Significance
Free-swimming larvae of many animals that inhabit the sea floor metamorphose in response to bacteria. However, the molecular mechanisms that underpin animal metamorphosis in response to bacterial triggers remain elusive. We investigated the developmental cascade induced by bacteria in a model tubeworm, Hydroides elegans, and identified a bacterial mutant and host signaling system critical for the initiation of and tissue remodeling during metamorphic development, respectively. Identifying the triggers for metamorphosis in Hydroides has implications for understanding basic principles of bacteria–animal interactions. Such interactions are also major causes of biofouling, which our research indicates may be controlled by manipulating bacterial inducers or MAPK activities that are essential for tubeworm metamorphosis.
Keywords: genome, phage, symbiosis, biofouling, development
Abstract
Diverse animal taxa metamorphose between larval and juvenile phases in response to bacteria. Although bacteria-induced metamorphosis is widespread among metazoans, little is known about the molecular changes that occur in the animal upon stimulation by bacteria. Larvae of the tubeworm Hydroides elegans metamorphose in response to surface-bound Pseudoalteromonas luteoviolacea bacteria, producing ordered arrays of phage tail-like metamorphosis-associated contractile structures (MACs). Sequencing the Hydroides genome and transcripts during five developmental stages revealed that MACs induce the regulation of groups of genes important for tissue remodeling, innate immunity, and mitogen-activated protein kinase (MAPK) signaling. Using two MAC mutations that block P. luteoviolacea from inducing settlement or metamorphosis and three MAPK inhibitors, we established a sequence of bacteria-induced metamorphic events: MACs induce larval settlement; then, particular properties of MACs encoded by a specific locus in P. luteoviolacea initiate cilia loss and activate metamorphosis-associated transcription; finally, signaling through p38 and c-Jun N-terminal kinase (JNK) MAPK pathways alters gene expression and leads to morphological changes upon initiation of metamorphosis. Our results reveal that the intricate interaction between Hydroides and P. luteoviolacea can be dissected using genomic, genetic, and pharmacological tools. Hydroides' dependency on bacteria for metamorphosis highlights the importance of external stimuli to orchestrate animal development. The conservation of Hydroides genome content with distantly related deuterostomes (urchins, sea squirts, and humans) suggests that mechanisms of bacteria-induced metamorphosis in Hydroides may have conserved features in diverse animals. As a major biofouling agent, insight into the triggers of Hydroides metamorphosis might lead to practical strategies for fouling control.
Free-swimming larvae of marine invertebrates must identify sites favorable for their settlement and subsequent metamorphosis into adults. But how do they identify these sites? Many species—including sponges (1), corals (2, 3), crabs (4), sea urchins (5), and ascidians (6)—use cues from bacteria (Fig. 1A) (7). This bacteria-mediated metamorphosis is essential for coral reef formation (8), and causes the costly accumulation of encrusting organisms on submerged surfaces such as the hulls of ships [i.e., biofouling (7, 8)]. Although in a few cases the bacterial cues (and the genes encoding them) have been identified (3, 9, 10), very little is known about how these cues mediate the resultant developmental cascade in animals and how developmental cues may be manipulated to promote or deter colonization.
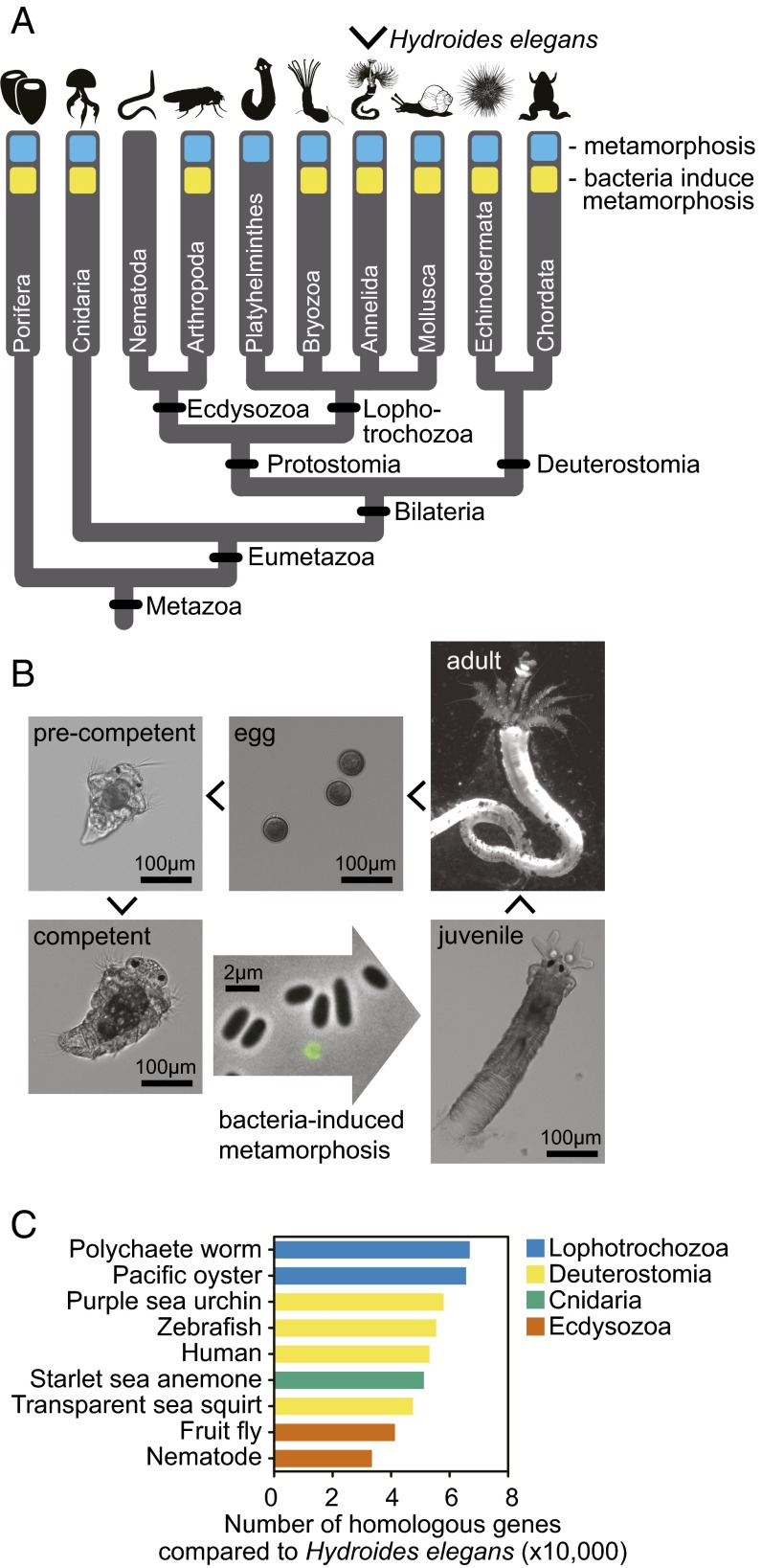
Fig. 1.
Bacteria-induced metamorphosis is widespread among animal taxa. (A) Many species from diverse taxa possess a biphasic lifestyle in which they undergo metamorphosis between larval and adult phases of life (blue). Nearly all of these taxa possess representative species that metamorphose in response to bacteria (yellow). (B) Hydroides developmental life cycle. RNA was sequenced from five stages of Hydroides development: precompetent larvae, competent larvae, larvae induced to metamorphose by the addition of MACs for 5 min and 30 min, and adult animals. A circular GFP-labeled MAC array is depicted in green. (C) The number of homologous genes identified between the Hydroides genome and the genomes of other model animals. The Hydroides (lophotrochozoan) genome is more similar to deuterostome animals (urchins, sea squirts, and humans) than to model ecdysozoan species. Polychaete worm, Capitella teleta; Pacific oyster, Crassostrea gigas; purple sea urchin, Strongylocentrotus purpuratus; zebrafish, Danio rerio; human, Homo sapiens; starlet sea anemone, Nematostella vectensis; transparent sea squirt, Ciona intestinalis; fruit fly, D. melanogaster; nematode, C. elegans.
Metamorphosis can be defined as the morphological, physiological, and behavioral transition from larva to juvenile (11, 12). During bacteria-induced metamorphosis, the sensing of bacterial cues must be coordinated with the regulation of development (7, 11, 12). It is hypothesized that metamorphosis evolved multiple times among metazoans, yet common signaling systems were independently coopted to coordinate the metamorphic transition (12–15). These systems include hormones (16–18), neurotransmitters (19–21), and nitric oxide (22–24). Additionally, diverse animals regulate metamorphosis at the transcriptional and posttranslational levels, such as differential expression of metamorphosis-associated genes (25–27) and MAPK signaling (28–31), respectively. Although a number of signaling systems and regulatory networks orchestrate metamorphosis in animals from diverse taxa, comparatively little is known about how bacteria stimulate these systems and networks.
The tubeworm Hydroides elegans (Haswell 1883; hereafter Hydroides) is a significant biofouling pest in tropical and subtropical harbors (32, 33), and its larvae are dependent on bacteria to initiate metamorphosis (9, 10, 34, 35). This makes it an appealing model organism in which to study the mechanisms of biofouling and how bacteria mediate animal development. In the laboratory, only certain bacterial species induce tubeworm metamorphosis (34–36), suggesting that some bacteria possess particular properties that serve as a metamorphosis cue. One such bacterium is Pseudoalteromonas luteoviolacea, a Gammaproteobacterium that induces metamorphosis of Hydroides (9, 34, 37), corals (2), and urchins (5). Recently, P. luteoviolacea was found to induce the metamorphosis of Hydroides by producing ordered arrays of bacteriophage tail-like structures, termed metamorphosis-associated contractile structures (MACs) (10). Similar phage tail-like structures possess bactericidal activity (38, 39) and mediate virulence in animals (40, 41). However, MACs are the first known phage tail-like structures to mediate a beneficial bacteria–animal interaction (10). When grown in rich media in the absence of Hydroides larvae, 2.4% of P. luteoviolacea cells produced MACs by a process of cell lysis (10). Although MACs are essential for Hydroides metamorphosis, we do not know what conditions stimulate MAC production in the environment or how MACs stimulate metamorphic development in Hydroides larvae.
To investigate the Hydroides developmental cascade induced by MACs, we used two bacterial mutations that block Hydroides settlement or metamorphosis at different stages. Sequencing Hydroides transcripts during five developmental stages identified groups of gene products that were key to metamorphosis. One such group comprised MAPK signaling proteins, whose roles were verified using inhibitors of MAPK phosphotransfer activity, indicating that metamorphosis relies on posttranslational modifications. These genomic, genetic, and pharmacological tools enabled us to dissect the sequence of events following the induction of tubeworm metamorphosis by a bacterial inducer.
Results
The Hydroides Genome Possesses a Conserved Gene Content.
To investigate the events that occur upon bacteria-stimulated metamorphosis, we sequenced the genome of Hydroides (48× average coverage) and transcripts at five developmental stages: (i) precompetent larvae; (ii) competent larvae (larvae capable of completing metamorphosis); competent larvae exposed to purified MACs for (iii) 5 min and (iv) 30 min; and (v) adult animals (Fig. 1B). The assembled draft genome is 1,026.1 Mb with an N50 (weighted median contig size) of 17.3 kb, and is predicted to encode 142,653 transcripts, corresponding to 113,410 loci, of which 93,636 are protein-coding; 33.94% of the genome is represented by repetitive sequences (SI Appendix, Table S1). Using the Core Eukaryotic Genes Mapping Approach (CEGMA) (42), the completeness of the assembly is estimated to be 89.1%. Additional information on the Hydroides genome and transcriptome can be found in SI Appendix, Results.
To gauge the relative gene conservation of Hydroides in relation to other model species that represent the breadth of multicellular animal life, we mapped all Hydroides genes against the genomes of several model organisms. Hydroides (a lophotrochozoan) and the fruit fly Drosophila melanogaster and nematode Caenorhabditis elegans (both ecdysozoans) group into the protostome clade (Fig. 1A). However, the Hydroides genome possesses more gene homologs with deuterostomes (urchins, sea squirts, zebrafish, and humans) and a nonbilaterian cnidarian (sea anemone) than it does with these model ecdysozoans (Fig. 1C). We further classified Hydroides genes into major metazoan groupings based on their homology [BLASTP against the non-redundant (nr) database, e-value cutoff of 10−10; SI Appendix, Fig. S1]. Of Hydroides' 61,540 genes with identified homologs, 6,087 genes (9.9%) are shared with deuterostomes and nonbilaterian animals only and not ecdysozoans. In comparison, 112 (0.2%) are shared with ecdysozoans and nonbilaterians only and not deuterostomes. These results suggest that Hydroides genome content has not evolved as quickly as the fruit fly and nematode genomes. Consistent with these findings, other annelids were found to possess relatively slowly evolving genomes (43, 44), and D. melanogaster and C. elegans are documented to possess more derived genomes with extensive gene loss and higher rates of molecular evolution (45, 46). Studying mechanisms that mediate bacteria-induced metamorphosis in Hydroides might provide a framework to understand bacteria–animal interactions in diverse animals.
Two Bacterial Loci Are Required for Different Stages of Metamorphosis.
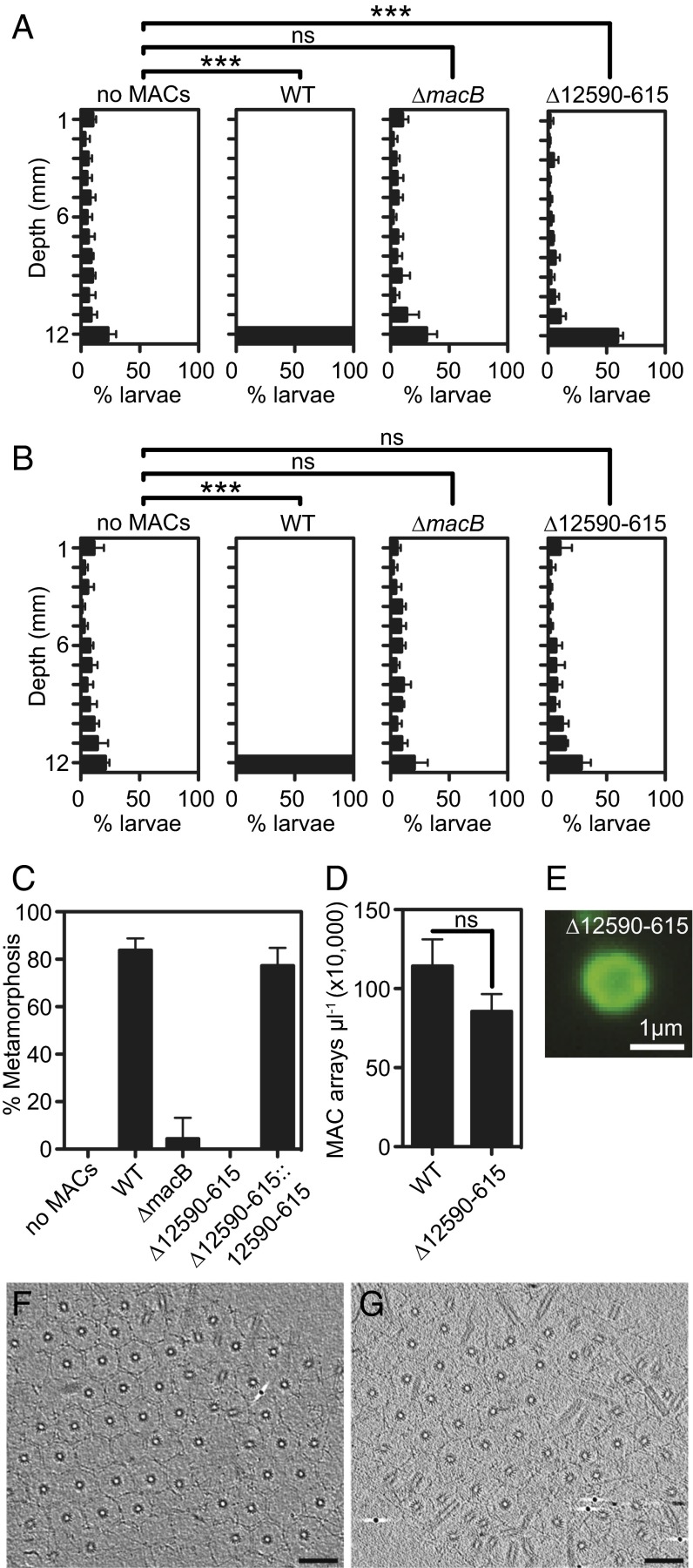
Settlement and metamorphosis of Hydroides in response to MACs can be classified into five stages: (i) swimming; (ii) initiation of settlement, indicated by slow swimming or crawling upon contact with MACs; (iii) irreversible initiation of metamorphosis, starting with the loss of cilia near the prototroch; (iv) body elongation and primary tube secretion; and (v) eversion of the collar, production of the secondary tube, and formation of branchial lobes (Movie S1). We quantified settlement by observing the vertical distribution of larvae at 5 and 30 min after exposure to MACs. Five minutes after larvae were exposed to wild-type MACs, larvae initiated settlement behavior (i.e., slowed or stopped swimming), causing them to collect at the bottom of the chamber (Fig. 2A). Larvae exposed to extract from a bacterial strain lacking the baseplate of MACs (∆macB), a key structural component of MACs (9, 10), did not initiate settlement behavior or begin metamorphosis.
Fig. 2.
Two bacterial loci are required for different stages of metamorphosis. (A and B) MAC extract from wild-type, ∆macB, or ∆12590–615 strains was added to competent larvae in polystyrene cuvettes in a total of 0.5 mL. The vertical distribution of larvae within the cuvette was quantified at 5 min (A) or 30 min (B) after addition of MACs. Artificial seawater without MACs served as a negative control. Error bars indicate SDs of six biological replicates. Twelve to 55 larvae were quantified per replicate. Student’s t test of mean depth: ns, not significant; ***P < 0.00001. (C) Metamorphosis of Hydroides exposed to MAC extract from wild-type, ∆12590–615, ∆macB, or ∆12590–615::12590–615. Error bars are SDs of four biological replicates. (D) Quantification of MAC arrays from extract of wild type or ∆12590–615. Error bars are SDs of three biological replicates. Student’s t test. (E) Image of a fluorescently tagged MAC from ∆12590–615 macB-sfgfp. (F and G) The wild type (F) and ∆12590–615 mutant (G) produce ordered arrays having comparable size and number of MACs. Shown are 9-nm slices through electron cryotomograms (Scale bars, 100 nm).
When exposed to MACs from a P. luteoviolacea mutant lacking a segment of the genome containing six genes (JF50_12590–JF50_12615, ∼8.2 kb; hereafter ∆12590–615), Hydroides larvae initiated settlement behavior but did not lose their cilia or continue to metamorphose compared with larvae exposed to wild-type MACs (Movie S2). After 5 min, these larvae were distributed significantly differently from larvae unexposed to MACs or larvae exposed to extract from a ∆macB strain (Fig. 2A). Thirty minutes after exposure, these larvae resumed a distribution similar to unexposed or ∆macB controls, whereas larvae exposed to wild-type MACs continued to metamorphose (Fig. 2B). All six genes deleted in the ∆12590–615 mutant encode hypothetical proteins and lie about 12 kb away from the closest characterized gene (macS) known to be essential for the structure and function of MACs (10). When these six genes were reintroduced into their original genomic location, the bacteria were again able to induce metamorphosis (Fig. 2C). MACs from the ∆12590–615 mutant were similar to MACs from P. luteoviolacea wild type in number (Fig. 2D), size, and structure (Fig. 2 E–G). These results suggest that the structure of MAC arrays produced by the ∆12590–615 mutant was largely intact and sufficient to initiate settlement. However, the deleted region is required for MACs to initiate the metamorphosis of Hydroides.
MACs Activate Tissue Remodeling-, Innate Immunity-, and Signaling-Associated Genes upon Stimulation of Metamorphosis.
To determine whether changes in gene expression accompany the initial events of Hydroides settlement and metamorphosis, we compared the transcriptomes of competent larvae unexposed to MACs with larvae exposed to MACs for 5 or 30 min. [These time points were chosen because at these times, larvae clearly responded differently to MACs from the wild-type and ∆12590–615 strains (Fig. 2).] When we compared transcripts from competent larvae unexposed to MACs with larvae exposed to MACs for 5 min, no transcripts were identified as significantly different. However, by 30 min, exposure to MACs resulted in significantly different transcription of 156 genes (P ≤ 0.05; 8 down-regulated and 148 up-regulated; Dataset S1).
Numerous genes were up-regulated upon the stimulation of metamorphosis with bacterial phage tail-like structures (SI Appendix, Fig. S2 and Dataset S1). Notably, MACs stimulated the expression of five astacin-family (47) matrix metalloproteinase (MMP)-encoding genes 4.9- to 21.0-fold after 30 min of induction and a collagen triple-helix (PF01391) domain-containing protein (up-regulated 6.0-fold). The up-regulation of MMP and collagen domain-containing genes is consistent with enzyme-mediated remodeling of tissues and structures accompanying metamorphosis (48). MACs stimulated the expression of two kazal-type serine protease inhibitor (SPI) domain-containing proteins (up-regulated 3.0- and 3.1-fold), known to have antimicrobial properties and implicated in host innate immunity in other invertebrate animals (49, 50). MACs induced the expression of 15 genes (up to 552.3-fold) encoding proteins with von Willebrand factor (VWF) domains, which can play important roles in cell adhesion (51). Nine genes encoding ankyrin repeats were up-regulated (up to 110.2-fold), which are often involved in protein–protein interactions (52). MACs stimulated a significant number of genes encoding nucleic acid-binding transcription factors, including a set that is typically downstream of MAPK signaling pathways (up-regulated 2.2- to 10.0-fold). Two genes encoding proteins with similarity to tumor necrosis factor receptor-associated factor 3, shown to function in MAPK signaling complexes in humans (53), were up-regulated 42.5- and 56.6-fold in response to MACs after 30 min. These results demonstrate that after 30 min of exposure to MACs, transcription of distinct sets of tissue remodeling-, innate immunity-, and signal transduction-associated genes was dramatically increased.
p38 and JNK MAPK Signaling Regulate Steps of Metamorphosis After Cilia Loss.
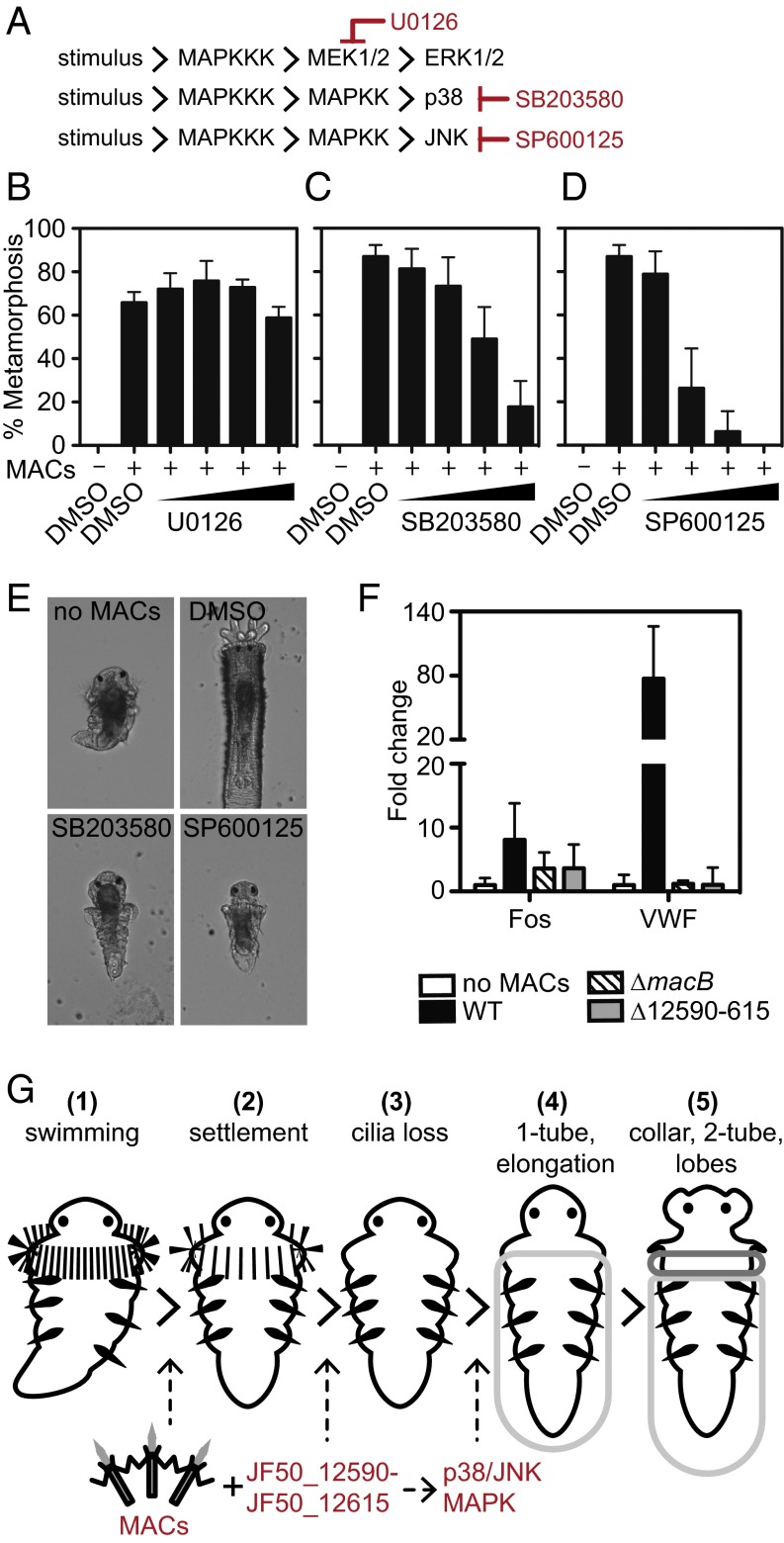
To understand the events following MAC-induced development, we asked whether MAPK signaling is important for Hydroides metamorphosis. We confirmed the presence of three extensively studied subgroups of MAPK genes (ERK, JNK, and p38) and associated signaling components in the Hydroides genome (Fig. 3A and SI Appendix, Table S2). Transcript abundances of most of the MAPK genes from competent larvae were high enough (>1 fragments per kb of transcript per million mapped reads) (SI Appendix, Table S2) to indicate that these genes were expressed upon induction of metamorphosis. We then asked whether metamorphosis proceeds in the presence of chemical inhibitors against each MAPK pathway. The U0126 inhibitor against mitogen/extracellular signal-regulated kinase (MEK; a kinase of ERK) did not prevent metamorphosis in response to MACs (Fig. 3B). However, the production of a primary and a secondary tube, larval body elongation, and development of branchial radioles were inhibited when larvae were pretreated for 2 h with the p38 inhibitor SB203580 or the JNK inhibitor SP600125 (Fig. 3 C–E). Although these inhibitors abrogated some developmental changes, larvae initiated settlement behavior (stopped swimming) and metamorphosis (lost cilia localized to the prototroch). Without the addition of MACs, the larvae appeared normal after 24 h in the presence of up to 10 µM all inhibitors tested, suggesting that the concentration of inhibitors used was not toxic. To determine whether the inhibitors were active against their intended targets, we measured the levels of specific phosphorylated components of the p38 and JNK pathways using antibodies raised against mammalian phosphoproteins with homologs in Hydroides (SI Appendix, Table S2). Hydroides protein extracts had increased levels of diphosphorylated p38 after treatment with SB203580 and decreased levels of diphosphorylated JNK after SP600125 treatment (SI Appendix, Fig. S3), similar to previous findings in human cells (54, 55). Upon stimulation, p38 phosphorylates MAPKAPK-2 and ATF-2, both of which were moderately increased after stimulation by MACs but not in the presence of SB203580 (SI Appendix, Fig. S3). Similarly, JNK phosphorylation of ATF-2 and p53 increased after exposure to MACs but was inhibited by SP600125 (SI Appendix, Fig. S3). These results identify p38 and JNK MAPK pathways as important phosphotransfer signaling systems in Hydroides metamorphic development.
Fig. 3.
Bacterial MACs and MAPK signaling regulate distinct steps of metamorphosis. (A) Components of the MAPK pathway identified in the genome of Hydroides. (B–D) Metamorphosis of Hydroides in the presence of the MEK inhibitor U0126 (B), p38 inhibitor SB203580 (C), or JNK inhibitor SP600125 (D). Larvae were pretreated with inhibitor for 2 h at 0.25, 1.0, 2.5, or 10 µM and induced to metamorphose with MACs. Solvent-only (DMSO) treatments are shown as controls. Error bars indicate SDs of eight biological replicates. (E) Representative images of larvae pretreated with the p38 inhibitor SB203580 or JNK inhibitor SP600125 for 2 h and induced to metamorphose with MACs for 24 h. Larvae without inhibitor and MACs (no MACs) or larvae treated with solvent and MACs (DMSO) are shown. (F) Fold change of Fos (TCONS_00100867) and VWF (TCONS_00123609) genes from larvae after a 30-min exposure to MACs from wild-type, ∆macB, or ∆12590–615 strains relative to unexposed larvae. (G) Timeline of metamorphic events in response to MACs: free-swimming competent larvae (1); MACs induce settlement behavior (2); with the JF50_12590–JF50_12615 region, MACs induce the irreversible initiation of metamorphosis (cilia loss) and activate metamorphosis-associated transcription (3); larvae produce a primary proteinaceous tube (1-tube) and their body elongates (4); larvae evert their collar, larvae produce a calcareous secondary tube (2-tube), and branchial lobes form (5). p38 and JNK MAPK signaling mediate body elongation, primary and secondary tube formation, and branchial lobe formation.
A Bacterial Locus Responsible for Metamorphosis-Associated Transcription.
We then sought to identify the developmental stage at which metamorphosis-associated transcription could be detected. Quantitative real-time PCR (qRT-PCR) robustly detected the up-regulation of Fos (TCONS_00100867) and VWF (TCONS_00123609) genes in response to wild-type MACs after 30 min (Fig. 3F and SI Appendix, Fig. S3). Pretreatment of larvae with the p38 and JNK inhibitors SB203580 or SP600125 partially repressed Fos and VWF expression compared with controls induced to metamorphose by MACs without an inhibitor (SI Appendix, Fig. S3). These results suggest that p38 and JNK signaling might act downstream of or in parallel to an initial signal of metamorphosis.
We then tested whether transcriptional changes occur coincident with the initiation of settlement or metamorphosis by using ∆12590–615 MACs or wild-type MACs, respectively, as inducers. Expression of Fos and VWF, measured by qRT-PCR, was not significantly activated in response to ∆12590–615 MACs compared with wild type (Fig. 3F), suggesting that metamorphosis-associated transcription and the activation of MAPK signaling require the JF50_12590–JF50_12615 genomic region of P. luteoviolacea. These results show that metamorphosis-associated transcription and MAPK signaling are activated coincident with the loss of cilia and establish a timeline of inductive and signaling events following MAC-initiated metamorphosis of Hydroides: MACs engage with tubeworm larvae, inducing them to initiate settlement behavior; then, particular properties or modification of MACs encoded by the JF50_12590–JF50_12615 region initiate metamorphosis (cilia loss) and activate metamorphosis-associated transcription; and, finally, p38 and JNK signaling are partially responsible for the regulation of gene expression and morphological remodeling events after cilia loss (Fig. 3G).
Discussion
We now know that interactions with bacteria can directly benefit animals (56, 57), and one of the most dramatic examples occurs when bacteria induce animal metamorphosis. Although representative species from diverse taxa undergo bacteria-induced metamorphosis (Fig. 1A), little is known about the processes that occur within the animal as they detect the cue produced by bacteria. To understand the developmental cascade in animals after bacteria induce metamorphosis, we sequenced the genome of Hydroides and analyzed changes in gene expression upon exposure to MACs. Although Hydroides (a lophotrochozoan), fruit flies, and nematodes (ecdysozoans) phylogenetically group as protostomes (58), the Hydroides genome shares more gene homologs with deuterostome (purple sea urchin, zebrafish, and human) and nonbilaterian (starlet sea anemone) genomes (Fig. 1C and SI Appendix, Fig. S1). Previous studies also reported relatively slow evolutionary rates of annelids compared with the model ecdysozoans D. melanogaster and C. elegans (43–46), suggesting that annelids might share more ancestral characteristics with distantly related model animals. Mechanisms underpinning the metamorphic response of Hydroides to bacterial inducers might therefore have conserved features present in other metazoans and serve as an example of how bacteria impact the development of an animal. In an applied context, such features are of interest because they might be used as targets for the general control of biofouling organisms.
In addition to the conservation of genome content, many genes that were differentially regulated upon the stimulation of Hydroides metamorphosis are homologous to genes associated with development in other model animals. Hydroides larvae up-regulated MMPs in response to MACs, important factors involved in the remodeling of tissue. The first MMP to be described was found to degrade fibrillar collagen in tadpole tails during metamorphosis (48). In the greater wax moth, both metamorphosis and collagen VI fragments were shown to stimulate an innate immune response (59), and this response is thought to be mediated by an endogenous MMP (60). Hydroides up-regulated SPI-encoding genes, which have potent bactericidal activity implicated in host innate immunity in Hydra (49) and black tiger shrimp (50). Similar differential expression of immunity genes during metamorphosis was observed in corals (25, 28) and ascidians (14, 27, 61). VWF domain-encoding genes were up-regulated and might be important for tube building. Proteins containing VWF domains were found in the organic matrix of oyster shells (62) and coral skeletons (63). Alternatively, VWF domain-containing genes could be involved in cell adhesion during tissue remodeling (51). Hydroides larvae up-regulated MAPK signaling genes, which were previously implicated in modulating metamorphosis of other marine organisms including corals (28), ascidians (30), and barnacles (31). The transcriptional signature of metamorphosis in Hydroides therefore parallels developmental processes identified in other animal species.
To understand how bacteria activate animal development, we dissected the events occurring upon bacteria-stimulated metamorphosis. Although transcription changed following MAC exposure, the initial signal reception and transduction orchestrating Hydroides metamorphosis are likely initially mediated by posttranslational mechanisms (64) and subsequently lead to the differential expression of metamorphosis-associated genes. Based on transcript sequencing and use of pharmacological inhibitors, we found that such posttranslational mechanisms include p38 and JNK MAPK signaling. A previous report showed the importance of p38 signaling in Hydroides metamorphosis in response to natural marine biofilms, but results were inconclusive for the JNK inhibitor SP600125 (29). Our results suggest that p38 and JNK signaling are necessary for the stages of MAC-induced metamorphosis involving morphological remodeling following cilia loss, including body elongation, primary and secondary tube formation, and branchial lobe formation (Fig. 3).
In addition to using genomics and pharmacological inhibitors, we used two MAC mutants to dissect metamorphosis; one is unable to initiate settlement (∆macB), and the other initiates settlement but does not induce metamorphosis (∆12590–615). Using these mutants, we determined that the irreversible step of metamorphosis starts with cilia loss, activation of metamorphosis-associated gene expression, and MAPK signaling. We hypothesize that the JF50_12590–JF50_12615 region encodes structures for the proper binding of MACs to larvae and components delivered to larvae by the MAC structure, or mediates metamorphosis by encoding chaperones essential for the proper assembly or conformation of MACs. Future work will reveal the mechanisms by which these and other loci affect settlement and metamorphosis of Hydroides.
In conclusion, we sequenced the genome and transcriptome of a model tubeworm and show how its settlement and metamorphic program respond to a bacterial inducer. As a member of the undersampled lophotrochozoan clade, the availability of the Hydroides genome will broaden the repertoire of tools available to study metazoan evolution. Identifying the molecular basis of bacteria-mediated metamorphosis will help us understand the molecular coevolution of bacteria and animals, which may have played an important role in generating the spectacular diversity in animal form and function that we see today. Our results indicate that bacterial factors and intracellular signaling systems may be controlled during surface colonization for applications, such as the management of biofouling, and to better understand processes that govern the recruitment and maintenance of marine animal populations.
Methods
Specimens of H. elegans were obtained from Marina Del Rey, California. Embryos were obtained and maintained as previously described (10, 32). Two libraries with average insert sizes of 300 and 600 bp were prepared using the New England Biolabs, Inc. Next Ultra DNA Library Prep Kit (NEB; E7370) according to the manufacturer’s instructions with minor modifications. Libraries were sequenced on an Illumina HiSeq 2500 instrument in paired-end mode with a read length of 150 nt to the depth of 100 million and 75 million reads for the 300- and 600-bp libraries, respectively. Detailed genome sequencing, assembly, and experimental procedures and datasets are provided in SI Appendix, Methods, and Datasets S2–S4.
Supplementary Material
Acknowledgments
We thank Heather Maughan for providing constructive feedback and editing the manuscript. The authors acknowledge support from Piotr Szwedziak, Peter Tittmann, and ScopeM at ETH Zürich. Experiments were carried out in the laboratories of N.J.S., M.P., D.K.N., and at the Millard and Muriel Jacobs Genome Facility at Caltech. This work was supported by grants from the Office of Naval Research (N00014-14-1-0340 to N.J.S. and D.K.N.; N00014-16-1-2135 to N.J.S.), Swiss National Science Foundation (31003A_152878 to M.P.), ETH Zürich (ETH-45 15-1 to M.P.), and the Helmut Horten Foundation (to M.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.Z.H. is a Guest Editor invited by the Editorial Board.
Data deposition: The raw sequence reads reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) database [accession nos. SRP067980 (genome) and SRP067899 (transcriptome)]. The whole-genome shotgun project reported in this paper has been deposited in the DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank database (accession no. LQRL00000000, version LQRL01000000).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603142113/-/DCSupplemental.
References
- 1.Whalan S, Webster NS. Sponge larval settlement cues: The role of microbial biofilms in a warming ocean. Sci Rep. 2014;4:4072. doi: 10.1038/srep04072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tran C, Hadfield MG. Larvae of Pocillopora damicornis (Anthozoa) settle and metamorphose in response to surface-biofilm bacteria. Mar Ecol Prog Ser. 2011;433:85–96. [Google Scholar]
- 3.Tebben J, et al. Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One. 2011;6(4):e19082. doi: 10.1371/journal.pone.0019082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson JA, Epifanio CE. Induction of metamorphosis in the Asian shore crab Hemigrapsus sanguineus: Characterization of the cue associated with biofilm from adult habitat. J Exp Mar Biol Ecol. 2009;382(1):34–39. [Google Scholar]
- 5.Huggett MJ, Williamson JE, de Nys R, Kjelleberg S, Steinberg PD. Larval settlement of the common Australian sea urchin Heliocidaris erythrogramma in response to bacteria from the surface of coralline algae. Oecologia. 2006;149(4):604–619. doi: 10.1007/s00442-006-0470-8. [DOI] [PubMed] [Google Scholar]
- 6.Roberts B, et al. A complement response may activate metamorphosis in the ascidian Boltenia villosa. Dev Genes Evol. 2007;217(6):449–458. doi: 10.1007/s00427-007-0157-0. [DOI] [PubMed] [Google Scholar]
- 7.Hadfield MG. Biofilms and marine invertebrate larvae: What bacteria produce that larvae use to choose settlement sites. Annu Rev Mar Sci. 2011;3:453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 8.Hadfield MG, Paul VJ. Natural chemical cues for settlement and metamorphosis of marine-invertebrate larvae. In: McClintock JB, Baker BJ, editors. Marine Chemical Ecology. CRC; Boca Raton, FL: 2001. pp. 431–461. [Google Scholar]
- 9.Huang Y, Callahan S, Hadfield MG. Recruitment in the sea: Bacterial genes required for inducing larval settlement in a polychaete worm. Sci Rep. 2012;2:228. doi: 10.1038/srep00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shikuma NJ, et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343(6170):529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop CD, et al. What is metamorphosis? Integr Comp Biol. 2006;46(6):655–661. doi: 10.1093/icb/icl004. [DOI] [PubMed] [Google Scholar]
- 12.Hadfield MG, Carpizo-Ituarte EJ, del Carmen K, Nedved BT. Metamorphic competence, a major adaptive convergence in marine invertebrate larvae. Am Zool. 2001;41(5):1123–1131. [Google Scholar]
- 13.Collins AG, Valentine JW. Defining phyla: Evolutionary pathways to metazoan body plans. Evol Dev. 2001;3(6):432–442. doi: 10.1046/j.1525-142x.2001.01048.x. [DOI] [PubMed] [Google Scholar]
- 14.Heyland A, Moroz LL. Signaling mechanisms underlying metamorphic transitions in animals. Integr Comp Biol. 2006;46(6):743–759. doi: 10.1093/icb/icl023. [DOI] [PubMed] [Google Scholar]
- 15.Bishop CD, Brandhorst BP. On nitric oxide signaling, metamorphosis, and the evolution of biphasic life cycles. Evol Dev. 2003;5(5):542–550. doi: 10.1046/j.1525-142x.2003.03059.x. [DOI] [PubMed] [Google Scholar]
- 16.Riddiford LM. Hormone receptors and the regulation of insect metamorphosis. Receptor. 1993;3(3):203–209. [PubMed] [Google Scholar]
- 17.Heyland A, Price DA, Bodnarova-Buganova M, Moroz LL. Thyroid hormone metabolism and peroxidase function in two non-chordate animals. J Exp Zool B Mol Dev Evol. 2006;306(6):551–566. doi: 10.1002/jez.b.21113. [DOI] [PubMed] [Google Scholar]
- 18.Heyland A, Hodin J, Reitzel AM. Hormone signaling in evolution and development: A non-model system approach. BioEssays. 2005;27(1):64–75. doi: 10.1002/bies.20136. [DOI] [PubMed] [Google Scholar]
- 19.Leitz T, Morand K, Mann M. Metamorphosin A: A novel peptide controlling development of the lower metazoan Hydractinia echinata (Coelenterata, Hydrozoa) Dev Biol. 1994;163(2):440–446. doi: 10.1006/dbio.1994.1160. [DOI] [PubMed] [Google Scholar]
- 20.Conzelmann M, et al. Neuropeptides regulate swimming depth of Platynereis larvae. Proc Natl Acad Sci USA. 2011;108(46):E1174–E1183. doi: 10.1073/pnas.1109085108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sutherby J, et al. Histamine is a modulator of metamorphic competence in Strongylocentrotus purpuratus (Echinodermata: Echinoidea) BMC Dev Biol. 2012;12:14. doi: 10.1186/1471-213X-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froggett SJ, Leise EM. Metamorphosis in the marine snail Ilyanassa obsoleta, yes or NO? Biol Bull. 1999;196(1):57–62. doi: 10.2307/1543167. [DOI] [PubMed] [Google Scholar]
- 23.Bishop CD, Bates WR, Brandhorst BP. Regulation of metamorphosis in ascidians involves NO/cGMP signaling and HSP90. J Exp Zool. 2001;289(6):374–384. doi: 10.1002/jez.1019. [DOI] [PubMed] [Google Scholar]
- 24.Bishop CD, Brandhorst BP. NO/cGMP signaling and HSP90 activity represses metamorphosis in the sea urchin Lytechinus pictus. Biol Bull. 2001;201(3):394–404. doi: 10.2307/1543617. [DOI] [PubMed] [Google Scholar]
- 25.Siboni N, et al. Using bacterial extract along with differential gene expression in Acropora millepora larvae to decouple the processes of attachment and metamorphosis. PLoS One. 2012;7(5):e37774. doi: 10.1371/journal.pone.0037774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams EA, et al. Widespread transcriptional changes pre-empt the critical pelagic-benthic transition in the vetigastropod Haliotis asinina. Mol Ecol. 2009;18(5):1006–1025. doi: 10.1111/j.1365-294X.2008.04078.x. [DOI] [PubMed] [Google Scholar]
- 27.Davidson B, Swalla BJ. A molecular analysis of ascidian metamorphosis reveals activation of an innate immune response. Development. 2002;129(20):4739–4751. doi: 10.1242/dev.129.20.4739. [DOI] [PubMed] [Google Scholar]
- 28.Siboni N, Abrego D, Motti CA, Tebben J, Harder T. Gene expression patterns during the early stages of chemically induced larval metamorphosis and settlement of the coral Acropora millepora. PLoS One. 2014;9(3):e91082. doi: 10.1371/journal.pone.0091082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Qian PY. Involvement of a novel p38 mitogen-activated protein kinase in larval metamorphosis of the polychaete Hydroides elegans (Haswell) J Exp Zool B Mol Dev Evol. 2010;314(5):390–402. doi: 10.1002/jez.b.21344. [DOI] [PubMed] [Google Scholar]
- 30.Chambon J-P, Nakayama A, Takamura K, McDougall A, Satoh N. ERK- and JNK-signalling regulate gene networks that stimulate metamorphosis and apoptosis in tail tissues of ascidian tadpoles. Development. 2007;134(6):1203–1219. doi: 10.1242/dev.002220. [DOI] [PubMed] [Google Scholar]
- 31.He L-S, et al. Evidence for the involvement of p38 MAPK activation in barnacle larval settlement. PLoS One. 2012;7(10):e47195. doi: 10.1371/journal.pone.0047195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nedved BT, Hadfield MG. 2009. Hydroides elegans (Annelida: Polychaeta): A model for biofouling research. Marine and Industrial Biofouling, Springer Series on Biofilms, eds Flemming H-C, Murthy PS, Venkatesan R, Cooksey K (Springer, Berlin), Vol 4, pp 203–217.
- 33.Hadfield MG, Unabia CC, Smith CM, Michael TM. 1994. Settlement preferences of the ubiquitous fouler Hydroides elegans. Recent Dev Biofoul Control, eds Thompson MF, Nagabhushanam R, Sarojini R, Fingerman M (Oxford and IBH Publishing Co, New Delhi) pp 65–74.
- 34.Huang SY, Hadfield MG. Composition and density of bacterial biofilms determine larval settlement of the polychaete Hydroides elegans. Mar Ecol Prog Ser. 2003;260:161–172. [Google Scholar]
- 35.Unabia CRC, Hadfield MG. Role of bacteria in larval settlement and metamorphosis of the polychaete Hydroides elegans. Mar Biol. 1999;133(1):55–64. [Google Scholar]
- 36.Lau SC, Mak KK, Chen F, Qian P-Y. Bioactivity of bacterial strains isolated from marine biofilms in Hong Kong waters for the induction of larval settlement in the marine polychaete Hydroides elegans. Mar Ecol Prog Ser. 2002;226:301–310. [Google Scholar]
- 37.Asahina AY, Hadfield MG. Draft genome sequence of Pseudoalteromonas luteoviolacea HI1, determined using Roche 454 and PacBio single-molecule real-time hybrid sequencing. Genome Announc. 2015;3(1):e01590-14. doi: 10.1128/genomeA.01590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84(5–6):499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 39.Uratani Y, Hoshino T. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol. 1984;157(2):632–636. doi: 10.1128/jb.157.2.632-636.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurst MR, Glare TR, Jackson TA. Cloning Serratia entomophila antifeeding genes—A putative defective prophage active against the grass grub Costelytra zealandica. J Bacteriol. 2004;186(15):5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang G, Dowling AJ, Gerike U, ffrench-Constant RH, Waterfield NR. Photorhabdus virulence cassettes confer injectable insecticidal activity against the wax moth. J Bacteriol. 2006;188(6):2254–2261. doi: 10.1128/JB.188.6.2254-2261.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 43.Raible F, et al. Vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science. 2005;310(5752):1325–1326. doi: 10.1126/science.1119089. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi T, et al. An EST screen from the annelid Pomatoceros lamarckii reveals patterns of gene loss and gain in animals. BMC Evol Biol. 2009;9:240. doi: 10.1186/1471-2148-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyder S, Kriventseva EV, Schröder R, Kadowaki T, Zdobnov EM. Quantification of ortholog losses in insects and vertebrates. Genome Biol. 2007;8(11):R242. doi: 10.1186/gb-2007-8-11-r242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kortschak RD, Samuel G, Saint R, Miller DJ. EST analysis of the cnidarian Acropora millepora reveals extensive gene loss and rapid sequence divergence in the model invertebrates. Curr Biol. 2003;13(24):2190–2195. doi: 10.1016/j.cub.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 47.Bond JS, Beynon RJ. The astacin family of metalloendopeptidases. Protein Sci. 1995;4(7):1247–1261. doi: 10.1002/pro.5560040701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gross J, Lapiere CM. Collagenolytic activity in amphibian tissues: A tissue culture assay. Proc Natl Acad Sci USA. 1962;48(6):1014–1022. doi: 10.1073/pnas.48.6.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Augustin R, Siebert S, Bosch TC. Identification of a kazal-type serine protease inhibitor with potent anti-staphylococcal activity as part of Hydra’s innate immune system. Dev Comp Immunol. 2009;33(7):830–837. doi: 10.1016/j.dci.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 50.Donpudsa S, Tassanakajon A, Rimphanitchayakit V. Domain inhibitory and bacteriostatic activities of the five-domain Kazal-type serine proteinase inhibitor from black tiger shrimp Penaeus monodon. Dev Comp Immunol. 2009;33(4):481–488. doi: 10.1016/j.dci.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Whittaker CA, Hynes RO. Distribution and evolution of von Willebrand/integrin A domains: Widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell. 2002;13(10):3369–3387. doi: 10.1091/mbc.E02-05-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13(6):1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Häcker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11(7):457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 54.Bennett BL, et al. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc Natl Acad Sci USA. 2001;98(24):13681–13686. doi: 10.1073/pnas.251194298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S, Jiang MS, Adams JL, Lee JC. Pyridinylimidazole compound SB 203580 inhibits the activity but not the activation of p38 mitogen-activated protein kinase. Biochem Biophys Res Commun. 1999;263(3):825–831. doi: 10.1006/bbrc.1999.1454. [DOI] [PubMed] [Google Scholar]
- 56.Gilbert SF, Bosch TCG, Ledón-Rettig C. Eco-Evo-Devo: Developmental symbiosis and developmental plasticity as evolutionary agents. Nat Rev Genet. 2015;16(10):611–622. doi: 10.1038/nrg3982. [DOI] [PubMed] [Google Scholar]
- 57.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493(7433):526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altincicek B, Vilcinskas A. Metamorphosis and collagen-IV-fragments stimulate innate immune response in the greater wax moth, Galleria mellonella. Dev Comp Immunol. 2006;30(12):1108–1118. doi: 10.1016/j.dci.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 60.Altincicek B, Vilcinskas A. Identification of a lepidopteran matrix metalloproteinase with dual roles in metamorphosis and innate immunity. Dev Comp Immunol. 2008;32(4):400–409. doi: 10.1016/j.dci.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 61.Woods RG, et al. Gene expression during early ascidian metamorphosis requires signalling by Hemps, an EGF-like protein. Development. 2004;131(12):2921–2933. doi: 10.1242/dev.01120. [DOI] [PubMed] [Google Scholar]
- 62.Ramos-Silva P, et al. The skeletal proteome of the coral Acropora millepora: The evolution of calcification by co-option and domain shuffling. Mol Biol Evol. 2013;30(9):2099–2112. doi: 10.1093/molbev/mst109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drake JL, et al. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci USA. 2013;110(10):3788–3793. doi: 10.1073/pnas.1301419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carpizo-Ituarte E, Hadfield MG. Transcription and translation inhibitors permit metamorphosis up to radiole formation in the serpulid polychaete Hydroides elegans Haswell. Biol Bull. 2003;204(2):114–125. doi: 10.2307/1543547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.