Abstract
Barley is one of the founder crops of Old world agriculture and has become the fourth most important cereal worldwide. Information on genome-scale DNA polymorphisms allows elucidating the evolutionary history behind domestication, as well as discovering and isolating useful genes for molecular breeding. Deep transcriptome sequencing enables the exploration of sequence variations in transcribed sequences; such analysis is particularly useful for species with large and complex genomes, such as barley. In this study, we performed RNA sequencing of 20 barley accessions, comprising representatives of several biogeographic regions and a wild ancestor. We identified 38,729 to 79,949 SNPs in the 19 domesticated accessions and 55,403 SNPs in the wild barley and revealed their genome-wide distribution using a reference genome. Genome-scale comparisons among accessions showed a clear differentiation between oriental and occidental barley populations. The results based on population structure analyses provide genome-scale properties of sub-populations grouped to oriental, occidental and marginal groups in barley. Our findings suggest that the oriental population of domesticated barley has genomic variations distinct from those in occidental groups, which might have contributed to barley’s domestication.
Food security is an urgent global issue for ensuring sustainable food supply for the current and future generations. Climate change adversely affects the global agricultural system, and thus increases the risk of food shortage1,2. Mining and biodiversity utilisation should be a potential promising strategy to prevent declines in crop production; this involves the systematic domestication of newly designed crops that have improved productivity3.
Barley (Hordeum vulgare ssp. vulgare) is the fourth most important cereal crop worldwide following wheat, rice, and maize (https://www.croptrust.org/crop/barley/). It is believed to be among the oldest crop species in the world and it was domesticated from the large-seeded wild barley (H. vulgare ssp. spontaneum)4,5. Archaeological evidences indicate that the seed size of wheat and barley began to increase during Pre-Pottery Neolithic A and early Pre-Pottery Neolithic B, dating approximately from 11,100 to 10,500 before present, at the sites Jerf el Ahmar in Syria6 and ZAD-2 in Jordan7,8. In many crops, including barley, multiple domestications have been hypothesised9,10,11,12,13,14,15, as multiple progenitor populations lead to numerous independent domestication processes with complex demographic histories16. In multiple domestication, many agronomical important mutations of independent origins could contribute simultaneously to the domestication processes17.
As barley is one of the most widely adapted crops, barley germplasm pools might possess genetic diversity associated with adaptability to various environmental conditions. They should be able to be used as a source for mining valuable allelic variations to improve barley productivity. The large ex situ barley seed collections allow the exploration of useful genes and allelic variations that can contribute to the improvement of productivity and sustainability of barley. Several studies investigated the extent of genetic variations and population structure of wild barley by using different seed collections18,19. Phylogeographical analysis performed using 318 wild barley accessions from the Wild Barley Diversity Collection suggested that the primary structured population differentiated into West and East populations across Zagros Mountains20. In domesticated barley, the population structure analysis based on five nuclear loci from >250 accessions showed a primary bisected population structure, which mainly represented a cluster consisting of landraces from Europe/North Africa, Ethiopia, and the Near East (occidental) and another cluster with landraces from Asia (oriental)21. This genetically differentiated pattern of domesticated barley was well correlated with the heritage research reported previously22. The Asian group was designated as “Oriental” barley whereas the “Occidental” group is distributed in the western part of Eurasia and North Africa. During the multiple domestications of barley, the eastern wild barley ancestors could have greatly contributed to the differentiation of Asian landraces, which indicates that valuable genes and allelic variations are involved in the adaptive traits of varieties to local environmental conditions in Asia11,23,24. Therefore, mining of genetic diversity from the Asian landraces might allow the detection of valuable variations that can be used to improve the adaptability of barley against various environmental conditions as well as improve our understanding on the evolutional history of this crop.
In-depth analysis of genes and alleles involved in crop domestication and adaptability to environments requires the use of comparative re-sequencing analyses and high-density single nucleotide polymorphism (SNP) genotyping25,26,27. In barley, genome-wide SNPs among accessions were identified using Sanger sequencing of cDNAs and pyrosequencing of genomic DNA using the genomic reduction method28,29. More recently, the array-based genotyping platform Infinium iSelect, consisting of 7,842 SNPs, was used to genotype 2,417 barley accessions sampled from the 33,176 accessions comprising the USDA National Small Grains Collection to assess their genetic diversity and population structure30. Although chip- or array-based genotyping has enabled the analysis of thousands of polymorphisms that were validated in advance, these methods involve the inclusion of ascertainment biases caused during the polymorphism discovery process31,32. The ascertainment biases can be avoided performing direct whole-genome re-sequencing, RNA sequencing (RNA-seq), or exome-sequencing using diverse panels of accessions to assess the genetic diversity in a germplasm set more accurately33,34. An exome capture kit with a 90.2 Mb capture space was designed for barley and applied to genotype three H. spontaneum and 13 H. vulgare accessions. The phylogenetic relationships of these barley accessions were obtained based on 122,940 bi-allelic SNPs identified using exome-sequencing analysis33. RNA-seq is a useful method to rapidly acquire comprehensive gene expression profiles and to generate genome-scale datasets of sequence polymorphisms35. Scalability and cost-effectiveness of RNA-seq methods allow in-depth analysis of polymorphisms found in the exome regions even in a species with a large and complex genome, such as barley.
In this study, to assess the genetic diversity of oriental barley landraces, we performed RNA-seq analysis of 19 diverse accessions of domesticated barley and one wild barley variety, H. vulgare ssp. spontaneum H602. Based on RNA-seq results, we obtained the global distribution of SNPs on the transcribed regions of the several barley landraces, including Asian barley. Using this SNP dataset, we determined the phylogenetic relationships among oriental and occidental barley landraces and wild barley. Furthermore, using this SNP dataset generated from RNA-seq and another generated using a publicly available exome-sequencing dataset provided by Mascher et al.33, we estimated the population structure of domesticated barley accessions. Based on the entire barley population, we revealed domesticated barley genomic diversity and population subdivision in a genomic context.
Results and Discussion
Transcriptome sequencing and transcribed region annotation
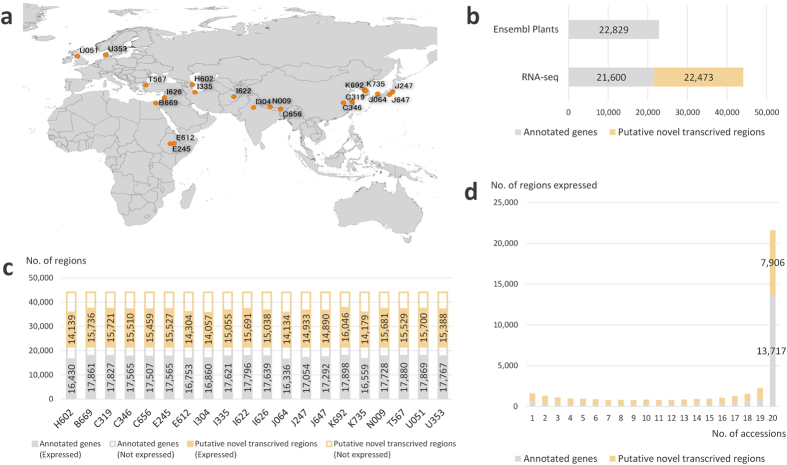
We selected 19 domesticated barley accessions (18 landraces and an improved variety) of several biogeographical origins, based on a previous analysis of molecular phylogenetics and growth habits21,36. The properties of these barley accessions are summarised in Supplementary Table 1. In addition, we used an ancestral variant of wild barley, H602, which is widely used as a reference species of H. spontaneum. The geographic origin of each barley accession is shown in Fig. 1a. Further, H. vulgare L. ‘Morex’ was used as a reference to construct the genomic framework for barley’s RNA-seq analysis.
Figure 1. Barley accessions and transcribed regions identified using RNA-seq analysis.
(a) Geographic origin of the barley accessions used in this study. The origins were plotted on a map data made with Natural Earth (http://www.naturalearthdata.com/) through the QGIS 2.8.3 Wein interface (http://qgis.org/en/site/index.html). (b) Number of transcribed regions identified using RNA-seq analysis, which were compared to those annotated using the Morex genome in Ensembl Plants. (c) Number of transcribed regions expressed in each accession (FPKM ≥ 1). (d) Range of regions expressed across the 20 barley accessions.
To rapidly generate genome-scale SNPs datasets for each barley accession from the transcribed regions, we sequenced mRNAs from the leaves and roots. Using an Ion Proton semiconductor sequencer based on the pyrosequencing method, we generated 278 million reads and mapped more than 94% (263 million reads) of these reads to the pseudomolecules of the Morex genome (Table 1). Using the gene annotation performed for Morex available from the Ensembl plants website (http://plants.ensembl.org/index.html), we predicted gene structure based on RNA-seq reads mapping to the Morex genome using Cufflinks. Comparing our gene structural annotation to that of Morex, 94.6% of the annotated genes had corresponding sequence reads and 22,473 newly identified transcribed regions showed sequence similarity to the protein sequences of at least one grass species (Fig. 1b). From 7,700 to 12,000 protein sequences showed sequence similarity to barley’s novel potentially transcribed regions (Supplementary Figure 1). These results suggested that a large number of transcriptional units (TUs) were not annotated in the current Morex genome annotation, and that the identification of such novel TUs might be facilitated using our RNA-seq reads. Comparative analysis between the transcripts of Brachypodium, a Pooideae model grass, and the Morex genome showed that many sequences expressed in Brachypodium were mapped to the inter-genic regions of the Morex genome37. Mascher et al.33 performed whole exome-sequencing of barley and conducted gene structural prediction by using RNA-seq contigs to identify as many TUs as possible33. Results of the present study suggest that comprehensive transcriptome sequencing and comparative analyses with related species are essential to further annotate TUs in the barley genome, which might provide a genomic foundation to explore genetic polymorphisms in the genic regions.
Table 1. Summary of the RNA-seq results for the 20 barley accessions.
| Species | Accession | Total No. of reads | Total No. of mapped reads | Percent of mapped reads | Average reads length (bp) |
|---|---|---|---|---|---|
| H. spontaneum | H602 | 10,022,606 | 9,358,952 | 93.38% | 136 |
| H. vulgare | B669 | 15,479,737 | 14,721,023 | 95.10% | 125 |
| C319 | 15,144,777 | 14,487,337 | 95.66% | 145 | |
| C346 | 15,176,183 | 14,345,822 | 94.53% | 146 | |
| C656 | 16,026,087 | 15,406,069 | 96.13% | 145 | |
| E245 | 15,341,723 | 14,679,342 | 95.68% | 149 | |
| E612 | 11,201,259 | 10,323,600 | 92.16% | 145 | |
| I304 | 9,081,490 | 8,402,871 | 92.53% | 145 | |
| I335 | 13,541,901 | 12,925,132 | 95.45% | 164 | |
| I622 | 16,406,298 | 15,659,799 | 95.45% | 152 | |
| I626 | 14,872,103 | 14,176,640 | 95.32% | 161 | |
| J064 | 15,403,340 | 14,384,534 | 93.39% | 152 | |
| J247 | 9,845,797 | 8,962,223 | 91.03% | 150 | |
| J647 | 10,068,027 | 9,195,339 | 91.33% | 145 | |
| K692 | 15,788,289 | 15,112,668 | 95.72% | 124 | |
| K735 | 11,214,410 | 10,408,421 | 92.81% | 160 | |
| N009 | 16,761,966 | 15,927,479 | 95.02% | 143 | |
| T567 | 15,294,760 | 14,574,021 | 95.29% | 142 | |
| U051 | 15,711,210 | 15,027,237 | 95.65% | 141 | |
| U353 | 15,519,320 | 14,754,353 | 95.07% | 160 | |
| Total | 277,901,283 | 262,832,862 | 94.33% | 147 |
We assessed the number of regions that are transcribed in each of the 20 barley accessions. In each accession, we analysed 30–33 K regions that showed expression levels measured as fragments per kilobase of exon per million mapped fragments (FPKM) values ≥1, which included about 16–17 K annotated genes and 14–16 K putative novel transcribed regions (Fig. 1c). We also investigated the levels of gene expression in the 20 barley accessions and found that more than 13 K annotated genes and 7 K putative novel transcribed regions were commonly expressed in all the 20 accessions (FPKM ≥ 1; Fig. 1d). Therefore, RNA-seq analysis appears to be a feasible approach to compare transcribed regions among accessions, and to allow a rapid and comprehensive acquisition of SNP data from accessions, without requiring pre-designed tools such as sequence capture probes or primer sets for targeted re-sequencing. For barley, in particular, that has a large genome and immature gene-structure annotation, RNA-seq analysis of several accessions might allow accumulating genome sequence variations to elucidate on the diversity of the population.
SNP discovery and quality control
Mapping RNA-seq reads into the genomic framework of Morex allowed identifying SNPs using the following programs: freebayes, glfMultiples, and samtools (Table 2). Although each program yielded a distinct number of SNPs, we extracted those that were commonly called by all programs, as these were considered the accurate SNPs38 (Table 2). Some studies have recently reported a significant difference between the set of variants called by different variant callers39,40. In our study, we also observed changes in the ranks of the accessions among the different programs, which suggests that the trend in variant calling is not only determined by a simple threshold but also by factors such as the sequencing method, sequencing depth, or quality of the reference sequences41, as well as calling argorithums and mathematical models42.
Table 2. Number of RNA-seq based SNPs called by the various tools in comparison with the genomic framework of barley.
| Species | Accession | No. of SNPs |
|||
|---|---|---|---|---|---|
| freebayes | glfMultiples | samtools | fgs* | ||
| H. spontaneum | H602 | 67,462 | 147,417 | 91,442 | 55,403 |
| H. vulgare | B669 | 64,472 | 210,241 | 108,105 | 49,094 |
| C319 | 100,387 | 257,743 | 150,167 | 79,903 | |
| C346 | 99,545 | 262,354 | 149,535 | 78,364 | |
| C656 | 100,366 | 257,088 | 148,563 | 77,253 | |
| E245 | 90,395 | 241,736 | 137,109 | 69,761 | |
| E612 | 62,054 | 155,797 | 88,866 | 49,325 | |
| I304 | 50,712 | 119,733 | 71,578 | 41,676 | |
| I335 | 85,453 | 229,912 | 131,492 | 66,382 | |
| I622 | 102,344 | 262,084 | 152,592 | 79,949 | |
| I626 | 92,970 | 243,682 | 141,367 | 72,405 | |
| J064 | 75,308 | 161,268 | 101,226 | 59,153 | |
| J247 | 51,806 | 130,889 | 73,312 | 38,729 | |
| J647 | 63,195 | 148,909 | 89,326 | 51,591 | |
| K692 | 93,428 | 242,556 | 139,799 | 75,444 | |
| K735 | 54,103 | 134,995 | 78,892 | 41,098 | |
| N009 | 95,951 | 244,587 | 144,688 | 75,198 | |
| T567 | 72,187 | 211,850 | 114,637 | 55,206 | |
| U051 | 83,207 | 230,728 | 128,448 | 64,319 | |
| U353 | 78,417 | 213,824 | 119,391 | 59,136 | |
*Intersection of the identified SNPs using the three SNP callers: freebayes, glfMultiples, and samtools.
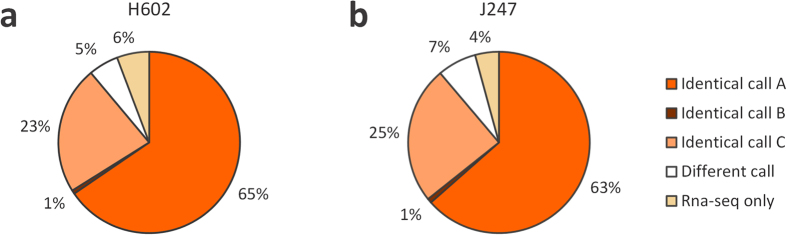
Our RNA-seq analysis and the exome-sequencing analysis performed by Mascher et al.33 enabled comparing the SNP datasets of H602 (OUH602) and J247 (Haruna Nijo), which were common to both studies. We applied the three programs to call SNPs in these exome-sequencing datasets, and identified 355,980 and 268,235 SNPs that were commonly called in the SNP call programs from H602 and J247, respectively (Table S2). Therefore, we assessed the accuracy of the RNA-seq-based SNPs in comparison with those from the exome-sequencing analysis. We also compared the 55,403 and 38,729 SNPs obtained for H602 and J247, respectively (Fig. 2, Table 2). Overall, 65% and 63% of the SNPs found for H602 and J247, respectively, in our RNA-seq analysis were also identified in the exome-sequencing analysis data, without ambiguity (Fig. 2. Identical call A: SNPs identically called by the three programs in both the RNA-seq and the exome-sequencing analyses). Further, 1% of the SNPs found in the RNA-seq dataset in H602 and J247 were shared by the exome-sequencing dataset (Fig. 2. Identical call B: SNPs identically called by the three programs in the RNA-seq and by one or two program(s) in the exome sequencing dataset). Twenty-three percent and 25% of the SNPs called for H602 and J247, respectively, were consistent with the raw dataset of SNP calls based on exome-sequencing (Fig. 2. Identical call C), which were filtered according to our threshold (see Methods). Among the RNA-seq-based SNPs, 5% in H602 and 7% in J247 were inconsistent with those in the exome-sequencing (Fig. 2. Different call). Of the remaining RNA-seq-based SNPs, 6% in H602 and 4% in J247 were only observed in RNA-seq (Fig. 2. RNA-seq only). Using the published exome-sequencing dataset, we could assess the accuracy of SNPs based on the RNA-seq analysis and determine the ideal combination of programs and thresholds for calling accurate SNPs. Our quality assessment suggested that intersecting the SNPs identified by the multiple calling programs provided further accurate SNPs with higher confidence. Table 2 summarises the number of SNPs identified based on RNA-seq analysis, ranging from 38.7 K (in J247) to 79.9 K (in I622) in each accession (Table 2).
Figure 2. Accuracy assessment of the RNA-seq-based SNPs and its comparison with exome-sequencing-based SNPs.
Proportion of RNA-seq-based SNPs also present in the exome-sequencing data obtained by Mascher et al.33 in H602 (a) and in J247 (b). Identical call A: SNPs consistent with exome-sequencing-based SNPs and called by all programs. Identical call B: SNPs consistent with exome-sequencing-based SNPs and called by one or two program(s). Identical call C: SNPs shared with raw data of the exome-sequencing dataset, which were filtered by our threshold. Different call: SNPs inconsistent between the RNA-seq and the exome-sequencing datasets. RNA-seq only: SNPs only observed in the RNA-seq dataset.
Several methods are currently available to carry out the discovery and genotyping of genome-scale polymorphisms. These methods are mainly classified into pre-designed and de novo approaches: SNP chips and exome-sequencing are representatives of the pre-designed approach, which require hybridisation probes or PCR primers to enrich targeted regions. High-density SNP chips like Illumina’sTM Infinium beads chip can support ~1 million SNPs in a custom designed panel. For exome-sequencing, the SeqCap EZ system (Roche), used for barley’s exome-sequencing, can cover up to 200 Mb of custom regions. If a high-quality and well-annotated reference genome is available, these pre-designed approaches allow genotyping validated SNPs and/or resequencing targeted regions with deeper coverage. On the other hand, whole genome re-sequencing, genotyping-by-sequencing (GBS), and RNA-seq are becoming popular approaches for de novo genotyping. Although recent price decline in sequencing cost has made it possible to perform whole genome re-sequencing in population analysis, it is still the most expensive approach, and shallow sequencing depth often becomes a problem, depending on the genome size of the target species. GBS is a cost effective approach for genome-scale genotyping, and has been widely used in several species, being feasible for large genomes such as those of wheat and barley. Although sequencing depth in GBS depends on the genome size of the target species, this method enables reducing genome representation, performing a cost-effective polymorphism discovery, and genotyping genic and intergenic regions. Because GBS does not always require a reference genome sequence, it can be used in population analysis and to construct an initial genetic map in organisms whose genome has not been sequenced. In this study, we applied RNA-seq to examine genome-scale polymorphisms among barley accessions. Although RNA-seq approach only covers SNPs on transcribed regions of expressed genes in analysed tissues, it is a cost-effective and well scalable method to analyse polymorphisms on expressed genes.
Although the range of SNPs identified in our RNA-seq analysis is lower than that obtained during deep exome-sequencing, it is similar or higher than those reported in previous GBS studies of Triticeae crops43,44. Such range of SNPs might be useful for genome-wide genotyping at middle-scale density, i.e. between amplicon-sequencing of panelled target genes and capture-based exome-sequencing densities. Our data suggested that sequencing a pooled RNAs sample is efficient to avoid tissue dependency and to increase the number of sequenced regions and identified SNPs. In addition, RNA-seq does not always require a reference genome, as a de novo transcriptome assembly might serve as proxy for a reference genome, enabling reducing genome representation without depending on its size and restriction sites. Therefore, RNA-seq and GBS appear to be reliable approaches to rapidly acquire genome-scale polymorphisms in organisms without reference genome information and mature structural annotation. RNA-seq might also be an efficient approach to rapidly acquire genome-scale polymorphism datasets at the population scale using a core-collection or sub-populations of landraces and mapping populations.
SNP profiles of the 20 barley accessions
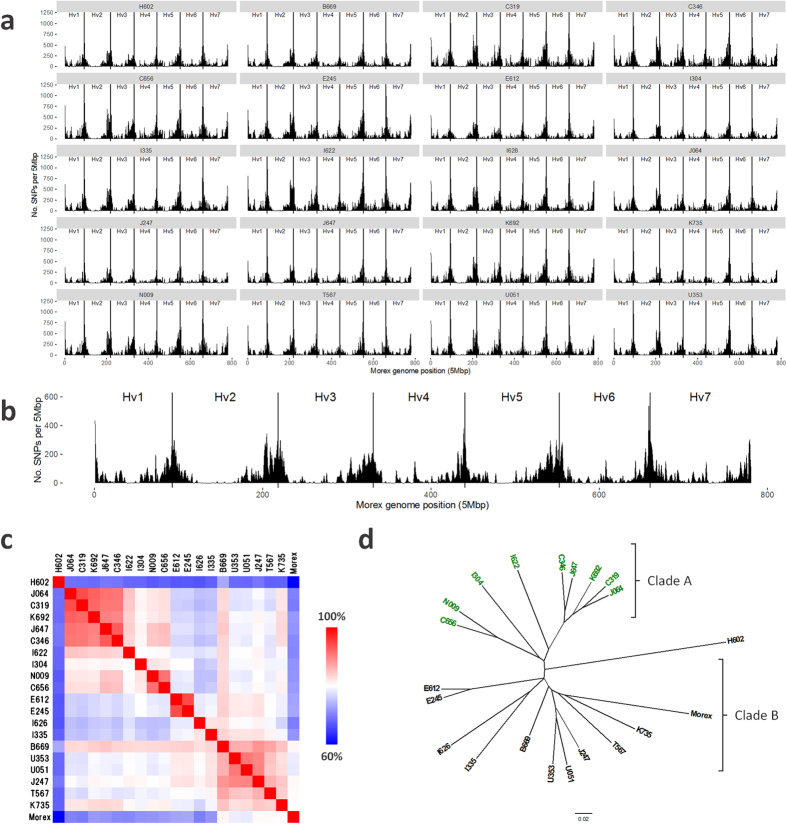
Using the SNP dataset described above, we identified the genome-wide SNP profiles of each barley accession (Fig. 3a). The density of SNPs was higher in the distal regions of each chromosome and was correlated with that of annotated genes in the barley genome. These profiles were in agreement with those determined using exome-sequencing33. For molecular phylogenetic analysis, we generated bi-allelic SNP datasets consisting of 37,774 SNPs with no missing data called from the 20 barley accessions and Morex (Fig. 3b).
Figure 3. Genome-wide distribution of the SNPs identified using RNA-seq analysis of the 20 diverse barley accessions.
(a) Distribution of the RNA-seq-based SNPs of the 20 barley accessions, using the Morex genome as a reference. The numbers of SNPs were computed for each chromosome using 5 million bases sliding windows. (b) Distribution of the bi-allelic RNA-seq-based SNPs. (c) Genome-wide sequence identity based on the bi-allelic SNP dataset across the 20 barley accessions and Morex. (d) Neighbour-joining tree of the 20 barley accessions and Morex based on the bi-allelic SNP dataset.
To estimate the genetic diversity among the 20 accessions and Morex, we calculated the pairwise proportion of identical nucleotides at the bi-allelic SNP sites. We found that approximately 60% of the bi-allelic SNPs were conserved between the wild barley, H602, and each of the remaining domesticated barley accessions (Fig. 3c). Using the bi-allelic SNP datasets, we determined the phylogenetic relationships among the accessions. In the phylogenetic tree, H602 was allocated to a distinct outer branch, as expected (Fig. 3d). Asian accessions were divided into two major groups: A, formed by nine Asian accessions; and B, comprising two Asian accessions (K735 and J247) and non-Asian accessions. K735 was originally collected from the Korean peninsula (approximately 37° N, 127° E) and is classified as the Manchurian type, which contributed to the breeding of North American six-rowed malting variety Morex, whereas J247 is Japanese modern variety for malting, developed by crossing European malting barley cultivars45. Thus, the phylogenetic relationships based on genome-scale polymorphisms reflect the genomic history of these accessions dictated by their modern breeding process. The topology of the tree and phylogenetic positions of each accession were in agreement with the previously reported population structure of domesticated barley21.
Barley molecular phylogeny based on genome-scale SNP datasets
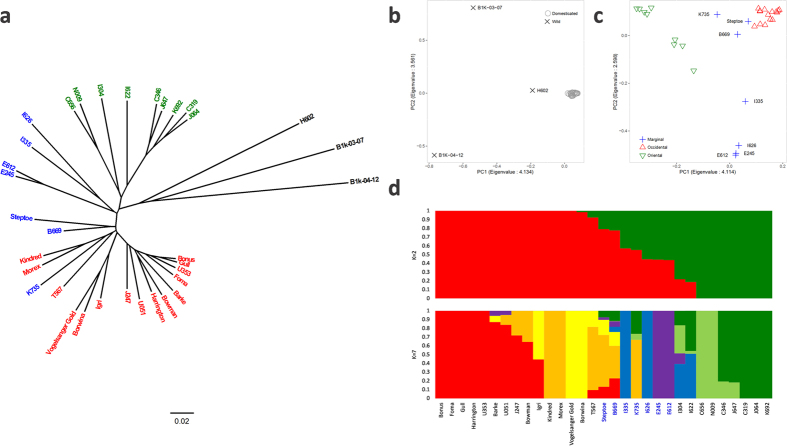
To conduct an in-depth analysis of barley divergence, we integrated the SNPs obtained from RNA-seq and exome-sequencing. We used the RNA-seq dataset of 20 barley accessions (Table 2) as well as the exome-sequencing datasets of 13 barley accessions comprised from two spontaneous and 11 domesticated barley accessions (mainly categorised to improved variety) (Table S2). The bi-allelic SNP datasets consisting of 40,364 SNPs with no missing data were generated from 33 barley accessions and Morex. Based on the bi-allelic SNP datasets, we determined the phylogenetic relationships among accessions (Fig. 4a). Within the clade comprising the 31 domesticated barley varieties, accessions were primarily classified into two groups with some intermediates. The nine Asian accessions were clustered into one clade (Fig. 4a), which corresponded to clade A in Fig. 3d and represented oriental accessions sequenced in this study. The other domesticated barley accessions were clustered into another clade that comprised the accessions grouped in clade B of Fig. 3d and all the cultivated barley accessions used in previously sequenced by exome-sequencing(Fig. 4a). A principal component analysis (PCA) using the bi-allelic SNP datasets of the 34 barley accessions evidenced the agglomeration of the domesticated accessions, all of which differed from the three wild barley accessions (Fig. 4b). The PCA based on the 30,547 bi-allelic SNPs called from the domesticated barley accessions only, showed a clear separation between the oriental and occidental barley accessions along the first principal component (PC1), with coastal Mediterranean accessions (n = 7) appearing between them (Fig. 4c). The phylogenetic tree and the PCA plots suggested that the presumed oriental sub-populations of domesticated barley possess particular variations that are different from those in the other accessions. In addition to phylogenetic and principal components analyses, we also examined sub-population structure in domesticated barley using the Bayesian clustering method. The number of populations (K) indicated by the delta K values46 the best-fitted model was two (Supplementary Figure 2). In this model, oriental and occidental barley were clearly classified into distinct clusters and the probability scores of some accessions suggested an admixture pattern (Fig. 4d). We also examined the model showing the second highest delta K value (K = 7) to elucidate the population structure of the domesticated barley accessions in a further classified model (Fig. 4d), where oriental and occidental populations being subdivided into two and three clusters, respectively. Although the Ethiopian (E245 and E612) and Near East (I335 and I626) landraces formed marginal clusters in the K = 2 model, these accessions formed differentiated clusters in the K = 7 model. Steptoe, B669, and K735 accessions showed mixed membership probabilities toward oriental and occidental populations in both models and in the PCA were plotted between oriental and occidental accessions, along PC1 (Fig. 4c), suggesting they form a marginal group. Based on these results, we classified domesticated barley accessions into three groups: oriental (C319, C346, C656, I304, I622, J064, J647, K692, and N009), occidental (Barke, Bonus, Borwina, Bowman, Foma, Gull, Harrington, Igri, J247, Kindred, Morex, T567, U051, U353, and Vogelsanger Gold), and marginal (B669, E245, E612, I335, I626, K735, and Steptoe).
Figure 4. Molecular phylogeny and population structure according to the integrated SNPs dataset derived from RNA and exome-sequencing in barley.
The domesticated barley accessions were classified into three groups based on geographic origins: occidental (red), oriental (green), and marginal (blue). (a) Neighbour-joining tree of 34 barley accessions based on the bi-allelic SNP dataset generated using the RNA-seq and exome-sequencing datasets (Mascher et al.33). (b) PCA plot of the 31 domesticated barley accessions and three wild barley accessions. (c) PCA plot of the 31 domesticated barley accessions. (d) Bayesian clustering of the 31 domesticated barley accessions using K = 2 (upper) and K = 7 (lower) populations.
Genomic diversity in domesticated barley sub-populations
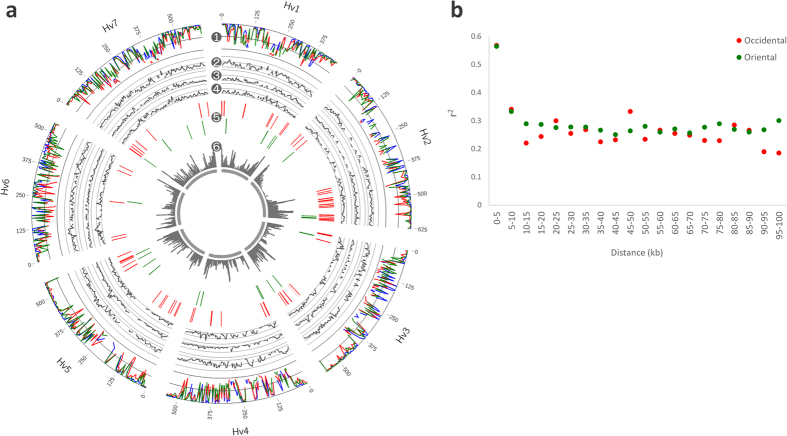
To elucidate the genomic diversity of the domesticated barley populations, we calculated haplotype diversity and pairwise FST47 using 5 million bases sliding windows for each of the three groups, i.e. oriental, occidental, and marginal. The marginal group showed higher average haplotype diversity than the oriental and occidental groups (Table 3). We also observed lower FST values between the occidental or oriental groups and the marginal group than between occidental and oriental groups (Table 3). These results indicated a greater divergence between oriental and occidental sub-populations than between any of them and the marginal group, suggesting each sub-population adapted to particular geographic properties throughout their demographic history. The genomic regions showing FST levels above the average level might have been the ones contributing the most to the divergence of domesticated barley sub-populations (Fig. 5a). We also estimated the linkage disequilibrium (LD) patterns of oriental and occidental sub-populations in a genome-scale. Oriental and occidental sub-populations showed LD decays between 5 and 10 kb (Fig. 5b), which were longer than that observed in maize (<1 kb)48 and Arabidopsis thaliana (L.) Heynh. (3–4 kb)49, but shorter than those of sorghum (10.3 kb for landraces and 19.7 kb for improved inbreds)50, rice (65 kb for Oryza sativa indica and 200 kb for O. s. japonica)51, and soybean (~75 kb and ~150 kb for wild and cultivated varieties, respectively)52. In contrast to the similar behaviour of the LD decays observed in the oriental and occidental sub-populations, distinct LD blocks were found among the differentiated barley groups (Fig. 5a). The results suggested that domesticated barley has selective genetic traits that contributed to the differences found between oriental and occidental accessions. Our RNA-seq based SNP analysis enabled us to identify SNPs on genic regions that are specific to oriental or occidental accessions (Table S3). Thus, these data might be useful to explore genomic regions and genes associated to particular traits in each sub-population, including the oriental and occidental accessions.
Table 3. Average haplotype diversity and average pairwise F ST between each sub-population of domesticated barley.
| Sub-populations | haplotype diversity | pairwise FST | ||
|---|---|---|---|---|
| Occidental | Oriental | Marginal | ||
| Occidental | 0.68 | |||
| Oriental | 0.73 | 0.33 | ||
| Marginal | 0.81 | 0.12 | 0.19 | |
Figure 5. Overview of the genomic diversity in domesticated barley sub-populations.
(a) Genome-scale statistics and LD blocks of domesticated barley sub-populations based on the bi-allelic SNP dataset. (1) Haplotype diversity calculated for the occidental (red), oriental (green), and marginal (blue) barley populations using sliding windows of 5 million bases. (2–4) Pairwise FST values calculated for the occidental and oriental groups (2), occidental and marginal groups (3) and oriental and marginal groups (4) using sliding windows of 5 million bases. (5) LD blocks (≥10 kb) of occidental (red) and oriental (green) barley sub-populations. (6) Density distribution of transcribed regions for the integrated dataset comprising annotated genes and the putative novel transcribed regions identified in the RNA-seq analysis. (b) LD decay plots for the occidental (red) and oriental (green) barley sub-populations. The average r2 values were plotted against physical distance.
Conclusions
The genetic patterns of differentiation of the Asian landraces of oriental barley might elucidate on the potential genetic adaptability of barley to the East Asian monsoon area. The genome-wide variation determined using the RNA-seq technology might become an established tool to address future food security issues. Our results suggest that oriental landraces and/or Asian barley might be important for the future increase of barley productivity.
Materials and Methods
Plant materials
Total RNA was extracted using the RNeasy plant mini kit (QIAGEN GmbH, Hilden, Germany) from the leaves and roots of 19 domesticated barley (Hordeum vulgare) accessions and one wild barley (H. spontaneum) accession, according to the manufacturer’s instruction. The accessions used were obtained from the domesticated barley collection preserved at the Institute of Plant Science and Resources, Okayama University, and from the publicly available Barley database (http://www.shigen.nig.ac.jp/barley). The identifiers, common name, and historical data of the barley accessions are summarised in Supplementary Table 1. The barley plants used for RNA extraction were grown using hydro-culture in a growth chamber. The growth chamber was maintained at 16-h light: 8-h darkness at 22 °C. First leaf blades (L1) and the remaining parts of shoots (L2), as well as roots (Rt), were separately harvested at 14 days after germination. The RNA extracted from L1, L2, and Rt from each barley accession was evenly pooled. All RNA samples were assessed using the Agilent 2100 bioanalyzer (Agilent Technologies, USA) using the Agilent RNA 6000 Pico Kit (Agilent Technologies, USA). Total RNAs were stored at −80 °C until library preparation for RNA sequencing.
RNA sequencing
Poly(A) + RNAs were purified using MicroPoly(A)Purist™ Kit (Life Technologies, USA), according to the manufacturer’s instructions, and assessed using the Agilent 2100 bioanalyzer (Agilent Technologies, USA) and the Agilent RNA 6000 Pico Kit (Agilent Technologies, USA). Libraries for strand-specific RNA sequencing were obtained in an Ion torrent sequencer using the Ion Total RNA-Seq Kit v. 2 (Life Technologies, USA), according to the manufacturer’s instructions, and Ion Xpress Barcode Adapters (Life Technologies, USA) developed for each accession. Templates for the 200 base-read libraries sequenced using the Ion Proton semiconductor sequencer (Life Technologies, USA) were prepared using the Ion P1 Template OT2 200 Kit v. 3 (Life Technologies, USA), Ion P1 Sequencing 200 Kit v. 3 (Life Technologies, USA), and Ion P1 Chip Kit v. 3 (Life Technologies, USA), according to the manufacturer’s instructions. Sequencing was performed using an Ion Proton sequencer (Life Technologies, USA) equipped with an Ion P1 chip.
Data analysis of RNA-seq reads
The pseudomolecular sequence of Morex was downloaded from Ensembl Plants FTP site (082214v1.25, ftp://ftp.ensemblgenomes.org/pub/plants/release-25/fasta/hordeum_vulgare/dna/) and used as a reference sequence to map the RNA-seq reads using the -n 16 -v -Y -u -o 2 stage1 map4 command in Tmap v. 3.4.1 (Life Technologies, USA). Binary Alignment/Map (BAM) files with mapped reads were generated using samtools53 and sorted BAM files were subjected to further analyses.
Putative novel transcribed regions were identified based on the RNA-seq data using the cufflinks and cuffmerge commands in the Cufflinks package v. 2.2.154 and the Morex gene structural annotation dataset retrieved from the Ensembl Plants FTP site (082214 v1.25, ftp://ftp.ensemblgenomes.org/pub/plants/release-25/gff3/hordeum_vulgare/). The cufflinks command with –g option was used to identify putative novel transcribed regions based on the RNA-seq data of each accession, in addition to the known genes annotated in the Ensembl Plants dataset. The cuffmerge command was used to merge the newly identified transcribed regions with the known genes. To confirm the shared homology between the novel transcribed regions and other known genes, sequences ≥ 200 bp were compared to the deduced proteome datasets of some grass species (Aegilops tauschii Coss., Brachypodium distachyon (L.) P. Beauv., O. sativa, Setaria italica (L.) P. Beauv., Sorghum bicolor (L.) Moench, Triticum aestivum L., T. urartu Thumanjan ex Gandilyan, and Zea mays L.) downloaded from Ensembl Plants FTP site (ftp://ftp.ensemblgenomes.org/pub/plants/release-30/fasta/), using the basic local alignment search tool and a translated nucleotide query (BLASTx, -e 1e -5). The cufflinks command with –G option was used to calculate FPKM values, based on the merged gene annotations including previously annotated genes and the putative novel transcribed regions for each accession. Regions with FPKM ≥ 1 were defined as expressed genes or regions.
SNP discovery using RNA-seq reads
BAM files of each barley accession were subjected to SNP calling using freebayes v. 1.0.1–2 g0cb269755, glfMultiples v. 2010-06-1656, and samtools v. 1.3, with default parameter settings. In each accession, the SNPs holding alternative allele frequency above 90% in depth and position read depth ≥5 were identified as putative homozygous SNPs. We omitted SNPs that were out of the threshold. The read depth at each locus was calculated using genomeCoberageBed in BEDTools v. 2.20.157. The intersectional SNPs identified by each of SNP callers that fulfilled the thresholds of read depth and allele frequency were subjected to further analyses as a set of reliable SNPs.
SNP discovery using public exome-sequencing reads
The exome-sequencing-based SNPs in barley33 (ERR271694-ERR271704, ERR271716-ERR271729, and ERR271735-ERR271737) were retrieved to compare them with the RNA-seq-based SNPs and integrate both SNPs datasets for further analyses. The exome-sequencing reads were trimmed using Trimmomatic v. 0.32 using the settings -thread 8 LEADING: 20 TRAILING: 20 SLIDINGWINDOW: 4:15 MINLEN: 36. The trimmed reads were mapped to the Morex pseudomolecule sequence using the BWA (mem) program v. 0.7.12-r103958 with a parameter setting of -t 8. The uniquely mapped and properly paired reads were used in further analysis. Possible duplicated reads were removed using the rmdup command in samtools. The SNPs of the exome-sequencing data were identified using the same methods as in RNA-seq-based SNPs discovery.
Bi-allelic SNP dataset construction
Bi-allelic SNP datasets were constructed based on the SNPs that met the following criteria: (i) The SNP was detected in at least one accession; (ii) read depth of the SNP position was≥ 5 in all accessions; and (iii) other nucleotides were found at the SNP position in <10% of all accessions.
Population analyses
The phylogenetic relationships between barley accessions were determined through phylogenetic analyses, PCA, and Bayesian clustering using the bi-allelic SNP datasets. Neighbour-joining trees were generated using MEGA v. 6.0 with a p-distance model59. PCA was performed using smartpca in EIGENSOFT v. 6.0.160 with default settings. PC values were plotted using R program v. 3.1.0. The population structure of the domesticated barley accessions was estimated in STRUCTURE v. 2.3.464 using 30,000 burn-in times and 60,000 replications. The number of populations (K) was set from 1 to 15, with three runs for each K value. To detect the optimal size of K, delta K values were calculated using Structure Harvester on the web (http://taylor0.biology.ucla.edu/structureHarvester/).
Haplotype diversity and F ST calculation
The genomic diversity between domesticated barley sub-populations, haplotype diversity, and pairwise FST were calculated using the PopGenome package62 in R v. 3.1.0 with 5 million bases sliding windows. These values, along with the barley chromosomes, were represented on the Morex reference genome using circos v. 0.67–563.
Linkage disequilibrium analysis
To detect LD decay in occidental and oriental barley sub-populations, correlation coefficient (r2) values were calculated using Haploview v. 4.264 with the setting -dprime -nogui. Average r2 values for each physical distance were calculated using original Perl scripts. LD blocks in occidental and oriental barley sub-populations were found using Haploview with the setting -blockoutput GAB -nogui. The LD blocks found were superimposed on the Morex reference genome using circos.
Additional Information
Accession codes: The read data have been submitted to DDBJ Sequence Read Archive [DDBJ: DRA003931].
How to cite this article: Takahagi, K. et al. Analysis of single nucleotide polymorphisms based on RNA sequencing data of diverse bio-geographical accessions in barley. Sci. Rep. 6, 33199; doi: 10.1038/srep33199 (2016).
Supplementary Material
Acknowledgments
The authors thank the National BioResource Project (NBRP: Barley) for providing barley materials. This work was supported by the IPSR Joint Usage/Research Center Research Grant (No. 2641) awarded to K.M and D.S and by Grants-in-Aid for Scientific Research (B) (Grant No. 15KT0038 awarded to K.M and D.S.) from the Japan Society for the Promotion of Science (JSPS). The authors also thank the research support grant for research staff with family responsibilities in RIKEN awarded to Y.U.Y.
Footnotes
Author Contributions D.S., K.M. and K.T. designed the experiments. D.S., K.M. and K.T. wrote the manuscript. K.T., K.M., T.Y. and T.S. performed bioinformatics analysis. D.S. prepared samples for RNA-seq analysis. Y.U.Y. performed RNA-seq analysis. K.S. contributed to biological interpretation of the results. All authors read and approved the final manuscript.
References
- Dawson I. K. et al. Barley: a translational model for adaptation to climate change. New Phytol 206, 913–931 (2015). [DOI] [PubMed] [Google Scholar]
- Wheeler T. & von Braun J. Climate change impacts on global food security. Science 341, 508–513 (2013). [DOI] [PubMed] [Google Scholar]
- McCouch S. et al. Agriculture: Feeding the future. Nature 499, 23–24 (2013). [DOI] [PubMed] [Google Scholar]
- Salamini F., Ozkan H., Brandolini A., Schafer-Pregl R. & Martin W. Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet 3, 429–441 (2002). [DOI] [PubMed] [Google Scholar]
- Pourkheirandish M. & Komatsuda T. The importance of barley genetics and domestication in a global perspective. Ann Bot 100, 999–1008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox G. Measuring grain size and identifying Near Eastern cereal domestication: evidence from the Euphrates Valley. J Archaeol Sci 31, 145–150 (2004). [Google Scholar]
- Edwards P. C., Meadows J., Sayej G. & Westaway M. From the PPNA to the PPNB: new views from the southern Levant after excavations at Zahrat adh-Dhra‘2 in Jordan. Pale´orient 30, 21–60 (2004). [Google Scholar]
- Meadows J. The earliest farmers? Archaeobotanical research at Pre-Pottery Neolithic A sites in Jordan. 119–128 (Department of Antiquities of Jordan, 2004). [Google Scholar]
- Londo J. P., Chiang Y. C., Hung K. H., Chiang T. Y. & Schaal B. A. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA 103, 9578–9583 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P. et al. Phylogeography of Asian wild rice, Oryza rufipogon: a genome-wide view. Mol. Ecol. 21, 4593–4604 (2012). [DOI] [PubMed] [Google Scholar]
- Morrell P. L. & Clegg M. T. Genetic evidence for a second domestication of barley (Hordeum vulgare) east of the Fertile Crescent. Proc. Natl. Acad. Sci. USA 104, 3289–3294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P. & Bliss F. A. F1 hybrid weakness in the common bean: differential geographic origin suggests two gene pools in cultivated bean germplasm. J. Hered. 76, 447–450 (1985). [Google Scholar]
- Gepts P., Osborn T. C., Rashka K. & Bliss F. A. Phaseolin-Protein Variability in Wild Forms and Landraces of the Common Bean (Phaseolus vulgaris): Evidence for Multiple Centers of Domestication. Economic Botany 40, 451–468 (1986). [Google Scholar]
- Bitocchi E. et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol 197, 300–313 (2013). [DOI] [PubMed] [Google Scholar]
- Bitocchi E. et al. Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. USA. 109, E788–E796 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Ibarra J. & Gaut B. S. Multiple domestications do not appear monophyletic. Proc. Natl. Acad. Sci. USA 105, E105, author reply E106 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T. Genes and mutations underlying domestication transitions in grasses. Plant Physiol. 149, 63–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob S. S. et al. Evolutionary history of wild barley (Hordeum vulgare subsp. spontaneum) analyzed using multilocus sequence data and paleodistribution modeling. Genome Biol. Evol. 6, 685–702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E., Beiles A. & Zohary D. Genetic resources of wild barley in the Near East: structure, evolution and application in breeding. Biol. J. Linn. Soc. Lond. 27, 355–380 (1986). [Google Scholar]
- Fang Z. et al. Two genomic regions contribute disproportionately to geographic differentiation in wild barley. G3 Bethesda) 4, 1193–1203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D. & Purugganan M. D. Molecular phylogeography of domesticated barley traces expansion of agriculture in the Old World. Genetics 177, 1765–1776 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R. The origin and evolution of cultivated barley. Vol. 7 227–266 (New York: Academic Press, 1955). [Google Scholar]
- Morrell P. L., Gonzales A. M., Meyer K. K. & Clegg M. T. Resequencing data indicate a modest effect of domestication on diversity in barley: a cultigen with multiple origins. J. Hered. 105, 253–264 (2014). [DOI] [PubMed] [Google Scholar]
- Poets A. M., Fang Z., Clegg M. T. & Morrell P. L. Barley landraces are characterized by geographically heterogeneous genomic origins. Genome Biol. 16, 173 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K. & Shinozaki K. Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol. 52, 2017–2038 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K. & Shinozaki K. Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol. 51, 497–523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P. L., Bukler E. S. & Ross-Ibarra J. Crop genomics: advances and applications. Nat Rev Genet 13, 85–96 (2012). [DOI] [PubMed] [Google Scholar]
- Sato K., Nankaku N. & Takeda K. A high-density transcript linkage map of barley derived from a single population. Heredity (Edinb.) 103, 110–117 (2009). [DOI] [PubMed] [Google Scholar]
- Fu Y. B. & Peterson G. W. Developing genomic resources in two Linum species via 454 pyrosequencing and genomic reduction. Mol. Ecol. Resour. 12, 492–500 (2012). [DOI] [PubMed] [Google Scholar]
- Munoz-Amatriain M. et al. The USDA barley core collection: genetic diversity, population structure, and potential for genome-wide association studies. PLoS One 9, e94688 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrechtsen A., Nielsen F. C. & Nielsen R. Ascertainment biases in SNP chips affect measures of population divergence. Mol. Biol. Evol. 27, 2534–2547 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z. et al. Comparative analyses identify the contributions of exotic donors to disease resistance in a barley experimental population. G3 (Bethesda) 3, 1945–1953 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M. et al. Barley whole exome capture: a tool for genomic research in the genus Hordeum and beyond. Plant J. 76, 494–505 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moragues M. et al. Effects of ascertainment bias and marker number on estimations of barley diversity from high-throughput SNP genotype data. Theor Appl Genet 120, 1525–1534 (2010). [DOI] [PubMed] [Google Scholar]
- Chepelev I., Wei G., Tang Q. & Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 37, e106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D., Ishii M., Hori K. & Sato K. Natural variation of barley vernalization requirements: implication of quantitative variation of winter growth habit as an adaptive trait in East Asia. Plant Cell Physiol. 52, 775–784 (2011). [DOI] [PubMed] [Google Scholar]
- Mochida K. et al. Large-scale collection and analysis of full-length cDNAs from Brachypodium distachyon and integration with Pooideae sequence resources. PLoS One 8, e75265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. P. et al. Genome diversity in Brachypodium distachyon: deep sequencing of highly diverse inbred lines. Plant J. 79, 361–374 (2014). [DOI] [PubMed] [Google Scholar]
- O’Rawe J. et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 5, 28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baes C. F. et al. Evaluation of variant identification methods for whole genome sequencing data in dairy cattle. BMC Genomics 15, 948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. et al. Steps to ensure accuracy in genotype and SNP calling from Illumina sequencing data. BMC Genomics 13 Suppl 8, S8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielczarek M. & Szyda J. Review of alignment and SNP calling algorithms for next-generation sequencing data. J Appl Genet 57, 71–79 (2016). [DOI] [PubMed] [Google Scholar]
- Poland J. A., Brown P. J., Sorrells M. E. & Jannink J. L. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7, e32253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. A high density GBS map of bread wheat and its application for dissecting complex disease resistance traits. BMC Genomics 16, 216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K. et al. Improvement of barley genome annotations by deciphering the Haruna Nijo genome. DNA Res. 23, 21–28 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S. & Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Slatkin M. & Maddison W. P. Estimation of levels of gene flow from DNA sequence data. Genetics 132, 583–589 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore M. A. et al. A First-Generation Haplotype Map of Maize. Science 326, 1115–1117 (2009). [DOI] [PubMed] [Google Scholar]
- Kim S. et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 39, 1151–1155 (2007). [DOI] [PubMed] [Google Scholar]
- Mace E. S. et al. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat Commun 4, 2320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30, 105–111 (2012). [DOI] [PubMed] [Google Scholar]
- Lam H. M. et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059 (2010). [DOI] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E. & Marth G. Haplotype-based variant detection from short-read sequencing. arXiv 1207, 3907v2 (2012). [Google Scholar]
- Genomes Project C. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr Protoc Bioinformatics 47, 11 12 11–11 12 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M. & Donnelly P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B., Wittelsburger U., Ramos-Onsins S. E. & Lercher M. J. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 31, 1929–1936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.