from phage-associated contractile tail operons and exclusively encoded simple baseplates (Dataset S5).
Phage-Related Components of Bacterial Tail-Like Apparatuses.
The bacterial T6SS clearly possesses proteins related to the contractile tail sheath, tube, BW1, BH2, and BS proteins (15), implying that this system is evolutionarily related to phage tails. Given that prophages provide the most abundant source of phage genes that could be adapted for other purposes by their bacterial hosts, it is probable the T6SS originally evolved from a prophage. As described above, all of the contractile-tailed prophages that we could detect encode simple baseplates. Thus, we expect the T6SS to encode homologs of the simple baseplate components. Consistent with this idea, Brunet et al. (16) recently reported that TssF and TssG, two conserved components of the T6SS, could be bioinformatically connected to BW2- and BW3-containing protein families. To further elucidate the evolutionary relationships between contractile tails and the T6SS, we analyzed operons encoding T6SSs and PVCs. PVCs are a separate class of contractile tail-related apparatuses that have been implicated in killing insect larva and tubeworm morphogenesis (17–19). We analyzed these because they are more closely related to phage tails than the T6SS, yet display features of a secretion system (3, 17). PVC operons were easily identified in bacterial genomes because they encode tail sheath and tube proteins, all six components of the simple contractile baseplate, as well as a distinctive AAA+ ATPase and a unique tail terminator protein (Pfam DUF4255) (3, 5, 20). In analyzing PVC baseplate components using HHpred, we discovered that their BW2 and BW3 proteins could often be strongly linked (probabilities > 90%) to the TssF and TssG protein families (Fig. 5 A and B). In addition, two PSI-BLAST iterations with a PVC BW3 protein (YP_003168847.1 from a Candidatus species) hit both phage BW3 proteins and T6SS TssG proteins. Similarly, searches with the BW2 protein from the same PVC (YP_003168848.1) yielded hits to both phage BW2 and T6SS TssF proteins. PSI-BLAST searches initiated with phage BW2 or BW3 proteins, or TssF or TssG proteins, did not reveal links between phage BW2 or BW3 proteins and the T6SS. HHpred searches with these phage and T6SS baseplate components yielded only low probability links for the BW2 proteins in some cases and no links for the BW3 proteins. These results indicate that the PVC BW2 and BW3 proteins are evolutionary intermediates between the phage and T6SS proteins. A striking distinction between contractile tails and the T6SS is that the BH2 protein is invariably fused to the T6SS tail spike (BS) (Dataset S5), whereas in phages with simple baseplates this rarely occurs, with only six examples in our survey (Dataset S2). Interestingly, BH2–BS fusions were observed in approximately half of the PVC operons (Dataset S5), consistent with these apparatuses being intermediate between phages and T6SSs. The BH1 protein is an example of a common baseplate feature encoded in all PVC and phage-tail operons yet entirely lacking in the T6SS.
Fig. 5.
Similarities between phage, T6SS, and PVC baseplate wedge proteins. HHpred-detected similarities between BW2 (A) and BW3 (B) proteins and protein families from the Pfam database are shown. Colored rectangles represent regions of similarity with the Pfam family HMMs indicated in the legend. Numbers in the rectangles indicate regions of the given HMM that matched. Hatched rectangles indicate matches with probabilities between 60% and 90%. Nonhatched rectangles represent matches with greater than 90% probability. Blue lines indicate matches to the structure of phage T4 gp6 (PDB ID code 5HX2), with dark blue indicating matches to residues 20–150 and light blue indicating matches to residues 300–450. The proteins shown are examples of TssF (YP_002811746.1), TssG (YP_002811747.1), BW2 (YP_003168848.1), and BW3 (YP_003168847.1) of a PVC, and of E. coli phages Mu and P2 (Datasets S2 and S5).
In 80% of contractile-tailed phage genomes, the genes encoding the BW1, BW2, and BW3 proteins were found adjacent to each other and in a conserved order (BW1, BW2, then BW3) (Dataset S2). In surveying 280 T6SS operons, we found the tssF and tssG genes located immediately adjacent in 99% of cases (Dataset S5) and the gene encoding the BW1 homolog (TssE) within three ORFs upstream of the tssF gene 93% of the time. Strong genetic linkage between these three genes in a smaller group of T6SS operons was previously reported (21). PVC operons also displayed the same conserved BW1-BW2-BW3 gene order, with these genes being adjacent greater than 90% of the time (Dataset S5). The similarly conserved syntenic relationships of the phage baseplate wedge genes, the T6SS tssE, tssF, and tssG genes, and the homologous PVC genes strongly support the evolutionary connection and shared functions of these genes.
Discussion
This work provides an experimental elucidation of the composition and assembly pathway of a simple contractile tail baseplate. The assembly pathway of phage Mu is likely applicable to all phages with simple baseplates because, despite their diversity, genes encoding the same components are invariably found across all types of these phages. We have shown that five proteins are required for baseplate assembly with BW1 (Mup46), BW2 (Mup47), and BW3 (Mup48) proteins comprising the baseplate wedge and BH1 (Mup43) and BH2 (Mup44) forming the central hub on which the wedges are recruited. Assembly of the baseplate wedge occurs through stepwise interaction of BW2 with BW3, followed by subsequent addition of BW1. BH1 and BH2 must assemble to form the central hub of the baseplate before binding to the baseplate wedges. However, we also found that complexes of wedges bound to BH1 readily dissociate from the complete baseplate once it has been formed, implying that BH1 forms stronger interactions with the wedges. A feature conserved in approximately half of simple baseplates is the presence of LysM domains. We previously showed that in E. coli phage P2 the LysM-containing protein, gpX, is located at the interface of the baseplate and tail sheath in the same region of the baseplate as the BW1 protein, gpW (9). In some phages, such as Mu, the LysM domain is fused to the BH1 protein, and in others it is fused to BW1. We hypothesized that the LysM domain is involved in joining the wedge to the baseplate hub by serving as part of the interface between BH1 and BW1. The newly discovered BH1a proteins, which are similarly sized to LysM domains and occur only in baseplates lacking these domains (with one exception), may also play a role in joining the wedges to the hub. Overall, our experimental and bioinformatic studies provide a role for each of the conserved components required for assembly of the simple contractile tail baseplate, and demonstrate that simple baseplates related to that of Mu predominate among the sequenced genomes of contractile-tailed phages and prophages.
The considerably more complex baseplates of T4-like phages share some components with the simple baseplates, possessing clear homologs of BW1 (gp25 in T4), BH2 (T4 gp27), and BS (T4 gp5) (5); however, no proteins directly related to the BH1 or BW3 protein families could be detected. Based on positioning in the T4 baseplate structure, it was previously speculated that BW3 might be related to T4 gp53 or gp7 (5). However, HHpred analyses of BW3 proteins revealed high-probability hits to the HMMs corresponding to BW2 proteins (Fig. 5B), indicating that these families are distantly related to each other and likely evolved through a gene duplication event. Assembly of the T4 wedge initiates with the sequential interactions of proteins gp10, gp7, and gp8 (13), none of which have equivalents in phage Mu. Similarity can be detected between BW2 proteins and the N-terminal half of T4 gp6 (Fig. 5A), which is incorporated into the wedge next. T4 gp53, added to the wedge after gp6, is a unique component of the T4 baseplate but does bear a LysM-like domain, which may play a similar role to LysM domains in simple baseplates. Complete T4 wedges assemble into a hexameric “star” conformation similar to the baseplate conformation observed in the contracted tail (13). In contrast, the Mu baseplate wedges did not assemble into stable ring-like structures in the absence of the hub proteins. Although the simple and T4-like baseplates are related through evolution, their components and modes of assembly have clearly diverged.
Features of the simple contractile baseplate bear similarities to the baseplates of noncontractile-tailed Lactococcus phages p2 and TP901-1 (22). These baseplates are composed of a central hub comprised of Dit and Tal, joined to at least six receptor-binding complexes. The N-terminal domain of Tal displays the same structure as the BH2 proteins Mup44 and gp27 of phages Mu and T4. The N-terminal domain of Dit possesses a “tail tube-fold” (23) and HHpred searches with Mup43/BH1 proteins result in high-probability hits to the Pfam HMM corresponding to the tail tube-fold region of Dit (PF05709: Sipho_Tail). Gene order analysis also supports a functional similarity between Dit and BH1 proteins as their genes generally lie between the genes encoding the tape-measure protein and BH2/Tal proteins. The ability of Mup43 to remain bound to the baseplate wedges after dissociation of Mup44 also parallels the noncontractile tails where a domain of Dit distinct from the tail tube-fold domain shows extensive interactions with the receptor-binding proteins, whereas Tal (BH2) displays minimal interactions with these proteins (24, 25). Although there is no detectable sequence or structural similarity between the receptor binding proteins of noncontractile tails and the baseplate wedges of contractile tails, the resemblance in the hubs of these two types of phages suggests an evolutionary connection.
Mup45, the baseplate spike protein, BS, is conserved, but was not required for assembly of the Mu baseplate; it associated weakly with the baseplate and could only be detected when protein cross-linker was added. The BS protein may attach more strongly to a complete tail through interactions with the tape-measure protein, which is likely assembled into the baseplate before tail tube polymerization. The weak association of the Mu BS protein with the baseplate parallels observations made for phages phi92 and T4. For example, in T4, dissociation of BH2 (gp27) and BS (gp5) can be triggered by lowering the pH, a condition that would be encountered upon contact with the periplasm (26). Weak binding of the central spike protein may reflect the requirement to give way upon sheath contraction to facilitate the breach of the bacterial cell envelope and allow genome injection through the tail tube. In contrast, T6SSs and PVCs may not need to eject their BS protein because they likely do not deliver cargo through the tube interior (2, 27). Consistent with this finding, the T6SS invariably features BH2–BS fusion proteins, and PVCs often do. Effectors for the T6SS are frequently fused to the BS or a PAAR (proline-alanine-alanine-arginine)-repeat protein bound to it (28). These distinctive properties may explain why BH2–BS fusion proteins are rare for phage tails and common for T6SSs and PVCs.
Prophages, which are found in most bacterial genomes, provide a huge reservoir of phage components that can be adapted for purposes advantageous to the bacterial host. In surveying prophages encoding contractile tails in more than 2,000 bacterial species, all that could be identified bore simple baseplates. Thus, we expect that all contractile tail-derived bacterial entities, including the T6SS, evolved from simple contractile tails, and that phages like Mu provide the best model for understanding these systems. In this work, we have demonstrated further similarities between the T6SS and simple baseplates. Our bioinformatic data, based on sequence similarity and genomic synteny analysis, strongly support the previously proposed evolutionary connection between BW2 and BW3 family proteins and the conserved T6SS proteins, TssF and TssG (16). In addition, our demonstration that BW2 and BW3 of phage Mu interact and the solubility of BW3 is dependent upon interaction with BW2, parallels the finding that TssF and TssG interact and are unstable unless produced together (16). Further solidifying the relationship between the simple baseplate and the T6SS, we have identified the PVC baseplate as an evolutionary intermediate between these two structures. Recognition of these evolutionary connections provides an important paradigm for the investigation of the phage tail-related entities of bacteria.
Materials and Methods
Protein Production, Purification, and Analysis.
The expression plasmids created for this study are summarized in Table S1. The details of protein purification and analysis are described in SI Materials and Methods.
Transmission Electron Microscopy.
Samples were applied to carbon-coated 400-mesh rhodium/copper grids and stained with 1% uranyl acetate. EM images were acquired using a Hitachi H-7000 electron microscope at an operational voltage of 75 kV and captured with a digital charge-coupled-device camera (ORCA-HR, Hamamatsu Photonics).
Bioinformatic Analyses.
The database used for all analyses was comprised of 755 tailed phage genomes and 2,119 bacterial genomes downloaded from the National Center for Biotechnology Information (NCBI) Refseq database in April 2013 (see SI Materials and Methods for details). Phage proteins of given function were preliminarily identified using iterated PSI-BLAST searches (29) and the HMMs are listed in Dataset S1. All hits with these HMMs to an e-value of 10 were collated into a single file and arranged according to the order of their Refseq accession numbers. In this way, clusters of hits comprising phage, PVC, or T6SS operons could be identified. HMM hits to proteins that did not lie in clusters were not analyzed further. This work was aided by a synteny viewing toolkit developed in our laboratory, which will be described elsewhere. HHpred searches were carried out using the online server (https://toolkit.tuebingen.mpg.de/hhpred) (30). HMM-based searches were carried out using HMMER3 (31) and analyzed by searching the Pfam (32) and TIGRfam (www.jcvi.org/cgi-bin/tigrfams/index.cgi) databases.
SI Materials and Methods
Protein Production, Isolation, and Analysis.
For protein production, Escherichia coli ER2566 cells containing expression plasmids were grown in LB medium with appropriate antibiotics at 37 °C to an OD600 of ∼0.8, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and incubated overnight at 20 °C. For in vivo cross-linking, 1:500 (vol/vol) of 37% (wt/vol) formaldehyde was added to the culture just before harvesting, incubation continued for 30 min at 37 °C, after which the reaction was quenched by adding 1:10 (vol/vol) of 2 M glycine. After centrifugation, cells were resuspended in buffer A [50 mM Tris⋅HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole, 5% (vol/vol) glycerol, 10 mM β-mercaptoethanol, 1× protease inhibitor mixture (PIC; Sigma)] with 0.01 mg/mL lysozyme, incubated on ice for 30 min, and then lysed by sonication. Proteins were affinity-purified using Ni-NTA agarose resin (Thermo Scientific) with 20 mM/250 mM imidazole in wash/elution buffers, respectively, and dialyzed into buffer B (20 mM Tris⋅HCl, pH 7.5, 200 mM/ 500 mM NaCl, 2.5% (vol/vol) glycerol, 1 mM DTT, 1× PIC). Purification of whole baseplates was performed in buffer C (20 mM Tris⋅HCl, pH 7.5, 500 mM NaCl, 2.5% (vol/vol) glycerol, 1–10 mM MgSO4, 1 mM CaCl2, 1 mM DTT, 1× PIC). Thioredoxin fusion proteins were cleaved using tabacco etch virus (TEV) protease. Protein complexes were analyzed using SEC on an ÄKTA Fast performance liquid-chromatography system (GE Healthcare) using Superdex 200 10/30 and 16/60 columns. Calibration was performed by fractionation of 100 μL of a sample protein, or a mixture of two well-resolved size-exclusion standards (Sigma-Aldrich).
Proteins were analyzed using 15% (wt/vol) discontinuous polyacrylamide gels in Tris-tricine SDS/PAGE. Coomassie Brilliant Blue G250-stained gels were scanned using ChemiDoc MP Imager and analyzed in ImageLab v4.1 (Bio-Rad). Electrospray ionization QqTOF spectra of purified Mu protein samples were collected on a linear ion-trap instrument (ThermoFisher LTQ).
Database Used for Bioinformatic Analyses.
Although many more genomes are now available, we continued our analysis on the genomes downloaded from the NCBI Refseq database in April 2013, because its manageable size allowed exhaustive and detailed analysis using both automated and manual approaches. In addition, the old Refseq accession numbers used in these genomes (i.e., those beginning with YP or NP) allowed genomic synteny to be easily assessed because these accession codes were numbered sequentially across individual genomes. This is not the case with the new NCBI Refseq accession numbers that start with WP.
Supplementary Material
Acknowledgments
We thank Battista Calvieri and Steven Doyle for technical advice on electron microscopy. This work was supported by Canadian Institutes of Health Research operating Grants MOP-136845 (to K.L.M.) and XNE-86943 and MOP-115039 (to A.R.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607966113/-/DCSupplemental.
References
- 1.Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:115–142. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- 2.Cianfanelli FR, Monlezun L, Coulthurst SJ. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016;24(1):51–62. doi: 10.1016/j.tim.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Sarris PF, Ladoukakis ED, Panopoulos NJ, Scoulica EV. A phage tail-derived element with wide distribution among both prokaryotic domains: A comparative genomic and phylogenetic study. Genome Biol Evol. 2014;6(7):1739–1747. doi: 10.1093/gbe/evu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10(9):688–693. doi: 10.1038/nsb970. [DOI] [PubMed] [Google Scholar]
- 5.Leiman PG, Shneider MM. Contractile tail machines of bacteriophages. Adv Exp Med Biol. 2012;726:93–114. doi: 10.1007/978-1-4614-0980-9_5. [DOI] [PubMed] [Google Scholar]
- 6.Kondou Y, et al. Structure of the central hub of bacteriophage Mu baseplate determined by X-ray crystallography of gp44. J Mol Biol. 2005;352(4):976–985. doi: 10.1016/j.jmb.2005.07.044. [DOI] [PubMed] [Google Scholar]
- 7.Harada K, et al. Crystal structure of the C-terminal domain of Mu phage central spike and functions of bound calcium ion. Biochim Biophys Acta. 2013;1834(1):284–291. doi: 10.1016/j.bbapap.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Haggård-Ljungquist E, et al. Bacteriophage P2: Genes involved in baseplate assembly. Virology. 1995;213(1):109–121. doi: 10.1006/viro.1995.1551. [DOI] [PubMed] [Google Scholar]
- 9.Maxwell KL, et al. Structural and functional studies of gpX of Escherichia coli phage P2 reveal a widespread role for LysM domains in the baseplates of contractile-tailed phages. J Bacteriol. 2013;195(24):5461–5468. doi: 10.1128/JB.00805-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundy FJ, Howe MM. Morphogenetic structures present in lysates of amber mutants of bacteriophage Mu. Virology. 1985;143(2):485–504. doi: 10.1016/0042-6822(85)90388-5. [DOI] [PubMed] [Google Scholar]
- 11.Kitazawa D, Takeda S, Kageyama Y, Tomihara M, Fukada H. Expression and characterization of a baseplate protein for bacteriophage Mu, gp44. J Biochem. 2005;137(5):601–606. doi: 10.1093/jb/mvi076. [DOI] [PubMed] [Google Scholar]
- 12.Kanamaru S, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415(6871):553–557. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- 13.Yap ML, Mio K, Leiman PG, Kanamaru S, Arisaka F. The baseplate wedges of bacteriophage T4 spontaneously assemble into hubless baseplate-like structure in vitro. J Mol Biol. 2010;395(2):349–360. doi: 10.1016/j.jmb.2009.10.071. [DOI] [PubMed] [Google Scholar]
- 14.Ghequire MGK, De Mot R. The tailocin tale: Peeling off phage tails. Trends Microbiol. 2015;23(10):587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Leiman PG, et al. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci USA. 2009;106(11):4154–4159. doi: 10.1073/pnas.0813360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet YR, Zoued A, Boyer F, Douzi B, Cascales E. The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 2015;11(10):e1005545. doi: 10.1371/journal.pgen.1005545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang G, Dowling AJ, Gerike U, ffrench-Constant RH, Waterfield NR. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol. 2006;188(6):2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hurst MRH, Glare TR, Jackson TA. Cloning Serratia entomophila antifeeding genes—A putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol. 2004;186(15):5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shikuma NJ, et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343(6170):529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rybakova D, et al. Role of antifeeding prophage (Afp) protein Afp16 in terminating the length of the Afp tailocin and stabilizing its sheath. Mol Microbiol. 2013;89(4):702–714. doi: 10.1111/mmi.12305. [DOI] [PubMed] [Google Scholar]
- 21.Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: What can be learned from available microbial genomic resources? BMC Genomics. 2009;10:104. doi: 10.1186/1471-2164-10-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spinelli S, Veesler D, Bebeacua C, Cambillau C. Structures and host-adhesion mechanisms of lactococcal siphophages. Front Microbiol. 2014;5:3. doi: 10.3389/fmicb.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cardarelli L, et al. Phages have adapted the same protein fold to fulfill multiple functions in virion assembly. Proc Natl Acad Sci USA. 2010;107(32):14384–14389. doi: 10.1073/pnas.1005822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sciara G, et al. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci USA. 2010;107(15):6852–6857. doi: 10.1073/pnas.1000232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campanacci V, et al. Solution and electron microscopy characterization of lactococcal phage baseplates expressed in Escherichia coli. J Struct Biol. 2010;172(1):75–84. doi: 10.1016/j.jsb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Kumar Sarkar S, Takeda Y, Kanamaru S, Arisaka F. Association and dissociation of the cell puncturing complex of bacteriophage T4 is controlled by both pH and temperature. Biochim Biophys Acta. 2006;1764(9):1487–1492. doi: 10.1016/j.bbapap.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Flaugnatti N, et al. A phospholipase A1 antibacterial type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol Microbiol. 2016;99(6):1099–1118. doi: 10.1111/mmi.13292. [DOI] [PubMed] [Google Scholar]
- 28.Shneider MM, et al. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature. 2013;500(7462):350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue) suppl 2:W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn RD, Clements J, Eddy SR. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue) suppl 2:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson AR, Sauer RT. Folded proteins occur frequently in libraries of random amino acid sequences. Proc Natl Acad Sci USA. 1994;91(6):2146–2150. doi: 10.1073/pnas.91.6.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.