Significance
Niemann–Pick C1 (NPC1) and Niemann–Pick C2 (NPC2) cooperate in the export of LDL-derived cholesterol from lysosomes; mutations in these proteins lead to Niemann–Pick type C disease. We present here the crystal structure of an NPC1–NPC2 complex and show that the amino acid residues that are important for this interaction in vitro are also important for cholesterol export from lysosomes of cultured cells. These data provide key information related to the mechanism by which these proteins catalyze cholesterol transport and form the basis for a molecular model for how cholesterol is transferred from NPC2 onto NPC1 protein.
Keywords: cholesterol trafficking, Ebola virus glycoprotein, Niemann–Pick type C disease, crystal structure
Abstract
Export of LDL-derived cholesterol from lysosomes requires the cooperation of the integral membrane protein Niemann–Pick C1 (NPC1) and a soluble protein, Niemann–Pick C2 (NPC2). Mutations in the genes encoding these proteins lead to Niemann–Pick disease type C (NPC). NPC2 binds to NPC1’s second (middle), lumenally oriented domain (MLD) and transfers cholesterol to NPC1’s N-terminal domain (NTD). Here, we report the 2.4-Å resolution crystal structure of a complex of human NPC1–MLD and NPC2 bearing bound cholesterol-3-O-sulfate. NPC1–MLD uses two protruding loops to bind NPC2, analogous to its interaction with the primed Ebola virus glycoprotein. Docking of the NPC1–NPC2 complex onto the full-length NPC1 structure reveals a direct cholesterol transfer tunnel between NPC2 and NTD cholesterol binding pockets, supporting the “hydrophobic hand-off” cholesterol transfer model.
Cholesterol is delivered to cells by LDL particles that carry cholesterol in esterified form to peripheral tissues (1). LDLs are recognized by cognate, cell-surface receptors and transported to endosomes and lysosomes by receptor-mediated endocytosis; there, acid lipase converts cholesterol esters to free cholesterol (2). Two lysosomal proteins are required for cholesterol export from lysosomes: Niemann–Pick C1 (NPC1) and Niemann–Pick C2 (NPC2) (3). Loss-of-function mutations in either of these proteins produce a fatal lysosomal storage disease that leads to premature death due to accumulation of cholesterol and glycosphingolipids in lysosomes throughout the body, especially in liver, spleen, lung, and brain. NPC1 is also an essential component for productive infection by Ebola and Marburg viruses (4, 5).
The NPC1 glycoprotein has 13 transmembrane domains and three relatively large, lumenally oriented domains. The first, N-terminal lumenal domain (NTD) contains a cholesterol binding site (6, 7); the second (middle) lumenal domain (termed here MLD) binds NPC2, presumably to assist in the transfer of cholesterol from NPC2 onto NPC1 protein (8). NPC1’s MLD is also important for cellular infection by Ebola and Marburg viruses because it interacts directly with the cleaved glycoprotein (GP) to permit viral escape from the lysosome into the cytoplasm (9–11).
NPC2 is a small, soluble protein that binds cholesterol from within the lysosome lumen (12) and transfers it to the sterol-binding pocket of NPC1’s NTD (13, 14). Cholesterol binds NPC2 with its isooctyl chain buried in the pocket and its hydroxyl group exposed (12). In contrast, cholesterol binds to NPC1’s NTD with its hydroxyl group buried (14). Infante et al. (13) showed that NPC2 binds and releases cholesterol rather quickly, whereas NPC1’s NTD does so more slowly, especially at 4 °C. Importantly, the rate of cholesterol binding to NPC1’s NTD increases more than 15-fold when the sterol is first bound to NPC2 and then transferred. These data support a “hydrophobic hand-off” transfer model in which NPC2 binds cholesterol in the lysosome lumen and then binds to NPC1 to deliver cholesterol to NPC1’s NTD; transfer of NTD-bound cholesterol to NPC1’s transmembrane domain follows, to achieve sterol export from lysosomes (14). Also supporting this transfer model was our finding that NPC2-bearing cholesterol can bind to NPC1’s MLD at acidic pH (8). The presence of a conserved, “sterol-sensing” domain within NPC1’s membrane spanning portions has implicated these sequences as possible sterol acceptors (15, 16). Indeed, recent crystal (17) and cryo-EM (18) structures of human NPC1 have provided a structural foundation to inform this transfer model.
Here, we present the crystal structure of the NPC1–MLD bound to NPC2 at 2.4-Å resolution. Docking of this structure onto full-length NPC1 reveals a cholesterol transfer tunnel between NPC2 and NPC1–NTD and provides a satisfying structural basis to explain cholesterol transfer between NPC2 and NPC1 proteins.
Results
We used bacterial expression to produce human NPC1–MLD (residues 374–620) as insoluble inclusion bodies. After denaturation and refolding, the resulting soluble protein eluted as a monodisperse peak upon size-exclusion chromatography. Full-length human NPC2 was expressed using the baculovirus-mediated gene transduction of mammalian cells (BacMam) system. Purified, glycosylated NPC2 protein migrated as a broad band upon SDS/PAGE. After deglycosylation, NPC2 migrated as a single, sharp band and behaved well upon gel filtration. NPC1–MLD and NPC2 were incubated together at a final concentration of 100 μM in the presence of 500 μM cholesteryl sulfate for 1 h, before crystallization screening. The complex structure was determined at a resolution of 2.4 Å by molecular replacement using previously determined NPC2 and NPC1–MLD structures (Table S1 and Fig. S1).
Table S1.
Data collection and refinement statistics
| NPC1–MLD:NPC2 | Native |
| Data collection | |
| Wavelength, Å | 0.979 |
| Space group | C2221 |
| Unit cell, Å; ° (a,b,c; α,β,γ) | 98.96, 109.69, 154.56; 90, 90, 90 |
| Resolution, Å | 50∼2.4 (2.47∼2.40)† |
| Rmerge, % | 9.8 (59.4) |
| I/σI | 10.14 (1.21) |
| Completeness, % | 97. 7 (89.8) |
| Redundancy | 3.0 (2.7) |
| Refinement | |
| Resolution, Å | 50∼2.4 |
| No. of reflections | 32,184 |
| Rwork/Rfree,‡ % | 19.39/24.94 |
| Molecules in an asymmetry unit | 2 |
| No. of atoms | |
| Protein/ligand/H2O | 5,633/120/105 |
| B-factors | |
| Protein/ligand/H2O | 56.02/69.71/49.08 |
| Ramachandran plot, % | |
| Favored/ allowed/ /disallowed | 96.15 /3.70/0.14 |
| Rmsd in: | |
| Bond distances, Å | 0.003 |
| Bond angles, ° | 0.684 |
Values in parentheses are for the highest-resolution shell.
Rfree was calculated with 5% of the reflections selected in the thin shell.
Fig. S1.
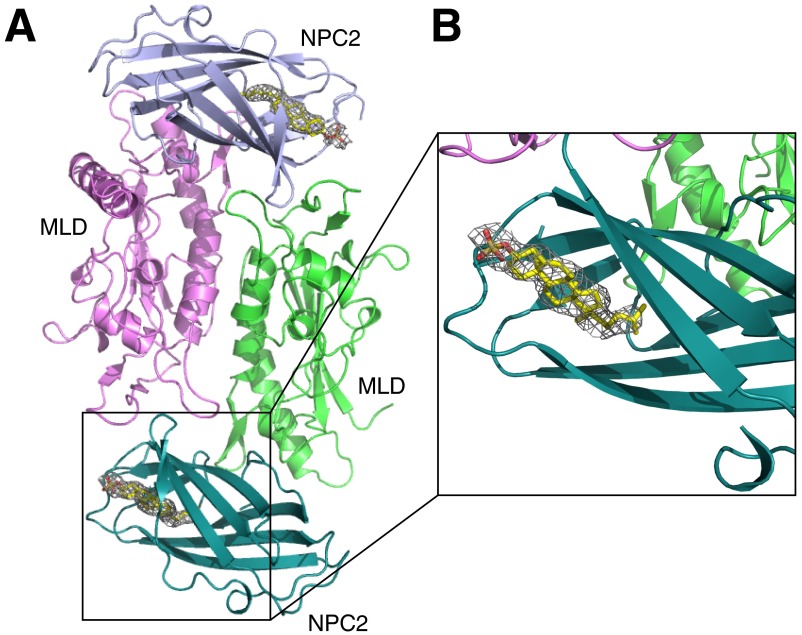
The Omit map for cholesteryl sulfate in NPC2. (A) Overall structure of MLD–NPC2 complex in one asymmetry unit. (B) Close-up view of SA-Omit map (Fo-Fc densities) for cholesteryl sulfate (gray mesh) contoured at σ level 2.0.
In the asymmetric unit, two MLDs form a homodimer and each MLD binds one NPC2 molecule (Fig. S1). The structure of the complex reveals that NPC1–MLD binds the top of the NPC2 sterol-binding pocket (Fig. 1 A and B and Fig. S2). Docking this complex onto the full-length NPC1 cryo-EM structure revealed spatial proximity between NPC2 and the NTD and a longer distance for NPC2 to reach the C-terminal, lumenal domain or transmembrane domain. There is ∼90 Å between NPC2 and the lysosome’s limiting membrane.
Fig. 1.
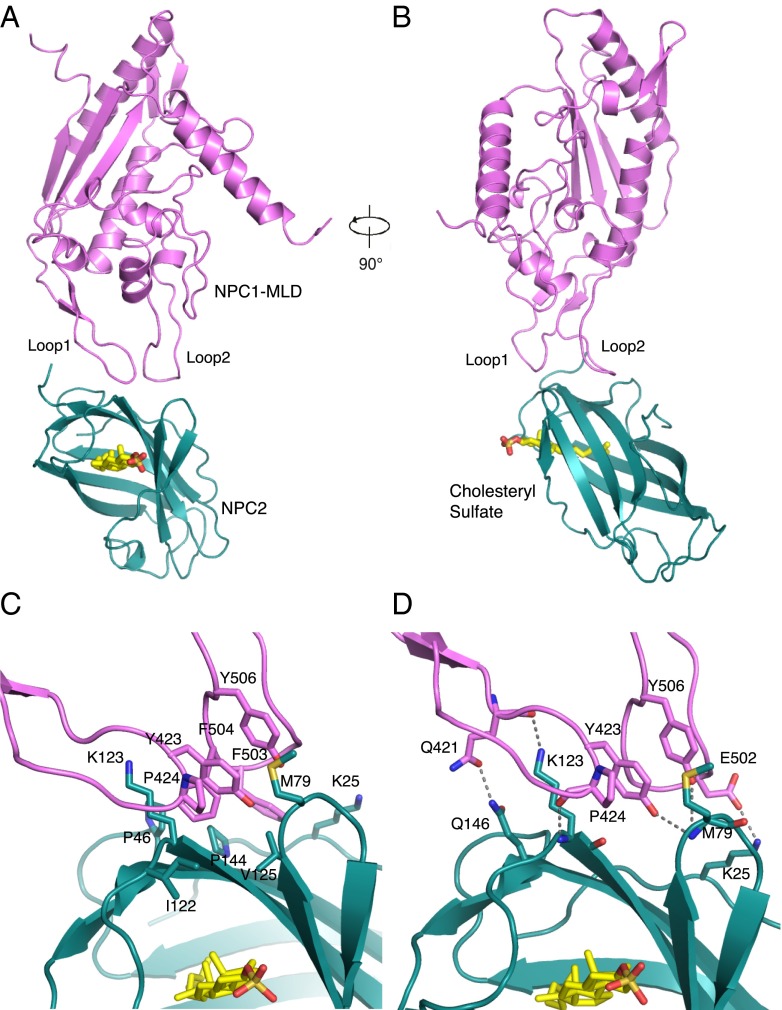
Structural overview of the NPC1 MLD–NPC2 complex. (A) NPC1–MLD (magenta), NPC2 (cyan), and cholesteryl sulfate (yellow) are shown. (B) Complex rotated 90° relative to A. (C) Close-up view of the interface. Residues involved in hydrophobic interactions are indicated. (D) Residues involved in hydrophilic interactions are indicated; dotted lines represent hydrogen bonds.
Fig. S2.
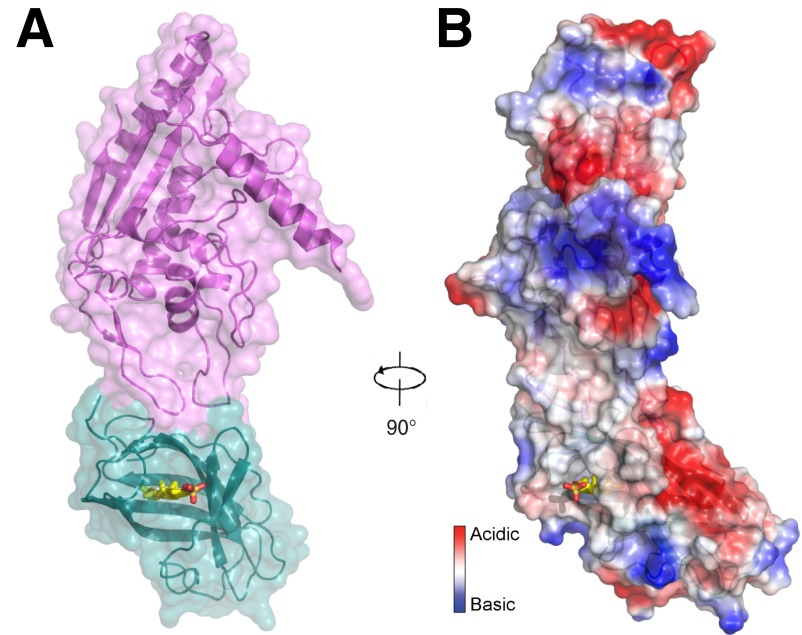
NPC1 MLD–NPC2 complex in a space-filling representation. (A) Surface is oriented as in Fig. 1A. (B) Electrostatic surface representation in the same orientation as Fig. 1B. Cholesteryl sulfate (yellow) is shown.
Two protruding loops of the NPC1–MLD comprise the predominant binding interface between this domain of NPC1 and NPC2 (Fig. 1 A and B and Fig. S3); the interaction surface area of the two proteins is about 500 Å2. Residue Q421 from protruding loop 1 forms a hydrogen bond with Q146 of NPC2 (Fig. 1D, at left); NPC1 residue Y423 interacts with NPC2 residue M79 (Fig. 1D). In NPC1 MLD’s protruding loop 2, two residues (F503 and F504) are the most significant contributors to hydrophobic interactions with NPC2. NPC1’s Y506 also contributes a hydrophobic interaction with M79 of NPC2 (Fig. 1D), a residue previously reported to be important for the interaction of NPC2 with NPC1’s NTD (19).
Fig. S3.
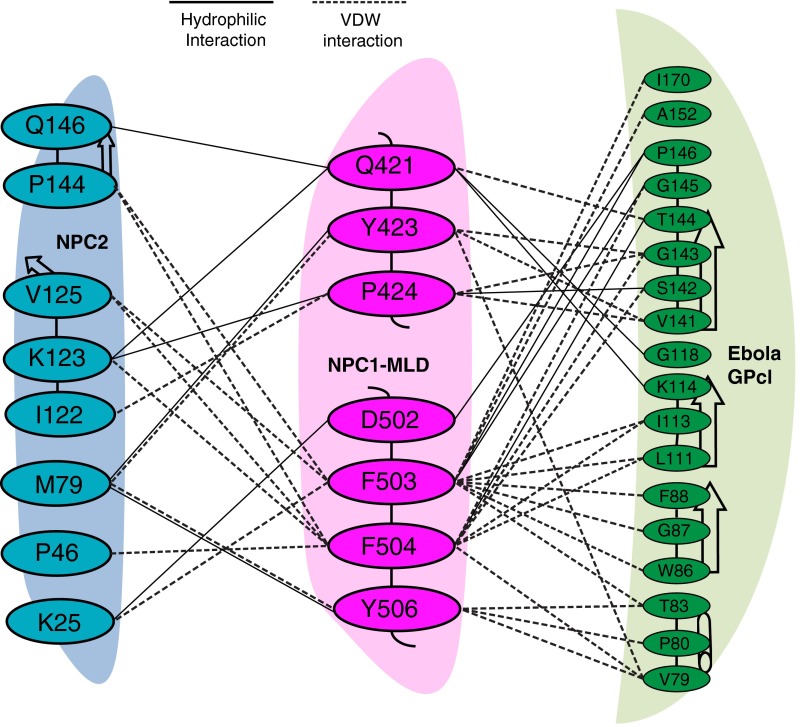
The interaction detail of NPC1–MLD and NPC2. Residues involved at the interface between NPC1–MLD (cyan), NPC2 (magenta), and Ebola GPcl (green) are shown. Black dotted lines indicate hydrophilic interactions; dotted lines indicate van der Waals interactions.
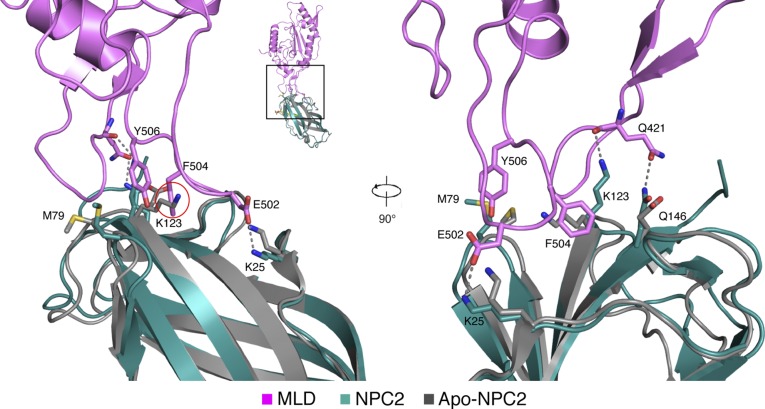
The NPC2–NPC1 MLD complex structure explains the previously reported cholesterol dependence of this interaction (8). Comparison of NPC2 structures with and without bound cholesterol sulfate reveals a significant shift in the orientations of the side chains of K25, M79, K123, and Q146 (12) (Fig. S4). These side chains are major contributors to the interface between NPC1–MLD and NPC2 (Fig. 1 C and D). It is noteworthy that in the absence of cholesteryl sulfate, the orientation of NPC2 K123 may have the potential to interfere with optimal NPC1–MLD interaction (Fig. S4). The orientations of NPC2 side chains K25, K123, and Q146 in the NPC1 MLD–NPC2–cholesteryl sulfate structure are similar to that seen in the structure of NPC2–cholesteryl sulfate (12) and indicate that cholesterol binding likely enhances interaction with NPC1 MLD via this interface.
Fig. S4.
Structural comparison of NPC2–cholesteryl sulfate in the NPC1–MLD–NPC2 complex versus apo-NPC2. NPC1–MLD (magenta), NPC2 (cyan), and cholesteryl sulfate (yellow) and apo-NPC2 (gray, without cholesteryl sulfate, PDB ID code 1NEP) are shown. K123 of NPC2 clashes with F504 of NPC1 (red circle) before binding cholesteryl sulfate. Upon binding cholesteryl sulfate, K25, M79, K123, and Q146 of NPC2 change their conformations significantly to bind Q421 and E502 of NPC1 (12).
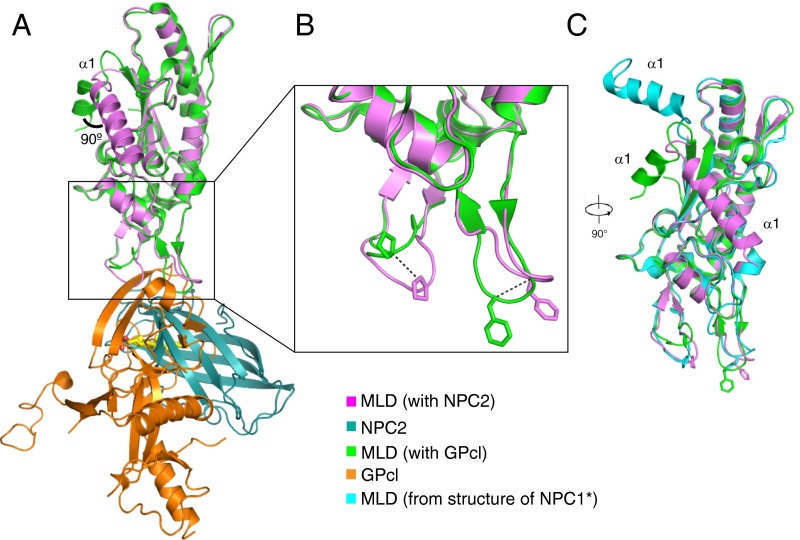
NPC1 uses a similar mode of interaction to bind NPC2 and Ebola GPcl (Fig. 2A and Fig. S3), although the interaction surface area of 500 Å2 for NPC2 is somewhat smaller than that used by GPc1 (685 Å2; refs. 10 and 11). In the MLD–GPcl complex, seven residues in loop 1 and six residues in loop 2 participate, creating an extensive interface that could explain why the MLD binding affinity for GPcl is approximately eightfold higher than that seen with NPC2 (18). The majority of the NPC1 MLD in the MLD–NPC2 complex can be aligned with the MLD–GPcl complex with an rmsd of 0.614 Å. Due to crystal packing, α-helix 1 is rotated by 90° (Fig. 2A). Another major difference between the two MLDs is seen when one compares the orientations of loops 1 and 2. The positions of loop 1 residue P424 Cα atoms differ by 5.8 Å between these structures; the Cα atoms of loop 2 residue F503 differ by ∼5 Å (Fig. 2B). Compared with the orientation of the MLD in the NPC1 crystal structure (17), MLD alpha helix 1 and the two loops also assume an alternate conformation (Fig. 2C). The conformation of the two MLD loops in the cryo-EM NPC1 structure (18) was modeled after MLD–GPcl and may not have detected this difference.
Fig. 2.
Comparison of NPC1 MLD structures. (A) Overlay of MLD–NPC2 and MLD–GPcl (PDB ID code 5F1B). (B) Close-up view of the conformational changes of the two protruding loops; the shift of Cα is indicated by dotted lines. (C) Overlay of NPC1 MLDs from MLD-NPC2, MLD–GPcl, and the NPC1* crystal structure (PDB ID code 5I31), rotated 90° relative to A.
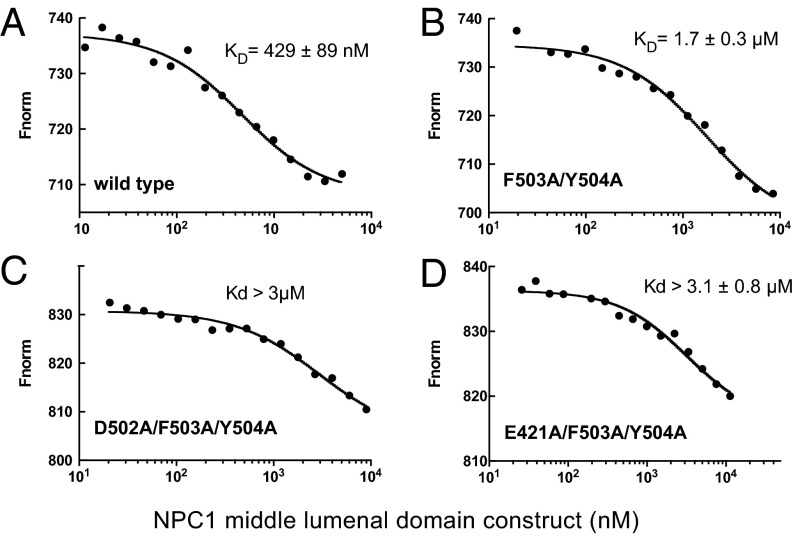
In previous work, we showed that NPC1’s MLD binds surface-immobilized NPC2 with an affinity of ∼2 µM as determined by surface plasmon resonance, at pH 5.5 in the presence of cholesterol sulfate that can bind to NPC2 (8). Here, we used microscale thermophoresis to measure the binding affinity between the murine NPC1 (mNPC1) MLD and NPC2 in solution, under otherwise similar conditions. These experiments used fluorescent, bovine NPC2 (NT647 dye-conjugated) and monitored its interaction with soluble, mammalian cell-expressed MLD constructs. Control experiments indicated that the bacterially and mammalian cell-expressed MLD’s bound NPC2 with essentially identical affinity.
Wild-type mNPC1 MLD bound NPC2 with a Kd of 429 ± 89 nM under these conditions (Fig. 3A). Mutation of residue Y506 did not alter the binding affinity significantly; however, mutation of residues F503 and Y504 (F504 in human) decreased binding affinity fourfold to 1.7 µM (Fig. 3B). Introduction of either of two additional mutations, E421A (Q421 in human) and D502A (E502 in human), yielded a protein with more than a sevenfold decrease in affinity for NPC2 protein (Fig. 3 C and D). These in vitro binding assays confirm the importance of the MLD loops in contributing to the NPC2 binding interface.
Fig. 3.
Microscale thermophoresis analysis of murine NPC1–MLD interaction with bovine NPC2–cholesteryl sulfate. (A) Wild type. (B) F503A/Y504A. (C) D502A/F503A/Y504A. (D) E421A/F503A/Y504A.
To verify the physiological importance of NPC1 MLD residues predicted to mediate NPC2 binding from the complex structure we introduced MLD mutations into the closely related, full-length mNPC1 membrane protein and tested whether the mutant proteins could rescue the cholesterol accumulation phenotype seen in NPC1−/− cells (20). In these experiments, cholesterol accumulation was monitored using Alexa 647-conjugated Perfringolysin O* that binds cholesterol directly (20).
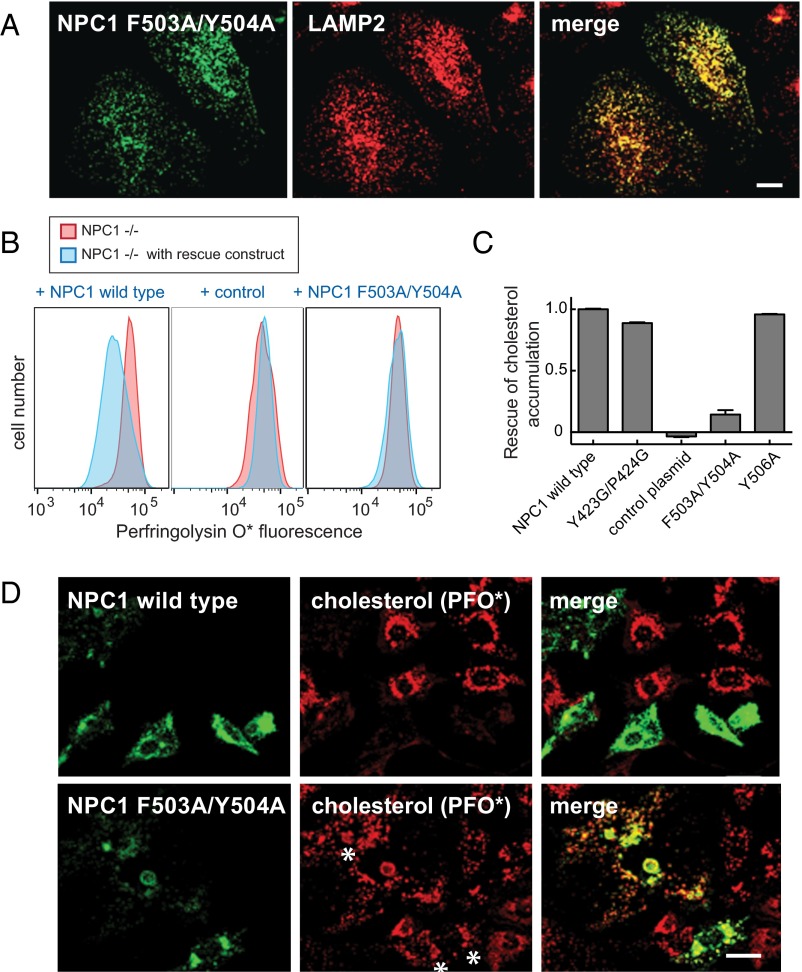
First, we verified that mNPC1-F503A/Y504A was correctly localized to lysosomes upon expression in HeLa cells (Fig. 4A). This is important, because many NPC1 mutations interfere with proper folding and export from the endoplasmic reticulum. Colocalization with endogenous lysosome-associated membrane protein 2 (LAMP2) confirmed that the mutant NPC1 protein was indeed correctly delivered to lysosomes. Despite its proper subcellular localization, mNPC1-F503A/Y504A could not rescue cholesterol accumulation seen in NPC1−/− cells, scored by quantitative flow cytometry (Fig. 4 B and C) or by immunofluorescence light microscopy (Fig. 4D). These data confirm the physiological importance of F503 and Y504 in NPC1 function in cells and correlate the ability of NPC1 MLD residues to bind NPC2 as part of the overall, NPC1-driven cholesterol export process.
Fig. 4.
Functional analysis of NPC1 MLD mutant proteins. (A) Confocal immunofluorescence microscopy analysis of the localization of mouse NPC1 F503A/Y504A and LAMP2 proteins in HeLa cells; a single stack is shown. (Scale bar, 20 µm.) (B and C) Flow cytometry of NPC1−/− CHO cells transfected for 48 h with GFP-tagged versions of either mouse wild-type NPC1, control plasmid encoding human GCC185 residues 1–889 (30), or NPC1 F503A/Y504A protein. Cells were labeled with AF-647 labeled perfringolysin O* (20) after fixation. More than 5,000 GFP-positive cells were analyzed; shown in C are mean values for the data presented in B, and error bars represent SEM. (D) Confocal immunofluorescence microscopy of the rescue experiment analyzed in B and C. NPC1−/− CHO cells were transfected with the indicated plasmids for 48 h before fixation; endogenous GFP fluorescence and AF647-PFO* labeling are shown. Asterisks denote regions that show mutant protein expression and residual cholesterol accumulation. Images represent maximum projections. (Scale bar, 20 µm.)
Discussion
The recent elucidation of the structure of NPC1, by protein crystallography (17) and cryoelectron microscopy (18), represents an important advance in terms of our determining the mechanism by which NPC1 might export cholesterol from lysosomes. Here, we have provided clues to how NPC1 may receive cholesterol from NPC2, a critical lysosomal cholesterol solubilizing and transport factor. Consistent with our previous cellular and biochemical studies implicating a role for NPC1’s middle lumenal domain in engaging NPC2 protein, the structure of a complex of the MLD with NPC2 protein-bearing cholesterol reported here has identified the precise interface needed for this interaction. Mutagenesis of the key residues has demonstrated their importance for NPC1–NPC2 interaction in vitro and NPC1’s ability to clear cholesterol from lysosomes in cultured cells. The interface between NPC2 and NPC1’s MLD explains why this interaction shows preference for NPC2 carrying bound cholesterol.
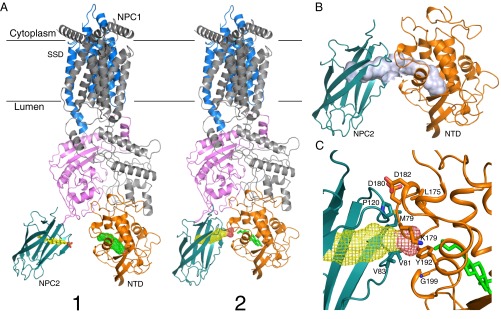
Previous work has provided the crystal structures of both NPC2 (12) and NPC1–NTD with bound sterol (14), and others have tried to model the interface between these proteins by model building and molecular dynamics simulation (19, 21). Instead, we have docked the crystal structure of the NPC1–MLD–NPC2 complex onto the NPC1 cryo-EM structure (Fig. 5A). In this model, the orientations of sterol molecules in the NPC2 and NPC1–NTD pockets are compatible with molecular transfer: the distance between the two pockets is about 15 Å (Fig. 5A, state 1) and NPC1 would be capable of receiving NPC2-bound cholesterol by first engaging NPC2 using NPC1–MLD sequences.
Fig. 5.
Hypothetical model for cholesterol transfer between NPC2 and NPC1. (A) Model of NPC2 and full-length NPC1 complex. State 1 was generated by alignment of major MLD structures from the NPC1 MLD–NPC2 complex and cryo-EM NPC1 structure (PDB ID code 3JD8); state 2 was generated by alignment of two protruding loops of both structures, creating a cholesterol transfer state. The sterol ligand is shown in stick representation, and the second sterol-binding site is shown in mesh. (B) Putative cholesterol transfer tunnel in state 2 (light purple); note that this tunnel is composed of two independent, adjacent cholesterol binding sites. (C) Potential interacting residues between NPC2 and NPC1-NTD in State 2.
Although the two NPC1–MLD protruding loops can present distinct conformations (Fig. 2 B and C), the Cα distances between Y423, P424, F503, and F504 are quite similar and suggest that they might move as a rigid body. Indeed, superposition of the two loops onto the full-length structure brings NPC2 closer to the NTD and may reflect a conformation competent for cholesterol transfer (Fig. 5A, state 2). In this state, the two cholesterol-binding pockets can be aligned to form a cholesterol-transfer tunnel (Fig. 5B), although the angle of this interaction is more acute than previous proposals (19, 21). The proximity and interaction of these pockets could trigger sterol transfer from NPC2 to NPC1’s NTD, consistent with biochemical data that suggest that NPC2–NPC1 interaction catalyzes this process (13). It is important to note that NPC2 binding to NPC1 may trigger a conformational change(s) that reorients NPC1 NTD into a more planar configuration in relation to NPC2 to accomplish actual cholesterol transfer, as previously modeled (19, 21).
Alanine scanning mutagenesis identified three NPC2 residues, M79, V81, and V83, as being needed for cholesterol transfer from NPC2 to NPC1’s NTD, but not for cholesterol binding; P120 influenced cholesterol binding (19). Similarly, NPC1–NTD mutations L175/L176 and E191/Y192 abolish cholesterol transfer from NPC2 to NPC1–NTD (14). In our model, NPC2’s P120 is close to NPC1 NTD’s K179, D180, and D182 (Fig. 5C). NPC2’s M79 could potentially bind NPC1–NTD residue L175. Moreover, NPC2’s V81 and V83 could provide a hydrophobic environment for interaction with an NPC1–NTD loop containing G199 and Q200. Interestingly, Y192 is in the center of the tunnel, which positions it in a perfect location to serve as a switch for cholesterol transfer (Fig. 5C).
Storch and coworkers have identified other regions of NPC2 that are also important for NPC2 function in cholesterol export (22, 23); these domains may interact with bis(monoacylglycerol)phosphate and/or other lipids within the lysosome lumen to facilitate initial cholesterol acquisition.
The structural observations reported here support further, the “hydrophobic hand-off” transfer model between NPC2 and NPC1 (14). After binding cholesterol, NPC2 undergoes a subtle conformational change (12) that enhances its binding to the NPC1–MLD. Additional interaction of NPC2 with NPC1’s NTD could then swing the two MLD protruding loops slightly. This would orient the NPC2 pocket directly adjacent to NPC1’s NTD cholesterol-binding pocket to form a cholesterol transfer tunnel (Fig. 5B). Cholesterol could then move through this tunnel, from NPC2 to NPC1’s NTD. After transfer, NPC2’s K25, M79, K123, and Q146 may revert to their apo-NPC2 conformations, which would release NPC2 for another round of cholesterol capture.
The cryo-EM structure was not able to provide information regarding the structures and positioning of either the first transmembrane domain (TM1) or the very long polypeptide linker that connects it to TM2 and the rest of the NPC1 protein (18). It is therefore possible that after cholesterol binding, the polyproline segment located between the NPC1–NTD and TM1 reorients in such a way as to make it possible for cholesterol delivery to the sterol binding pocket identified in the sterol sensing domain (17). Altogether, the NPC2–NPC1 transfer system can facilitate cholesterol transfer across the lysosomal glycocalyx (∼8 nm; ref. 20) and initiate a “pocket brigade” for cholesterol transport (17).
Finally, our structure has shown that NPC2 interacts with NPC1 using an interaction interface that is similar to that used by the Ebola virus GP. At slightly acidic pH (∼6), the viral GP binds to NPC1 approximately eightfold more tightly than to NPC2 in vitro (18). This feature may permit the virus to outcompete NPC2 to achieve productive infection and might also block cholesterol export in infected cells. How the sterol-related compound, U18666A, and other small molecules that bind NPC1 and block its function (5, 24, 25) interfere with Ebola virus entry will be important to determine in future studies.
Experimental Procedures
Protein Expression and Purification.
Human NPC2 (GI: 48429027) including its native signal peptide (residues 1–19) was cloned into pEG BacMam with a C-terminal His6-tag. The protein was expressed using baculovirus-mediated transduction of mammalian HEK-293S GnTI− cells. At 72 h postinfection at 30 °C, cells were disrupted by sonication in buffer A (20 mM Hepes, pH 7.0, 150 mM NaCl, 1 mM PMSF, and 5 μg/mL each of leupeptin and aprotinin). After high-speed centrifugation, the supernatant was loaded onto a Ni2+-NTA column (Qiagen). After two washes, the protein was eluted in 20 mM Hepes, pH 7.0, 300 mM NaCl, and 300 mM imidazole. The eluted protein was applied to Hi-trap Q (GE Healthcare); the glycosylated protein was collected in the flow through and dialyzed into 50 mM MES, pH 5.5, plus 10 μg/mL EndoHf (NEB) at room temperature overnight. The deglycosylated protein was purified by Superdex-200 chromatography (GE Healthcare) in Buffer B containing 20 mM MES, pH 5.5, and 100 mM NaCl. Peak fractions were concentrated for crystallization. Human NPC1 (GI: 83305902) residues 374–620 were cloned into pET21b with a C-terminal His6-tag. NPC1–MLD proteins were expressed in Escherichia coli strain BL21 (DE3) as inclusion bodies and then refolded in vitro. Briefly, inclusion bodies were dissolved in 6 M guanidine, 50 mM Tris⋅Cl, pH 8.0, 2 mM EDTA, and 100 mM NaCl. The ∼15 mg denatured protein was added to 500 mL cold refolding buffer: 100 mM Tris⋅Cl, pH 8.0, 2 mM EDTA, 400 mM l-arginine, 0.1 mM PMSF, 6.5 mM cysteamine, and 3.7 mM cystamine. Refolding was at 4 °C with mixing for at least 1 h before dialysis with 20 mM Tris⋅Cl, pH 8.0; the buffer was changed after 24 h. After 48 h, refolded proteins were applied to Hi-Trap Q (GE Healthcare) and further purified by Superdex-200 chromatography (GE Healthcare) in Buffer B. Bovine NPC2 and mammalian cell expressed MLD (domain 2) were purified as previously described (8). The mutant proteins were generated by standard molecular biology techniques.
Crystallization.
Before crystallization, NPC2 and NPC1–MLD were mixed at a final concentration of 100 μΜ each, plus 500 μM cholesterol-3-O-sulfate (Sigma-Aldrich), at room temperature for 1 h. Crystals were grown at 20 °C by sitting-drop vapor diffusion. Crystals appeared in 3 d in well buffer containing 0.1 M MES, pH 6.5, 0.1 M NaCl, and 30% (vol/vol) PEG400. The crystals in space group C2221 have unit cell dimensions a = 98.956 Å, b = 109.686 Å, and c = 154.560 Å. Each asymmetric unit contains two molecules of complex (∼50% solvent content). All crystals were flash-frozen in a liquid nitrogen stream with well buffer for cryoprotection.
Data Collection and Structure Determination.
The data were collected at the Advanced Photon Source beamline ID24-E at 100K. The dataset was processed using HKL2000 (26). The complex structure was solved by molecular replacement method using Phaser from the CCP4 program suite (Collaborative Computational Project) with the previously reported NPC1–MLD structure [Protein Data Bank (PDB) ID code 5F1B] and NPC2 structure (PDB ID code 1NEP) as search models. The initial model was built in Coot (27) manually. The structure was refined with PHENIX.REFINE (28) at 2.4-Å resolution. Model validation was performed with MolProbity (29). All figures were generated with PyMOL.
Microscale Thermophoresis.
Experiments were performed on a Monolith NT.115 instrument (Nanotemper Technologies). Bovine NPC2 protein was labeled using the RED-NHS (Amine Reactive) Protein Labeling Kit (Nanotemper Technologies). Labeled NPC2 (75 nM) was incubated with 1 μM cholesteryl sulfate for 15 min at 30 °C. Next, it was mixed with either wild-type or mutant soluble mNPC1–MLD–FLAG-HIS6 in a final buffer composed of 50 mM MES, pH 5.5, 150 mM NaCl, and 0.004% Nonidet P-40. Reactions were analyzed using premium capillaries and contained 16 twofold serial dilutions of wild type, F503A/Y504A, E421A/F503A/Y504A, or D502A/F503A/Y504A mutants starting with 5, 8.5, 11, or 9 µM protein. Analysis was at 60% microscale thermophoresis power for 20 s, followed by 5 s of cooling. The dissociation constant Kd was obtained by plotting the normalized fluorescence Fnorm against the logarithm of the different concentrations of the dilution series according to the law of mass action.
Light Microscopy and Flow Cytometry.
Confocal microscopy and flow cytometry were carried out as described (20). NPC1−/− CHO ldlD cells were generated by CRISPR techniques (20) and grown in 100 µM GalNAc and 8 µM galactose in MEM-alpha medium supplemented with 7.5% (vol/vol) FBS.
Acknowledgments
We thank Jiawei Wang for help with the structure determination and the staff of the Northeastern Collaborative Access Team at the Advanced Photon Source for assistance with data collection. This research was supported by the Howard Hughes Medical Institute (G.B.), the Ara Parseghian Medical Research Fund, the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases Grant 037332 (to S.R.P.), and American Diabetes Association Grant 7-12-MN-67 (to P.S.). X. Li is a Gordon and Betty Moore Foundation fellow of the Life Sciences Research Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5KWY).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611956113/-/DCSupplemental.
References
- 1.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232(4746):34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem. 1975;250(21):8487–8495. [PubMed] [Google Scholar]
- 3.Rosenbaum AI, Maxfield FR. Niemann-Pick type C disease: Molecular mechanisms and potential therapeutic approaches. J Neurochem. 2011;116(5):789–795. doi: 10.1111/j.1471-4159.2010.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477(7364):340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Côté M, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477(7364):344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Infante RE, et al. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J Biol Chem. 2008;283(2):1052–1063. doi: 10.1074/jbc.M707943200. [DOI] [PubMed] [Google Scholar]
- 7.Infante RE, et al. Purified NPC1 protein: II. Localization of sterol binding to a 240-amino acid soluble luminal loop. J Biol Chem. 2008;283(2):1064–1075. doi: 10.1074/jbc.M707944200. [DOI] [PubMed] [Google Scholar]
- 8.Deffieu MS, Pfeffer SR. Niemann-Pick type C 1 function requires lumenal domain residues that mediate cholesterol-dependent NPC2 binding. Proc Natl Acad Sci USA. 2011;108(47):18932–18936. doi: 10.1073/pnas.1110439108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller EH, et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31(8):1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, et al. Ebola viral glycoprotein bound to its endosomal receptor Niemann-Pick C1. Cell. 2016;164(1-2):258–268. doi: 10.1016/j.cell.2015.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao Y, Ren J, Harlos K, Stuart DI. Structure of glycosylated NPC1 luminal domain C reveals insights into NPC2 and Ebola virus interactions. FEBS Lett. 2016;590(5):605–612. doi: 10.1002/1873-3468.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu S, Benoff B, Liou HL, Lobel P, Stock AM. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J Biol Chem. 2007;282(32):23525–23531. doi: 10.1074/jbc.M703848200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Infante RE, et al. NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proc Natl Acad Sci USA. 2008;105(40):15287–15292. doi: 10.1073/pnas.0807328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwon HJ, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137(7):1213–1224. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuwabara PE, Labouesse M. The sterol-sensing domain: Multiple families, a unique role? Trends Genet. 2002;18(4):193–201. doi: 10.1016/s0168-9525(02)02640-9. [DOI] [PubMed] [Google Scholar]
- 16.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124(1):35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 17.Li X, et al. Structure of human Niemann-Pick C1 protein. Proc Natl Acad Sci USA. 2016;113(29):8212–8217. doi: 10.1073/pnas.1607795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong X, et al. Structural insights into the Niemann-Pick C1 (NPC1)-mediated cholesterol transfer and Ebola infection. Cell. 2016;165(6):1467–1478. doi: 10.1016/j.cell.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ML, et al. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 2010;12(2):166–173. doi: 10.1016/j.cmet.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Deffieu MS, Lee PL, Saha P, Pfeffer SR. Glycosylation inhibition reduces cholesterol accumulation in NPC1 protein-deficient cells. Proc Natl Acad Sci USA. 2015;112(48):14876–14881. doi: 10.1073/pnas.1520490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estiu G, Khatri N, Wiest O. Computational studies of the cholesterol transport between NPC2 and the N-terminal domain of NPC1 (NPC1(NTD)) Biochemistry. 2013;52(39):6879–6891. doi: 10.1021/bi4005478. [DOI] [PubMed] [Google Scholar]
- 22.Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J Biol Chem. 2006;281(42):31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- 23.McCauliff LA, et al. Multiple surface regions on the Niemann-Pick C2 protein facilitate intracellular cholesterol transport. J Biol Chem. 2015;290(45):27321–27331. doi: 10.1074/jbc.M115.667469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu F, et al. Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife. 2015;4:e12177. doi: 10.7554/eLife.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhein BA, Maury WJ. Ebola virus entry into host cells: Identifying therapeutic strategies. Curr Clin Microbiol Rep. 2015;2(3):115–124. doi: 10.1007/s40588-015-0021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski ZM, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung PY, Limouse C, Mabuchi H, Pfeffer SR. Protein flexibility is required for vesicle tethering at the Golgi. eLife. 2015;4:e12790. doi: 10.7554/eLife.12790. [DOI] [PMC free article] [PubMed] [Google Scholar]