Abstract
Steroid hormone 20-hydroxyecdysone is known as the systemic regulators of insect cells; however, how to impact the fate and function of mature and stem cells is unclear. For the first time, we report ecdysone regulatory cascades in both mature midgut cell and stem cell fractions related to developmental events by using histological, immunohistochemical, biochemical and gene expression analysis methods. Ecdysone receptor-B1 (EcR-B1) and ultraspiracle 1 (USP-1) mRNAs were detected mainly in mature cells during programmed cell death (PCD). Lowered E75A and probably BR-C Z4 in mature cells appear to provide a signal to the initiation of PCD. E74B, E75B and BR-C Z2 seem to be early response genes which are involved in preparatory phase of cell death. It is likely that βFTZ-F1, E74A and BR-C Z1 are probably associated with execution of death. EcR-A and USP2 mRNAs were found in stem cells during remodeling processes but EcR-B1, USP1 and E74B genes imply an important role during initial phase of metamorphic events in stem cells. BHR3 mRNAs were determined abundantly in stem cells suggesting its primary role in differentiation. All of these results showed the determination the cell fate in Bombyx mori (Linnaeus) midgut depends on type of ecdysone receptor isoforms and ecdysone-related transcription factors.
Keywords: Bombyx mori, ecdysone-related gene, midgut, programmed cell death, stem cell
Insect steroid hormone, 20-hydroxyecdysone (20E) regulates metamorphic processes via binding a heterodimeric nuclear hormone receptor, ecdysone receptor (EcR) and its partner molecule ultraspiracle (USP). Homologues of Drosophila EcR-A and EcR-B1 isoforms were characterized in Bombyx mori (Linnaeus). (Swevers et al. 1996). A small set of early genes which are also transcription factors, are activated by the hormone receptor complex. Appearance of these transcription factors is a function of hemolymph ecdysteroid concentration coordinates the response of target tissue to the 20E. These transcriptional regulatory hierarchy leads to proliferation and differentiation of tissues like wing discs, pupal midgut and programmed cell death of larval tissues like silk gland, salivary glands and larval midgut. Degeneration in salivary gland of D. melanogaster is triggered by rising ecdysone level at the end of the last larval instar and then ecdysone binds its heterodimeric receptor complex, EcR and ultraspiracle (USP) which activates the early genes, E93, the zinc finger transcription factor Broad complex (BR-C), the ETS family transcription factor E74 and another transcription factor E75. Expressions of the genes regulate early late genes including Drosophila hormone receptor (DHR3), βFTZ-F1 and E78. BRC gene expresses four related proteins which are distinct as consequence of alternative splicing. These proteins are named as Z1, Z2, Z3 and Z4 which share common core domain but they differ by zinc finger domains. Ijiro et al. (2004) cloned Z1, Z2 and Z4 isoforms of BR-C in B. mori but the Z3 zinc finger sequence was found in the 3′-UTR of the Z2 isoforms. E74 is one of the early genes induced by 20E during metamorphosis of D. melanogaster. It has two alternative splicing forms—E74A and E74B—with an isoform specific N terminus and a DNA binding Ets domain at the C terminus. The E75 gene encodes three isoforms: E75A, E75B, and E75C, which have a common C-terminus and unique N-terminal region (Segraves and Hogness 1990). The products of early genes trigger the expression of late or effector genes which perform the metamorphic changes like PCD of larval tissues, growth and differentiation of imaginal cells into adult tissues (Jiang et al. 1997).
Midgut structures and functions dramatically change during metamorphosis of holometabolus insects because of the changing food habit. Midgut remodeling, which is controlled by 20E and paracrine factors (Smagghe et al. 2005), involves both PCD of mature midgut cell and formation of new pupal midgut by proliferation and differentiation of stem cells (Lee et al. 2002). The PCD mechanisms of midgut in different species including Aedes aegypti (Wu et al. 2006), Heliothis virescens (Tettamanti et al. 2007) and B. mori (Goncu and Parlak 2011) have been studied in detail. Previous studies in different insect species generally focused on ecdysone-regulated gene activation related to PCD processes but little is known about the regulation of stem cell proliferation and differentiation during pupal midgut formation. In addition to this, there is no information about the 20E-triggered sequential gene activation in both mature midgut cells and stem cells during midgut remodeling of B. mori.
In this study, we investigated the hierarchical gene activation in both mature midgut cells and stem cells, separately. Although morphology and processes during midgut remodeling are well known, we needed to evaluate morphologic and biochemical features of remodeling processes in our experimental animals, because, qRT-PCR analysis were discussed associated with morphological developmental and biochemical events. Obtained data provided important information about how same ecdysone signal induces two different responses in two different cell types of the same tissue.
Materials and Methods
Insect Rearing and Staging
Japanese × Chinese hybrid races of the silkworm, B. mori, were reared on fresh mulberry leaves at 25 ± 1 °C, 75–80% relative humidity and (LD 12:12) photoperiod conditions. The final (fifth) larval instar lasts 10 d: 7 d for feeding, followed by 3 d of spinning to build the cocoon (Prepupal period). Experiments were performed every 12 h from day 7 larvae of the fifth instar to 24 h after pupation. We divided experiment period into four stages according to morphologic evaluations. Feeding stage: Insects show feeding activity during day 7 of fifth instar. They exhibit normal larval stage characteristics and healthy larval midgut. Early prepupal stage: Insects stop feeding activity at the beginning of day 8 and gut purge occurs. Larvae exhibit wandering behavior then spinning begins on this day. On day 9, larvae are completely surrounded by the cocoon but spinning activity progress. Late prepupal stage: It lasts during day 10 of fifth instar. Spinning activity almost finishes and larvae became completely quiescent (Pharate pupae). Early pupal stage: It lasts from larval-pupal ecdysis to 24 h pupae.
Stem Cell Isolation
Isolation of the stem cells from midguts was performed according to Hakim et al. (2009). Very high percentages (>95%) of stem cells can be separated by using this method (Hakim et al. 2009), because, they lie loosely associated among the bases of the epithelial cells. Ten insects were used for isolation every 12 h. Separated midgut cells were classified according to their morphological features (Cermenati et al. 2007). Clear and round shape stem cells were observed and counted by using haemocytometer slide under inverted microscope. Cells were counted in three randomly chosen fields each measuring 1 mm2. Every calculation was repeated 3 times. After isolation of stem cells, the remaining tissue fragments were used as a mature cell fraction. During late prepupal stage, stem cells began to organize as monolayers surrounding the midgut lumen. The larval midgut was expelled into the lumen which is called as yellow body. For this reason, the contents of midgut lumen isolated after day 9 of fifth larval instar were collected into sterile conical tube, then, centrifuged at 400 × g, 5 min. Pellet was used as yellow body fractions and the remaining tissue was used as the stem cell fractions.
Preparation of Cross-Section of Midguts for Staining
Midgut tissues were dissected every 12 h from day 7 larvae of fifth instar to 24 h after pupation. Following fixation in Bouin’s solution for 5–6 h at 4 °C, tissues processed for embedding in paraffin wax. Five micrometer-thick sections were stained with hematoxylin and eosin (H&E) and periodic acid & Schiff (PAS) reagent using routine protocol. Sections were examined under a Leica DM3000 (Leica microsystems, Wetzlar, Germany) microscope and photographed with a digital camera.
Acid Phosphatase (AP) Histochemistry
Gomori (1950) procedure was used for detecting AP activity in midgut tissue. Five micrometer-thick midgut sections were placed in incubating mixture containing, 30 ml of 0.01 M sodium β-glycerophosphate in 30 ml of 0.05 M acetate buffer (pH 5.0) and 10 ml of 0.004 M lead nitrate at 37 °C for 30 min. The reaction was terminated by immersing slides in distilled water. Tissue AP activity was demonstrated by submerging sections in 2% aqueous yellow ammonium sulphide for 2 min. All chemicals were purchased from Sigma-Aldrich (St Louis, M.O). Sections were examined under a Leica DM3000 microscope and photographed with a digital camera. Images were analyzed by using image J analysis (http://imagej.nih.gov/ij/).
Acid Phosphatase (AP) Assay
Acid phosphatase activities in the midguts were measured using paranitrophenol phosphate (Sigma-Aldrich, St Louis, MO) as a substrate according to Bergmeyer (1974). The measurements were repeated on three different series of animals. The optical density of the liberated paranitrophenol was read at 405 nm by using a spectrophotometer.
BrdU Labelling and TUNEL Assay
For in vivo labeling with BrdU (5-bromo-2′-deoxyuridine), 10 µl of the labeling reagent per 1 g body weight was injected into the haemocoel of staged larvae/pupae. Midguts were dissected 4 h later and fixed immediately in 10% neutral buffered formalin for 8 h. Paraffin sections of midgut samples were processed using In Situ Cell Proliferation Kit, Fluos (Roche, Penzberg, Germany) according to manufacturer instructions.
DNA fragmentation in apoptotic cells was identified by using the In Situ Cell Death Detection Kit, Fluorescein (Roche, Penzberg, Germany). Nuclear staining was done with DAPI (4,6-diamidino-2 phenyl indol, Invitrogen). Sections were examined under a Leica DM3000 microscope and photographed with a digital camera. Images were analyzed by using image J analysis (http://imagej.nih.gov/ij/).
Western-Blot Analysis
Cleaved caspase 3 and cytochrome c were detected in midgut tissues by immunoblotting. Two different buffers were used to detect cleaved caspase 3 and cytosolic cytochrome c. Midguts were homogenized a buffer containing, 100 mM mannitol, 10 mM HEPES–Tris pH 7.2, and protease inhibitor cocktail (Complete Mini, Roche, Penzberg, Germany) for cleaved caspase 3 and 20 mM HEPES (pH 7.5), 250 mM sucrose, 1 mM EGTA, 1 mM EDTA, 100 mM KCl, 1 mM DTT, 0.1 mM PMSF, 2 μg/ml pepstatin, containing protease inhibitor cocktail (Complete Mini, Roche, Penzberg, Germany) for cytochrome c. The homogenates were centrifuged at 15,000×g for 15 min at 4°C and the supernanats were collected. Total protein concentration was determined with the bicinchoninic acid (BCA) protein assay kit (Pierce, Thermo Fisher Scientific, Waltham, Massachusetts, USA) employed according to manufacturer’s instructions. Twenty micrograms of total proteins were separated by sodium dodecylsulfate(SDS)–polyacrylamide gel electrophoresis in a gel running buffer (25 mMTris, 192 mM glycine, 0.1% SDS, pH 8.3) using a Bio-Rad vertical electrophoresis system (California, USA). Proteins were electrotransferred onto a nitrocellulose membrane (88018, Pierce, ABD) using a Bio-Rad Transblot cell. Membranes were placed in blocking solution (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA and 0.1% Tween-20 (TBST) containing 1% bovine serum albumin and 5% dried non-fat milk overnight at 4°C. They were then incubated with a 1:1000 dilution of 6B7 anti-cleaved caspase 3 antibody (ASP 175, cell signaling technology, Danvers, Massachusetts, USA) and cytochrome c antibody (4272, Cell Signaling Technology, Danvers, Massachusetts, USA) during overnight at 4 °C followed by 2-h incubation with horseradish peroxidase conjugated secondary antibody (7074, Cell Signaling Technology, Danvers, Massachusetts, USA). Detection was performed by chemiluminescence (ECL Western blotting substrate, 32106, Pierce) according to manufacturer’s instructions. The results were analyzed with a Chemidoc (Biorad, Hercules, California, USA) imaging system.
RNA Isolation and cDNA Synthesis
Stem cell fractions and mature midgut cell fractions were pooled and collected in Tripure Isolation Reagent (Roche, Penzberg, Germany) for every 12 h. Samples were homogenized in Tripure reagent and total RNA was isolated according to manufacturer’s instructions. Concentration of total RNA and purity were evaluated by using Nanodrop UV/VIS spectrophotometer. cDNA synthesis by reverse transcription was performed using 1 µg of RNA and High Fidelity cDNA synthesis kit (Roche, Penzberg, Germany) in a 20-µl reaction volume. It was used a template in the PCR reactions.
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
Relative expression of selected genes was evaluated by qRT-PCR using lightcycler 480 real-time PCR detection systems (Roche, Penzberg, Germany) as previously described (Gibson et al. 1996). qRT-PCR was performed in a total reaction volume of 10 µl reaction containing 2 µl of cDNA, 0.5 µl each of forward and reverse sequence specific primers, 2.7 µl H2O, 0.2 µl Tprobe and 5 µl enzyme. PCR conditions were 95 °C for 10 min followed by 45 cycles of 95 °C for 10 s, 60 °C for 30 s, 72 °C for 1 s and 40 °C for 30 s. The primers used for PCR that were derived from the sequences of the Bombyx genes and references are listed in Table 1. Primer probe design was performed by using clustal W align and oligo7 software. Specificity of obtained primers and probes was controlled by using blast program. In addition, specificity of RT-PCR products were demonstrated in a gradient cycler (Techne TC-400, ST15 OSA, UK) and carried out in a total volume 45 µl containing 1.8 µl Faststart high fidelity reaction buffer (Roche diagnostics, Penzberg, Germany), 5 µl cDNA, 0.4 µM forward and reverse primers and 2.5 U/µl Faststart High Fidelity Enzyme. Amplification cycles were as follows: 95 °C for 30 s, 55 °C for 30 s and 72 °C for 50 s. The 35 amplification cycles were preceded by a primary denaturation step 95 °C for 2 min. RT-PCR products were determined with 4% high resolution agarose gel electrophoresis containing 4 µg/ml GelRed and analyzed with Chemidoc MP imaging system (Biorad, California, USA). Bombyx mori Actin 3 was used as an endogenous control. Mean and standard errors were obtained from the averages of three independent sample sets.
Table 1.
Forward and reverse primers used in qRT-PCR
| Gene | Forward primer Reverse primer | Accession no | upl prob no |
|---|---|---|---|
| Bombyx mori actin 3 | 5′-GCTCCCTCGAGAAGTCTTACG-3′ | U49854 | 9 |
| 5′-CTGGGCAACGGAATCTTTC-3′ | |||
| EcR-A | 5′-CATCCGGTCAACGGACAC-3′ | D87118 | 141 |
| 5′-ACCGTAGCTGCCTGAGGATA-3′ | |||
| EcR-B1 | 5′-ACTTGGCAGTCGGATGAAG-3′ | L35266 | 153 |
| 5′-CGTCATCTCCGTGATCTGG-3′ | |||
| USP-1 | 5′-TCAAATAGGCAACAAACAGATAGCCGCTC-3′ | U06073 | 150 |
| 5′-CAGGAACTCCATAGACCG-3′ | |||
| USP-2 | 5′-CAGTGTCACATGTAGAGTGCAAAGA-3′ | AB182582 | |
| FAM-GTTCAACGACCTTGTTGCTGACAGGTTC-Tamra | |||
| 5′-CCACTTTCATAGAACAGTTCAGTTGC-3′ | Taqman probe | ||
| E74A | 5′-CCACCTATCGAGATAAAGCAAGA-3′ | Q1KLS0 | 141 |
| 5′-CTTGCAGTTGACTGCTGGAC-3′ | |||
| E74B | 5′-ACCCGAGTGACTACGTGAGG-3′ | Q1KLR9 | 95 |
| 5′-CGGAGTCTGTGCCTGAGTCT-3′ | |||
| E75A | 5′-TCGGTCGAGCTTGAGTGAG-3′ | AB024904 | 59 |
| 5′-GATGAAGGTCGCTTGTCTCG-3′ | |||
| E75B | 5′-GGACAGCTCTCAAAGACGTGA-3′ | AB024905 | 91 |
| 5′-CGCACCATTCACTACACTCG-3′ | |||
| BR-C Z1 | 5′-TCTGCAGAGTCCTCTCGCTTC-3′ | AB166725 | 36 |
| 5′-TACACGCGCTGGCAAATG-3′ | |||
| BR-C Z2 | 5′-TCTGCAGAGTCCTCTCGCTTC-3′ | AB166726 | 99 |
| 5′-GTGTATATGTGCGTCATCAGGGA-3′ | |||
| BR-C Z4 | 5′-TCTGCAGAGTCCTCTCGCTTC-3′ | AB166727 | 56 |
| 5′-TCTTGTGGTTGTTGAGCGAGTT-3′ | |||
| βFTZ-F1 | 5′-TTCCGCAAGTATCATCATTGAC-3′ | D10953 | 141 |
| 5′-CTTGTCGTGAGTTGGTGGTG-3′ | |||
| BHR3 | 5′-GGGATGCAAAGGATTCTTCA-3′ | AB024902 | 99 |
| 5′-GCGAGGACACTGGTAGTTCAC-3′ | |||
| BmATG8 | 5′-TCCGGAAACGTATTCACCTG-3′ | FJ416330 | 124 |
| 5′-GGGTGGAATGACATTGTTCAC-3′ | |||
| BmATG12 | 5′-CCTGTATGTGAATCAGACTTTTGC-3′ | FJ416329 | 9 |
| 5′-CCGAAGCACTCATAAAGATTCC-3′ | |||
Results
Morphologic and Biochemical Events During Midgut Remodeling of Experimental Animals
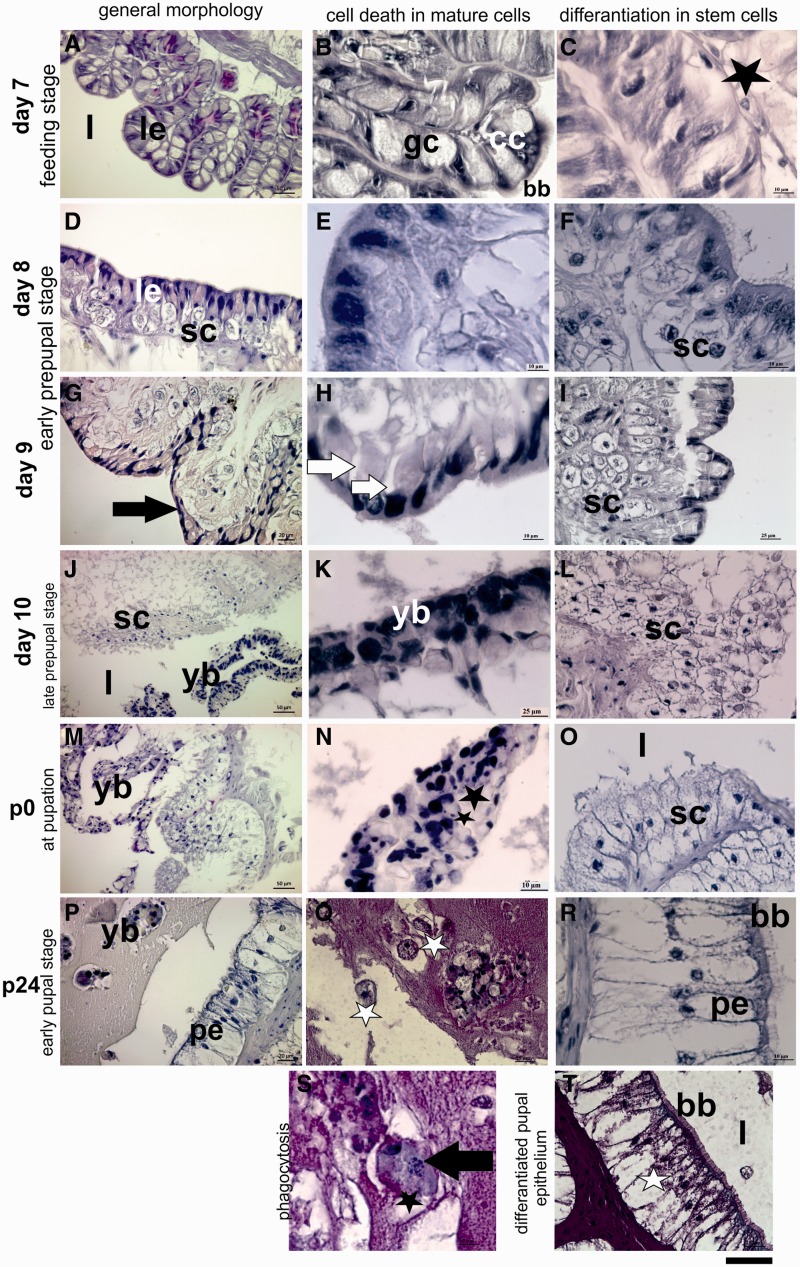
Midguts from day 7 larvae are composed of columnar epithelial cells, goblet cells and few stem cells located in the basal region of the epithelium (Fig. 1A–C). The morphology and the location of midgut stem cells were reported by Turbeck (1974) and Baldwin and Hakim (1991) as they are roughly conical- or spindle-shaped cells with a large nucleus and are located at the base of the epithelium. Additionally, Loeb and Hakim (1996) indicated that they are the only cell type that undergoes proliferation. According to these previous findings, we detected a few spindle-shape stem cells located the basal region of epithelium during feeding stage (Fig. 1C). First morphologic signs of PCD especially cell shrinkage, nuclear and cytoplasmic condensations in mature cells were observed in tissue sections after gut purge on day 8 (Fig. 1D and E). At the same time, stem cells enlarged and started to proliferate (Fig. 1F). Dying mature midgut cells began to separate locally from basal lamina (Fig. 1G) and also detached from each other on day 9 (Fig. 1H). Stem cell niches under the degenerated larval epithelium were clearly seen on this day (Fig. 1I). During late prepupal stage (day 10), larval epithelium was pushed into the midgut lumen (Fig. 1J) and this newly formed structure is called as yellow body (Fig. 1K). When larval midgut separated from the basal membrane, organization of proliferated stem cells resembled multilayer epithelium around the lumen on day 10 (Fig. 1L). The cellular structure of yellow body completely degenerated at pupation time (Fig. 1M and N). The morphology of proliferated stem cells began to change as a columnar like cell shape (Fig. 1O). Degeneration in yellow body proceeded (Fig. 1P) in addition; presence of insect phagocytic cells—granulocytes—around the yellow body (Fig. 1R) and phagocytosis of degenerated cell bodies by these cells (Fig. 1T) were demonstrated 12 h after pupation. At the same time, stem cells completely differentiated and organized as pupal midgut epithelium. They gained columnar like shape and typically arranged as a single layer epithelium (Fig. 1M). Brush border membrane which is an indicator of differentiation also shaped in these newly formed epithelial cells (1s). PAS staining of these cells clearly indicated their carbohydrate content which suggests their absorptive function (Fig. 1U).
Fig. 1.
Morphologic changes in the midgut of experimental animals during remodeling were analyzed by using hematoxylin & eosin staining. Healthy larval midgut epithelium (le) during feeding stage on day 7 of fifth instar (A). Columnar cells (cc), goblet cells (gc) and brush border membrane (bb) was easily discerned (B). Stem cell (black star) lies under the larval epithelium (C). First degenerative changes in mature midgut cells (le) (D and E). And, enlarged stem cells (sc) under the larval epithelium on day 8 (D and F). Black arrow shows detachment of larval epithelium from basal membrane (G), white arrows exhibit nuclear condensation and cellular detachment (H). Stem cell niches (sc) were seen on day 9 (I). Larval midgut epithelium pushed into the midgut lumen is called as yellow body on day 10 (yb) (J and K). Proliferated stem cells (sc) organize as multilayer epithelium on day 10 (L). Degenerative changes in yellow body (yb) at pupation time (M). Black stars show nuclear fragmentation (N). Differentiated columnar pupal epithelium surrounds the midgut lumen (O). Reduced yellow body (yb) and granulocyte like cells (white stars) were detected at 24 h pupae (R). Pupal midgut epithelium (pe) completely formed at 24 h after pupation. Brush border membrane (bb) develops in these cells (S). Black arrow shows fragments of dead cells within granulocyte like cell (T). Periodic acid & Schiff (PAS) staining showed the glycogen content of pupal midgut cells (U). Scale bar is 110 µm for (A), (J) and (M); 55 µm for (B), (D), (G), (I), (K), (L), (O), (P), (R) and (U) and 22 µm for (C), (E), (F), (H), (N), (S) and (T).
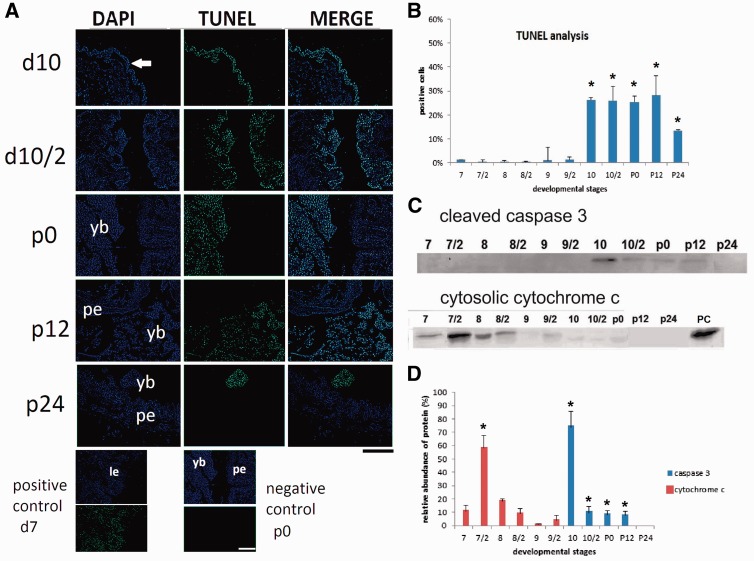
Apoptosis specific DNA fragmentation was detected by using TUNEL staining (Fig. 2). TUNEL positive cells increased significantly on day 10 compared with levels in day 7 (P ≤ 0.05) (Fig. 2A). The percentage of TUNEL positive cells maintained 26.29 ± 2.3% during day 10 and early pupal stage (Fig. 2B). Positive signals decreased to 13.30 ± 2.3% at 24 h after pupation. The release of cytochrome c from mitochondria to the cytosol as an apoptosis initiator and cleaved caspase 3 as a critical executioner of apoptosis were shown by using Western blotting (Fig. 2C). Despite healthy appearance of midgut, cytosolic cytochrome c was detected in the first half of day 7 and its relative abundance was 9.8%. The most abundant cytosolic cytochrome c was found in the second half of day 7 just before cessation of feeding and it was significantly high compared with levels in the first half of day 7 (P ≤ 0.05). Positive signals were detected every 12 h of day 8 and relative abundances were 17.12% and 9.29%, respectively. Cleaved caspase 3 was first detected in the first half of day 10 and calculated relative abundance was 70%. When compared with levels in day 7, significantly high levels of cleaved caspase 3 were maintained until 12 h after pupation (P ≤ 0.05) (Fig. 2D).
Fig. 2.
Apoptosis specific DNA fragmentation and biochemical changes in mature midgut cells. Experiments were performed every 12 h. Second half of each day was shown as day/2. White arrow shows positive signals located the detached larval epithelium on day 10 (d10). Positively stained nuclei were detected in yellow body (yb) in the second half of day 10 (d10/2), at pupation (p0) and 12 h after pupation (p12), then, reduced 24 h after pupation (p24). DNase treated day 7 samples were used as positive control, p0 samples were used as negative control. Pupal epithelium (pe). Scale bar = 250 µm (A). Intense of TUNEL positive signals were analyzed by using image J software (http://imagej.nih.gov/ij/). Mean ± SDs for five independent sections are shown. (B). Cytosolic cytochrome c and cleaved caspase 3 were detected by using Western blotting (C). Relative abundance of them was calculated by using Image Lab software (Biorad) (D). Mitochondrial fraction was used as positive control (PC). The differences between two dependent groups were evaluated with the paired samples Student’s T test. The results are presented as means ± SD and were considered significant when P≤ 0.05.
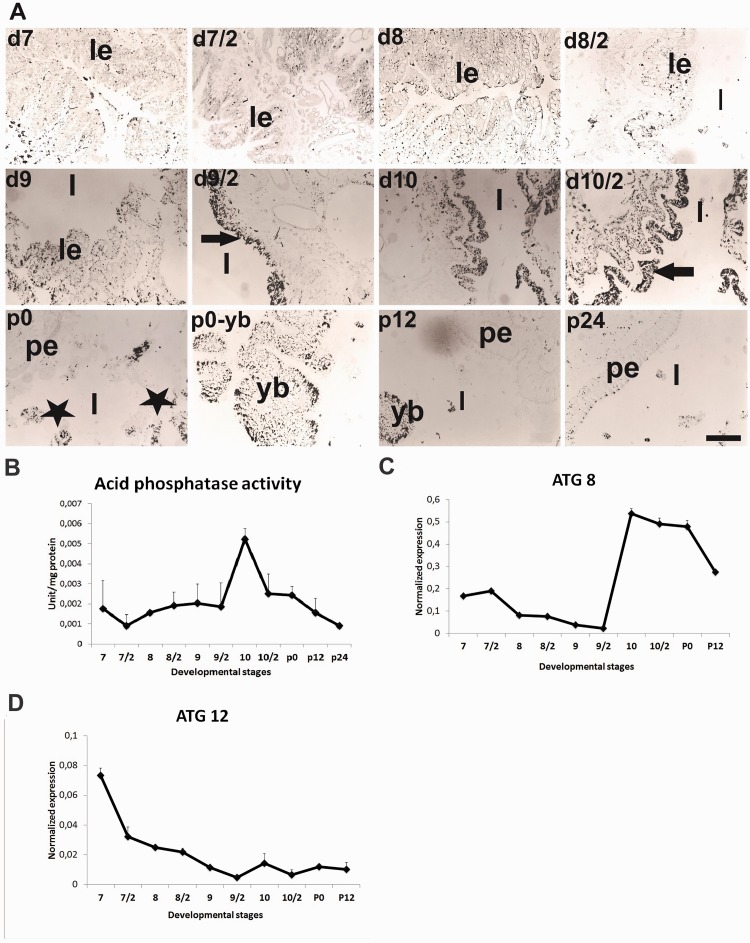
Acid phosphatase (AP) activity as an indicator of autophagy was analyzed by both immunohistochemically (Gomori 1950) (Fig. 3A) and spectrophotometrically (Fig. 3B). In transvers sections of midgut, staining intensities were low until the second half of day 9. After this day, AP content of the tissue slightly increased (Fig. 3A) and intense staining was localized especially in degenerated larval epithelium. The highest enzyme content was determined on day 10 during late prepupal period. After larval-pupal ecdysis, the yellow body in midgut lumen showed positive staining and only a few pupal midgut cells positively stained (Fig. 3A). The highest AP enzyme activity was found on day 10 then decreased gradually (Fig. 3B). Expressions of autophagy-related genes—ATG8 and ATG12—were determined by using qRT-PCR. ATG8 expression in mature cells was almost undetectable levels until day 9 then its expression sharply increased on day 10 and remained steady level until 12 h after pupation (Fig. 3C). In contrast to ATG8, the highest ATG12 mRNA level was determined on day 7 then its expression began to decrease and low mRNA levels continued until 12 h after pupation (Fig. 3D).
Fig. 3.
Autophagy specific biochemical changes and autophagy-related genes—ATG8 and ATG12—were detected in mature midgut cells. Black spots within cells and tissue sections show localization of acid phosphatase activity (A). Black arrow shows the acid phosphatase contents of larval epithelium (le) (A). Positive signal (black stars) located in yellow body (yb) after day 9. Pupal midgut epithelium (pe) did not exhibit acid phosphatase content. Acid phosphatase enzyme activity was also measured by using spectrophotometer (B). Expression profile of autophagy-related genes, ATG 8 (C) and ATG 12 (D) were demonstrated. Lumen (l). Mean ± SD for the three independent experiments are shown. Scale bar = 200 µm.
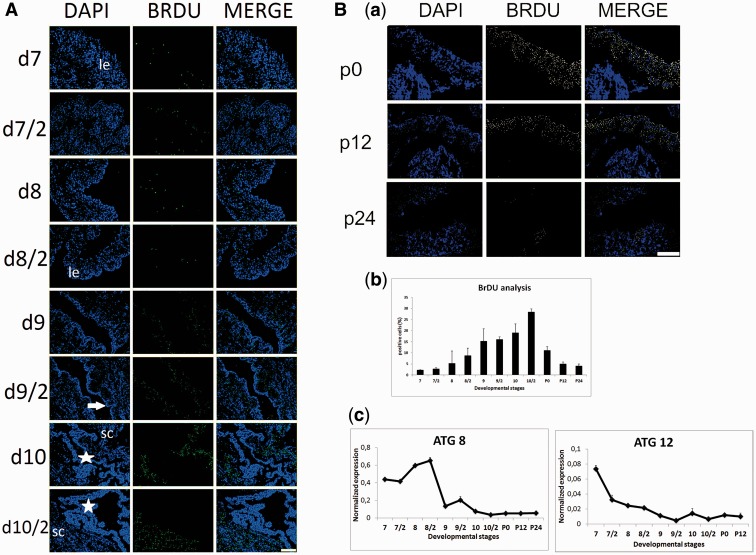
The cell proliferative activity in the midgut was examined by using 5-bromodeoxyuridine (BrdU) incorporation (Fig. 4). Slight to moderate numbers of BrdU incorporated cells were observed in the basal layer of larval epithelium until day 10 (Fig. 4A). Proliferation rate of stem cells began to increase from day 9 (Fig. 4A) and reached the maximum level in the second half of day 10 just before pupation (Fig. 4B, panel a). Image J analysis revealed that 2.3% of cells on day 7, 5.21% of cells on day 8 and 15.2% of cells on day 9 were BrdU positive. Percentage of maximum BrdU incorporated cells was determined as 28.43% just before larval-pupal ecdysis. Proliferation rate of cells rapidly decreased during early pupal period (Fig. 4B, panel b). Autophagy-related genes—ATG8 and ATG12—expressions were analyzed in stem cells to survey if any possible role of autophagy during metamorphic processes of stem cells (Fig. 4B, panel c). ATG8 transcripts started to increase at the end of the feeding period and peaked in the second half of day 8. Expression of ATG8 suddenly decreased on day 9 and remained at very low levels until 24 h after pupation. Moderate ATG12 mRNA levels until day 10 suddenly decreased and continued low levels during early pupal stage (Fig. 4B, panel c).
Fig. 4.
Proliferation rate of stem cells was determined by using BrDU staining. Nuclear staining was carried out by DAPI. According to merge images positive signals were located the basal surface of larval epithelium (le). White arrow shows separated larval epithelium from basal lamina. Stars show yellow body. (A). After larval-pupal ecdysis, proliferation rate of stem cells significantly decreased (B; panel a). Stem cells (sc). Percentage of positive staining nuclei was determined by using Image J software (http://imagej.nih.gov/ij/) (B; panel b). Autophagy-related gene expressions—ATG8 and ATG12—were determined by using qRT-PCR (B; panel c). Mean ± SD for the three independent experiments are shown. Scale bar = 250 µm.
Identification of Stem Cells After Isolation
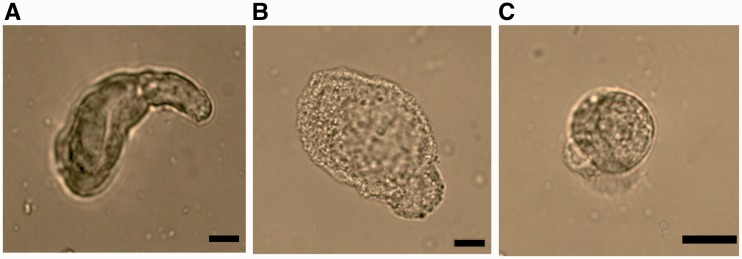
For investigation the hierarchical gene activation in both mature midgut cells and stem cells, separately, stem cells were isolated according to Hakim et al. (2009). Stem cells from the insect midgut can be easily removed from the tissue because they are not connected to the other cells by junctions (Cermenati et al. 2007). After isolation procedure, midgut cells were classified according to their morphologic feature (Turbeck 1974; Cioffi 1979; Baldwin and Hakim 1991). Columnar cells were detected as cylindiral shape cells with centrally located nucleus (Fig. 5a). Goblet cells were observed chalice-like cells by the presence of their typical vide cavity and basally located nucleus (Fig. 5b). Round shape cells were determined as stem cells (Fig. 5c). To determine whether the isolation procedure of stem cells was successful, we counted the numbers of each cells by using hemocytometer. The stem cells represented the 88.6 ± 5.3% of the total cells and the remaining cells were 5.6 ± 2.7% of columnar cells and 5.8 ±2.6% of goblet cells.
Fig. 5.
Identification of midgut cell types after stem cell isolation. Columnar cell (A), goblet cell (B) and stem cell (C). Scale bars = 20 µm.
Expression of Ecdysone Receptors and Ecdysone-Related Transcription Factors Differs in Both Mature Midgut Cells and Stem Cells
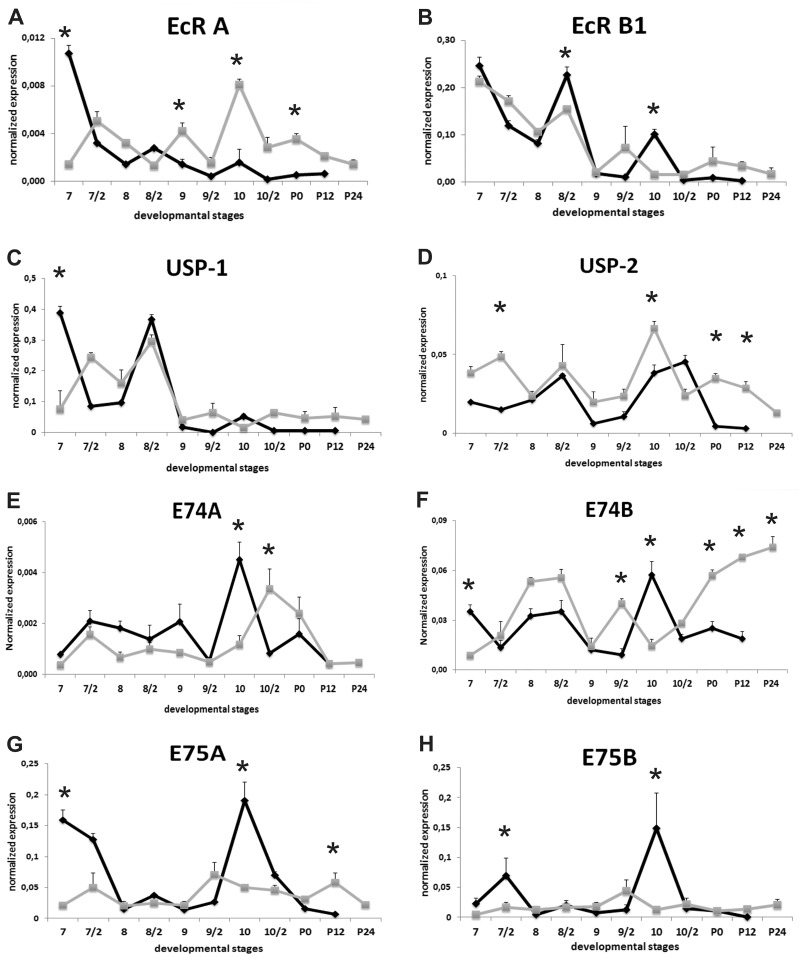
The developmental expression patterns of ecdysone receptors and ecdysone-related transcription factors were analyzed by using qRT-PCR (Fig. 6). Specificity of RT-PCR products was detected with high resolution gel electrophoresis and resulted in a single product with desired length (EcR A, 135 bp; EcR B1, 66 bp; USP1, 157 bp; USP2, 134 bp; E74A, 70 bp; E74B, 87 bp; E75A, 69 bp; E75B, 72 bp; BR-C Z1, 192 bp; BR-C Z2, 213 bp; BR-C Z4, 218 bp; βFTZ-F1, 110 bp; BHR3, 61 bp; actin, 75 bp; ATG8, 68 bp and ATG12, 69 bp).
Fig. 6.
Expression of ecdysone receptors and ecdysone-related transcription factors during remodeling of Bombyx mori midgut in mature cell and stem cell fractions. mRNA levels of these genes were quantified by qRT-PCR from day 7 of 5th instar to 24h after pupation. Experiments were performed every 12 h. Second half of each day was shown as day/2. EcR A (A), EcR B (B), USP 1 (C), USP 2 (D), E74A (E), E74B (F), E75A (G), E75B (H), BR-C Z1 (I), BR-C Z2 (J), BR-C Z4 (K), βFTZ-F1 (L) and BHR3 (M). The significance of means was tested by a two-tailed paired “t”-test. Stars: significance at *P ≤ 0.05. Mean ± SD for the three independent experiments are shown. Black line with closed diamonds denotes mature cells and grey line with closed squares indicates stem cells.
The levels of transcripts for both EcR isoforms in mature midgut cells were high in the first half of day 7 and then started to decrease until day 8 (Fig. 6A and B). EcR A mRNA levels in mature cells remained at rather low levels during experiment (Fig. 6A), although, EcR B1 expression showed two distinct peaks, the first, a large peak in the second half of day 8, the second on day 10 (Fig. 6B). Its mRNA levels were 80-fold higher than that of EcR-A on second half of day 8 and 60-fold higher on day 10. Expression patterns of USP isoforms were considerably different from each other. USP 1 expression profile was reflecting to EcR B1 (Fig. 6C). USP 2 mRNA levels exhibited two peaks in the second half of day 8 and day 10, respectively (Fig. 6D).
In the stem cells, fluctuated expression pattern of EcR A reached the highest level on day 10. After gut purge, EcR A mRNA levels were significantly higher than mature cells (Fig. 6A). Relatively high EcR B1 mRNA levels in stem cells began to decrease on day 9 and maintained low levels at the end of the experiment (Fig. 6B). Expression profiles of USP isoforms were different in stem cells. USP 1 expression patterns were similar to EcR B1 (Fig. 6C), besides, USP2 expression pattern resembled to EcR expression profile (Fig. 6D).
Both E74 isoforms in mature cells showed similar expression profiles during experiment but the relative abundance of E74B was 10-fold higher than E74A. The mRNA levels of both isoforms reached the highest level on day 10 and then decreased the low levels at the end of the experiment (Fig. 6E and F). In contrast to mature cells, E74 isoforms exhibited distinct expression patterns in stem cells. Low levels of E74A mRNA until day 10 started to increase and peaked in the second half of day 10 followed by a drop and remained low levels during early pupal period (Fig. 6E). Fluctuations in E74B mRNA levels were seen until day 10 and then increased in a stepwise manner throughout the early pupal stage (Fig. 6F).
Other early transcription factors E75A (Fig. 6G) and E75B (Fig. 6H) mRNA levels decreased very low level after cessation of feeding in both cell fractions. In the mature midgut cells, both E75 isoforms temporarily expressed on day 7 then levels decreased and remained low levels until second half of day 9. The mRNA levels of both isoforms in mature cells suddenly increased and peaked on day 10 followed by a decline in expressions thereafter (Fig. 6G and H). Low expression levels of E75A and E75B in stem cells did not show any significant changes throughout the experiment (Fig. 6G and H).
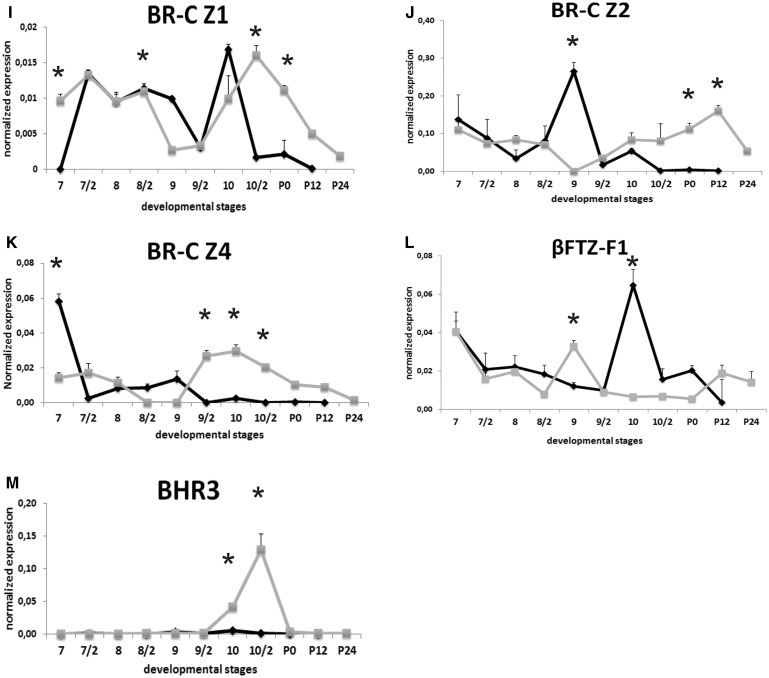
The temporal expression patterns for mRNA of three BR-C isoforms, BR-C Z1, Z2 and Z4, exhibited three distinct patterns in mature midgut cells. BR-C Z1 mRNA levels in both mature and stem cells were among the lowest levels in the BR-C isoforms examined (Fig. 6I). BR-C Z1 mRNA levels were sharply increased in mature cells after cessation of feeding activity and persisted moderate level until day 9, then, decreased in the second half of day 9 of fifth instar. Its transcripts sharply increased and peaked in the first half of day 10, subsequently, decreased to almost undetectable levels. In midgut stem cells, moderate BR-C Z1 mRNA levels until day 9 decreased to almost undetectable levels following with a transient peak in the second half of day 10, then, began to decrease in stepwise manner at the end of the experiment (Fig. 6I). The most abundant isoform was BR-C Z2 (Fig. 6J) in both mature and stem cells. BR-C Z2 mRNA showed two peaks, a broad peak on day 9 and a second, small peak on day 10 and then abruptly decreased to undetectable levels. Low BR-C Z2 levels in stem cells until second half of day 8 decreased to undetectable levels on day 9 followed by gradually rising in mRNA levels until 12h after pupation (Fig. 6J). The highest BR-C Z4 mRNA level in mature cells was found during actively feeding period. After cessation of feeding, its expression sharply decreased and remained very low levels until second half day 9, then declined undetectable levels at the end of the experiment. BR-C Z4 mRNA levels in midgut stem cells started to increase on day 9 and peaked on day 10 then decrease gradually (Fig. 6K).
Orphan nuclear receptor βFTZ-F1 was detected in both mature and stem cells during remodeling period (Fig. 6L). Moderate βFTZ-F1 mRNA levels in mature cells decreased progressively until second half of day 9 and then sharply increased and exhibited a large peak on day 10. Following this day, βFTZ-F1 mRNA levels decreased to low levels. Low βFTZ-F1 mRNA levels in stem cells exhibited one small peak around first half of day 9 and it was 2-fold higher than those in mature cells. After this day, its mRNA level abruptly decreased until pupation then slight increment was seen during early pupal stage (Fig. 7L).
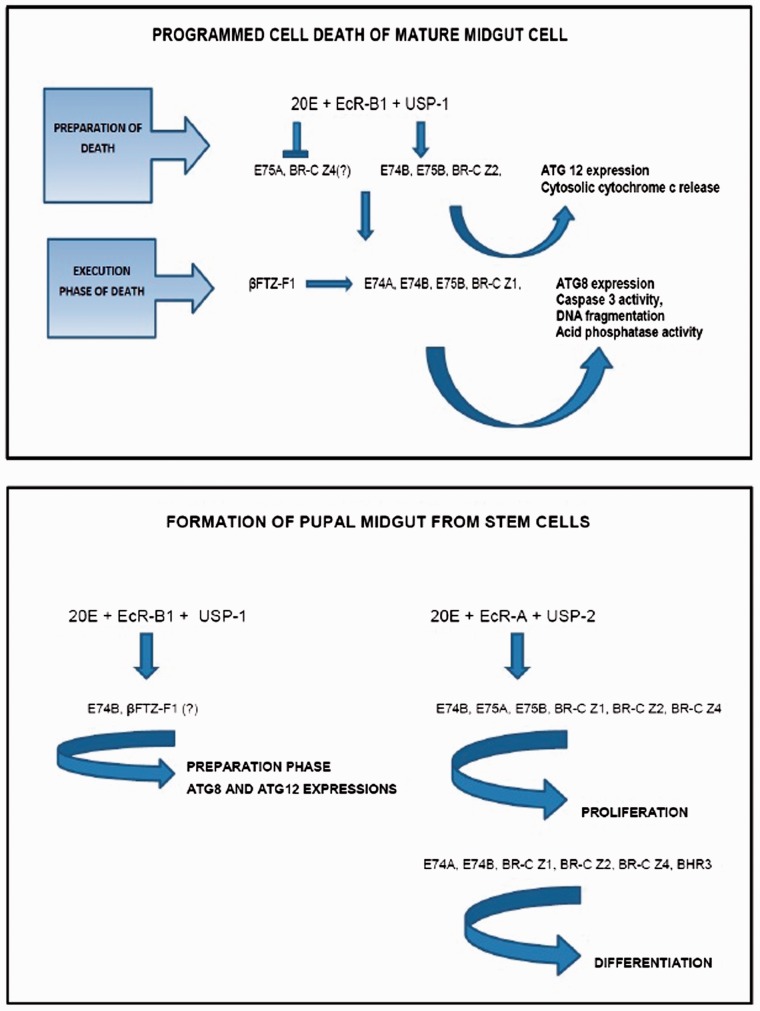
Fig. 7.
Proposed gene regulation cascades during midgut remodeling. (A) Gene regulation cascade during programmed cell death in mature midgut cells and (B) gene regulation cascade during formation of pupal midgut from stem cells.
BHR3 trancripts were almost undetectable levels in mature cells during experiment. Similarly, we did not detect BHR3 mRNA in stem cells until second half of day 9. Its mRNA levels began to increase from day 10 and exhibited single sharp peak in the second half of the day. BHR3 mRNA level in stem cells was found 180-fold higher than those in mature cells. Its transcripts sharply decreased to undetectable levels during early pupal period (Fig. 6M).
Discussion
As the objective of our study was to correlate midgut remodeling to ecdysone-related gene expression patterns, it was important that we evaluate remodeling events morphologically and biochemically in our system. Previous studies (Goncu and Parlak 2011; Franzetti et al. 2012) and our results suggested that larval midgut cells degenerate via association with apoptosis and autophagic cell death. Involvement of cytosolic cytochrome c signaling during midgut PCD was firstly demonstrated in this study. Varkey et al. 1999 reported the function of cytochrome c in apoptotic cell death in Drosophila and found cytochrome c precedes all signs of PCD. Inhibition of cytochrome c expression significantly affected apoptosis and activation of caspase (Liu et al. 2012). The last stage of apoptosis—phagocytosis—of yellow body fragments by a few number of granulocytes were also observed during early pupal stage. Shinohara et al. (2008), firstly demonstrated the granular cells in midgut lumen during larval-pupal ecdysis of B. mori and proposed the phagocytic function of them during degeneration process. However, due to lack of sufficient phagocytic cells, autophagy is an important process during degeneration of larval midgut epithelium. According to different expression patterns of autophagy-related genes—ATG8 and ATG12—they probably show different functions in larval midgut cell death. In the HEK293 cells, mitochondrial apoptosis is closely related with ATG12 proteins. Release of cytochrome c from mitochondria into cytosol is substantially reduced in ATG12 depleted HEK293 cells (Rubinstein et al. 2011). Timing of ATG12 expression in larval midgut cells was coincident with the release of cytochrome c from mitochondria. Therefore, we proposed that, ATG12 may a candidate molecule which provides crosstalk between apoptosis and autophagic cell death in mature midgut cells. In addition, low levels of ATG12 expression when higher ATG8 expression and AP activity occurred indicated that autophagosome formation in the larval midgut cells predominantly dependent on ATG8-PE ubiquitin-like conjugation system like those in degenerating anterior silk gland (Li et al. 2011). Rising ATG8 expression and AP activity during late prepupal stage and presence of cleaved caspase 3 at the same time may show the overlapping between apoptosis and autophagic cell death after second half of day 9.
Midgut epithelial stem cells are one of the multipotent stem cell types in insects (Corley and Lavine 2006). According to recent studies, autophagy is important the maintenance of unique properties of stem cells like self-renewal, pluripotency and differentiation (Vessoni et al. 2012). Consideration of ATG8 and ATG12 gene expressions in Bombyx midgut stem cells during early prepupal stage suggests that autophagy may also be important for proliferation and reprogramming of midgut stem cells during metamorphosis and this information offers the first evidence about the role of autophagy in midgut stem cells.
Expressions of ecdysone receptors and transcription factors were compared to midgut mature cells and stem cells. EcR A mRNA levels correlated with hemolymph 20E levels (Sakurai et al. 1998) in stem cells, however, similar upregulation was not detected in mature cells. In contrast, expression of EcR B1 was triggered by the rising 20E levels (Sakurai et al. 1998) in both cells fractions until day 9. In Drosophila, EcR-B1 mRNA expression is found dominantly in degenerating larval tissues by PCD while EcR A mRNA is detected in proliferated and differentiated tissues during larval-pupal metamorphosis (Talbot et al. 1993). According the co-expression profiles of EcR-B1 and USP-1 isoforms, this heterodimer is involved in early events of PCD in mature cells during prepupal period and execution phase of death during late prepupal period. Typically peaks of EcR B1 expression coincided with PCD-related events like cytoplasmic and nuclear condensation, cell shrinkage DNA fragmentation and caspase 3 activation in mature midgut cells. Parthasarathy and Palli (2007) reported that EcR-B/USP-A heterodimer regulates PCD in larval midgut and EcR-A/USP-B heterodimer regulates formation of pupal/adult midgut in Aedes aegypti. When stem cells were analyzed in terms of morphologic changes and autophagy-related genes expressions, EcR-B1 probably associated with preparation of stem cells for reprogramming and enhances the inductive effects of ecdysone for proliferation and differentiation. Co-expression patterns of EcR-A and USP-2 and their expressions at higher levels in stem cells suggested that this heterodimer is involved in stem cell proliferation and beginning of differentiation during remodeling period.
Drosophila melanogaster E74A mutants and E74B loss-of-function mutants are defective in puparium formation and die during prepupal stage (Fletcher et al. 1995). These results suggest that E74 isoforms play pivotal roles in pupal commitment. In our study, timing of E74A and E74B expressions in mature midgut cells and high hemolymph 20E levels during late prepupal period (Sakurai et al. 1998) indicated that expressions of both isoforms were triggered by large 20E levels and associated with execution of cell death. Sekimoto et al. (2006) reported a similar rising in E74A expression in the anterior silk glands of B. mori correlates with the increasing ecdysteroid level and death execution however E74B is maximally expressed until PCD is began. When considered the E74A expression profile in stem cells together with developmental events at the same time suggested that E74A might involve differentiation processes. The relative abundance of E74B in stem cells was generally higher than those in larval cells. Rising hemolymph 20E levels during prepupal period (Sakurai et al. 1998) seemed to induce E74B expression in stem cells, however, the main upregulation occurred after big 20E peak just before pupation. In terms of timing of expression, E74B isoform is related to preparation of stem cells for remodeling. Stilwell et al. (2003) reported that similar expression pattern of MsE74B mRNA in the epidermis of Manduca sexta is connected with pupal commitment. Hence, it is likely that gradually rising E74B transcripts during formation of pupal midgut is also important for differentiation of stem cells.
E75 plays an important role in controlling ecdysone response in the late third instar salivary gland of D. melanogaster. Decreasing the expression of E75A in mature cells after termination of feeding shows its relation with preparation of death. The highest 20E level in hemolymph just before pupation (Sakurai et al. 1998) induced the E75 isoform peaks on day 10 and these sharp peaks probably associated with beginning of degenerative changes and execution phase of death, respectively. Based on the study in Manduca epidermis, rising E75A expression is involved in both sustaining larval characters and promoting metamorphosis (Zhou et al. 1998; Keshan et al. 2006). In terms of expression patterns of E75 isoforms, their role(s) in metamorphic changes of stem cells were not clear, however, these genes may play a role for proliferation.
Expression of BR-C Z1 isoform coincided with rising hemolymph 20E levels (Sakurai et al. 1998) in both cell fractions after termination of feeding and the Z1 mRNA peaks were detected when hemolymph 20E levels reached the highest levels just before pupation (Sakurai et al. 1998). Cakouros et al. (2002) reported that BR-C Z1 expression during prepupal period is important for larval salivary gland degeneration in Drosophila and it is involved in the regulation of the initiator caspase DRONC transcription. Similarly, rising BR-C Z1 mRNA levels in mature cells after cessation of feeding suggests its possible role in PCD process. Especially, its expression peak on day 10 coincident with the caspase activation and supported the view that the Z1 isoform may be important for caspase regulation process or its activation during larval midgut degeneration. When considered in conjunction with the appearance of PCD before and after the peak of BR-C Z2, PCD processes in mature midgut cells proceeded rapidly after the peak level. Therefore, this isoform is probably involved in promoting of death in mature midgut cells. Similar with our conclusion, Kuch rov-Mahmood et al. (2002) reported that BR-C Z2 may function as a death inducing factor at low ecdysone level. In contrast to Z2 isoform, high level of BR-C Z4 mRNA suddenly decreased after cessation of feeding in B. mori. Kuchárová-Mahmood et al. (2002) suggested that BR-C Z4 expression in salivary gland during third instar larvae retards degeneration process but accelerates PCD during prepupal period. Therefore high BR-C Z4 level during feeding period may prevent death in mature cells and it might be downregulated for preparation of death process. Expression levels of Z1, Z2 and Z4 isoforms in the midgut stem cells began to increase after day 9 of fifth instar. According to previous study on Drosophila ovarian development, BR-C is a major effector for niche formation and cell differentiation of germ line stem cells (Gancz et al. 2011). BR-C expression patterns in stem cells suggested that all BR-C isoforms are important for morphogenesis of B. mori pupal midgut. Based on the morphologic evidence, Z1 and Z4 isoforms appear to be involved in cell proliferation and niche formation but the Z2 isoform is probably require for differentiation to the pupal midgut epithelium.
Caspase 3 activation and high ATG8 expression were found when βFTZ-F1 expression in mature cells peaked on day 10 which suggests that βFTZ-F1 expression could regulate execution phase of PCD. Previous studies on βFTZ-F1 expression in D. melanogaster exhibited that the ability of late genes like DRONC to respond to the ecdysone is mediated through βFTZ-F1 transcription factor and mid-prepupal expression of βFTZ-F1 in Drosophila salivary glands is pivotal for execution of PCD (Broadus et al. 1999; Lee et al. 2002). Expression profile and timing of βFTZ-F1 in stem cells suggested that it may be important for stem cell’s responses to ecdysone at the beginning of remodeling process.
HR3 is categorized as an early late gene which is induced by a higher concentration of 20E. Almost undetectable levels of BHR3 expression in mature cells suggested that BHR3 gene did not have any function in mature cells. According to Lan et al. (1999), presence of USP-2 with EcR-B1 and USP-1 inhibited MHR3 promoter activation. Therefore high USP-2 expression in mature midgut cells during late prepupal stage probably inhibits BHR3 gene activation through this process. Siaussat et al. (2005) reported that ecdysteroids are able to coordinate cell proliferation and differentiation via regulation of cell cycle and cytoskeleton proteins through ecdysone-related transcription factors EcR, USP and HR3. When hemolymph ecdysone titer increases at the end of last larval instar, imaginal cells are arrested in G2/M phase of mitosis which coincides with the beginning of pupal differentiation. During this cell cycle arrest, USP, EcR and HR3 were found high in Plodia interpunctella. Critical timing of BHR3 mRNA levels in the midgut stem cells suggested that it may directly regulate differentiation of stem cells into pupal midgut epithelium.
According to our results, two different responses to 20E are caused by different expression patterns of ecdysone receptor isoforms and ecdysone-related transcription factors. EcR-B1/USP-1 heterodimer complex, E74B, E75B and BR-C Z2 transcription factors were first responsive genes to the ecdysone in mature cells and these genes are probably responsible for preparation of PCD process in mature midgut cells. In addition, down-regulation of E75A and BR-C Z4 genes expressions strongly suggested their possible roles in specifying the preparation of death process of mature cells. βFTZ-F1 likely mediate ecdysone stimulation effects on second responsive genes—E74A, E74B E75B and BR-C Z1. In respect to their expression profiles, these transcription factors presumably regulate the execution phase by controlling ATG8 expression, caspase 3 and AP activation (Fig. 7A). During the formation of pupal midgut both EcRs and partners appeared to be effective for metamorphic response. According to timing of expression of EcR-B1 and USP-1, this heterodimer probably important for pupal commitment and autophagy-related gene expressions in stem cells. Proliferation and differentiation processes of midgut stem cells are carried out via EcR-A and USP-2 heterodimer complex. Expression pattern of E74B gene suggested that this gene is important for reprogramming of stem cells for metamorphic process. In addition to E74B; E75A, E75B, BRC-Z4 and βFTZ-F1 were first responsive genes that prepare stem cells for proliferation and the second responsive genes E74A, E74B, BR-C Z1 and BHR3 influence both proliferation and differentiation. Timing of βFTZ-F1 peak in stem cells showed its possible role(s) for onset of proliferation (Fig. 7B).
Acknowledgments
The research was supported by The Scientific & Technological Research Council of Turkey (111T880) and Ege University Science and Technology Centre Technology Transfer Office (2004 BIL 010) from Ege University.
References Cited
- Baldwin K. M., Hakim R. S. 1991. Growth and differentiation of the larval midgut epithelium during molting in the moth Manduca sexta. Tissue Cell 23: 411–422. [DOI] [PubMed] [Google Scholar]
- Bergmeyer H. U., Gawehn K., Grassl M. 1974. In Methods of Enzymatic Analysis (Bergmeyer H.U.) Volume I, 2nd ed., 495–496, Academic Press, Inc, New York, NY. [Google Scholar]
- Broadus J., McCabe J. R., Endrizzi B., Thummel C. S., Woodard C. T. 1999. The Drosophila βFTZ-F1 orphan nuclear receptor provides competence for stage-specific responses to the steroid hormone ecdysone. Mol. Cell. 3: 143–149. [DOI] [PubMed] [Google Scholar]
- Cakouros D., Daish T., Martin D., Baehrecke E. H., Kumar S. 2002. Ecdysone-induced expression of the caspase DRONC during hormone-dependent programmed cell death in Drosophila is regulated by Broad-Complex. J. Cell Biol. 157: 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermenati G., Corti P., Caccia S., Giordana B., Casartelli M. A. 2007. Morphological and functional characterization of Bombyx mori larval midgut cells in culture. Invert. Surv. 4: 119–126. [Google Scholar]
- Cioffi M. 1979. The morphology and fine structure of the larval midgut of a moth (Manduca sexta) in relation to active ion transport. Tissue and Cell. 11: 467–479. [DOI] [PubMed] [Google Scholar]
- Corley L. S., Lavine M. D. 2006. A review of insect stem cell types. Semin. Cell. Dev. Biol. 17: 510–517. [DOI] [PubMed] [Google Scholar]
- Fletcher J. C., Burtis K. C., Hogness D. S., Thummel C. S. 1995. The Drosophila E74 gene is required for metamorphosis and plays a role in the polytene chromosome puffing response to ecdysone. Development 121: 1455–1465. [DOI] [PubMed] [Google Scholar]
- Franzetti E., Huang Z. J., Shi Y. X., Xie K., Deng X. J., Li J. P., Li Q. R., Yang W. Y., Zeng W. N., et al. 2012. Autophagy precedes apoptosis during the remodeling of silkworm larval midgut. Apoptosis 17: 305–324. [DOI] [PubMed] [Google Scholar]
- Gancz D., Lengil T., Gilboa L. 2011. Coordinated regulation of niche and stem cell precursors by hormonal signaling. PLoS Biol. 9: e1001202.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson U. E., Heid C. A., Williams P. M. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6: 995–1001. [DOI] [PubMed] [Google Scholar]
- Gomori G. 1950. An improved histochemical technic for acid phosphatase. Biotech. Histochem. 25: 81–85. [Google Scholar]
- Goncu E., Parlak O. 2011. The influence of juvenile hormone analogue, fenoxycarb on the midgut remodeling in Bombyx mori (L., 1758)(Lepidoptera: Bombycidae) during larval–pupal metamorphosis. Turk. J. Entomol. 35: 179–194. [Google Scholar]
- Hakim R. S., Caccia S., Loeb M., Smagghe G. 2009. Primary culture of insect midgut cells. In Vitro Cell. Dev. Biol. Anim. 45: 106–110. [DOI] [PubMed] [Google Scholar]
- Ijiro T., Urakawa H., Yasukochi Y., Takeda M., Fujiwara Y. 2004. cDNA cloning, gene structure, and expression of Broad-Complex (BR-C) genes in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 34: 963–969. [DOI] [PubMed] [Google Scholar]
- Jiang C., Baehrecke E. H., Thummel C. S. 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683. [DOI] [PubMed] [Google Scholar]
- Keshan B., Hiruma K., Riddiford L. M. 2006. Developmental expression and hormonal regulation of different isoforms of the transcription factor E75 in the tobacco hornworm Manduca sexta. Dev. Biol. 295: 623–632. [DOI] [PubMed] [Google Scholar]
- Kuchárová-Mahmood S., Raška I., Mechler B. M., Farkaš R. 2002. Temporal regulation of Drosophila salivary gland degeneration by the Broad-Complex transcription factors. J. Struct. Biol. 140: 67–78. [DOI] [PubMed] [Google Scholar]
- Lan Q., Hiruma K., Hu X., Jindra M., Riddiford L. M. 1999. Activation of a delayed-early gene encoding MHR3 by the ecdysone receptor heterodimer EcR-B1–USP-1 but not by EcR-B1–USP-2. Mol. Cell. Biol. 19: 4897–4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Simon C. R., Woodard C. T., Baehrecke E. H. 2002. Genetic mechanism for the stage-and tissue-specific regulation of steroid triggered programmed cell death in Drosophila. Dev. Biol. 252: 138–148. [DOI] [PubMed] [Google Scholar]
- Li Q., Deng X., Huang Z., Zheng S., Tettamanti G., Cao Y., Feng Q. 2011. Expression of autophagy-related genes in the anterior silk gland of the silkworm (Bombyx mori) during metamorphosis. Can. J. Zool. 89: 1019–1026. [Google Scholar]
- Liu K., Yang H., Peng J. X., Hong H. Z. 2012. Cytochrome c and insect cell apoptosis. Insect Sci. 19: 30–40. [Google Scholar]
- Loeb M. J., Hakim R. S. 1996. Insect midgut epithelium in vitro: an insect stem cell system. J. Insect Physiol. 42: 1103–1111. [Google Scholar]
- Parthasarathy R., Palli S. R. 2007. Stage-and cell-specific expression of ecdysone receptors and ecdysone-induced transcription factors during midgut remodeling in the yellow fever mosquito,Aedes aegypti. J. Insect Physiol. 53: 216–229. [DOI] [PubMed] [Google Scholar]
- Rubinstein A. D., Eisenstein M., Ber Y., Bialik S., Kimchi A. 2011. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol. Cell. 44: 698–709. [DOI] [PubMed] [Google Scholar]
- Sakurai S., Kaya M., Satake S. 1998. Hemolymph ecdysteroid titer and ecdysteroid-dependent developmental events in the last-larval stadium of the silkworm, Bombyx mori: role of low ecdysteroid titer in larval–pupal metamorphosis and a reappraisal of the head critical period. J. Insect Physiol. 44: 867–881. [DOI] [PubMed] [Google Scholar]
- Segraves W. A., Hogness D. S. 1990. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 4: 204–219. [DOI] [PubMed] [Google Scholar]
- Sekimoto T., Iwami M., Sakurai S. 2006. Coordinate responses of transcription factors to ecdysone during programmed cell death in the anterior silk gland of the silkworm, Bombyx mori. Insect Mol. Biol. 15: 281–292. [DOI] [PubMed] [Google Scholar]
- Shinohara Y., Ishii N., Takahashi S., Okazaki T. 2008. Appearance of apoptotic cells and granular cells in the silkworm midgut lumen during larval-pupal ecdysis. Zoolog. Sci. 25: 139–145. [DOI] [PubMed] [Google Scholar]
- Siaussat D., Bozzolan F., Queguiner I., Porcheron P., Debernard S. 2005. Cell cycle profiles of EcR, USP, HR3 and B cyclin mRNAs associated to 20E‐induced G2 arrest of Plodia interpunctella imaginal wing cells. Insect Mol. Biol. 14: 151–161. [DOI] [PubMed] [Google Scholar]
- Smagghe G., Vanhassel W., Moeremans C., Wilde D., Goto S., Loeb M. J., Blacburn M. B., Hakim R. S. 2005. Stimulation of midgut stem cell proliferation and differentiation by insect hormones and peptides. Ann. N. Y. Acad. Sci. 1040: 472–475. [DOI] [PubMed] [Google Scholar]
- Stilwell G. E., Nelson C. A., Weller J., Cui H., Hiruma K., Truman J. W., Riddiford L. M. 2003. E74 exhibits stage-specific hormonal regulation in the epidermis of the tobacco hornworm, Manduca sexta. Dev. Biol. 258: 76–90. [DOI] [PubMed] [Google Scholar]
- Swevers L., Cherbas L., Cherbas P., Iatrou K. 1996. Bombyx EcR (BmEcR) and Bombyx USP (BmCF1) combine to form a functional ecdysone receptor. Insect Biochem. Mol. Biol. 26: 217–221. [DOI] [PubMed] [Google Scholar]
- Talbot W. S., Swyryd E. A., Hogness D. S. 1993. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell. 73: 1323–1337. [DOI] [PubMed] [Google Scholar]
- Tettamanti G., Grimaldi A., Casartelli M., Ambrosetti E., Ponti B., Congiu T., Ferrarese R., Rivas-Pena M. R., Pennacchio F., De Eguileor M. 2007. Programmed cell death and stem cell differentiation are responsible for midgut replacement in Heliothis virescens during prepupal instar. Cell Tissue Res. 330: 345–359. [DOI] [PubMed] [Google Scholar]
- Turbeck B. O. 1974. A study of the concentrally laminated concretions, ‘spherites’, in the regenerative cells of the midgut of lepidopterous larvae. Tissue Cell 6: 627–640. [DOI] [PubMed] [Google Scholar]
- Varkey J., Chen P., Jemmerson R., Abrams J. M. 1999. Altered cytochrome c display precedes apoptotic cell death in Drosophila. J. Cell Biol. 144: 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vessoni A. T., Muotri A. R., Okamoto O. K. 2012. Auto phagy in stem cell maintenance and differentiation. Stem Cells Dev. 21: 513–520. [DOI] [PubMed] [Google Scholar]
- Wu Y., Parthasarathy R., Bai H., Pallii S. R. 2006. Mechanisms of midgut remodeling: Juvenile hormone analog methoprene blocks midgut metamorphosis by modulating ecdysone action. Mechanisms of Development. 123: 530–547. [DOI] [PubMed] [Google Scholar]
- Zhou B., Hiruma K., Jindra M., Shinoda T., Segraves W. A., Malone F., Riddiford L. M. 1998. Regulation of the Transcription Factor E75 by 20-Hydroxyecdysone and Juvenile Hormone in the Epidermis of the Tobacco Hornworm, Manduca sexta, during Larval Molting and Metamorphosis. Dev. Biol. 193: 127–138. [DOI] [PubMed] [Google Scholar]