Abstract
Mechanically interlocked supramolecular assemblies are appealing building blocks for creating functional nanodevices. Here we describe the multi-step assembly of massive DNA origami rotaxanes that are capable of programmable structural switching. We validated the topology and structural integrity of these rotaxanes by analyzing the intermediate and final products of various assembly routes using electrophoresis and electron microscopy. We further demonstrated two structural switching behaviors of our rotaxanes, both mediated by DNA hybridization. In the first mechanism, the translational motion of the macrocycle can be triggered or halted at either terminus. In the second mechanism, the macrocycle can be elongated after the completion of rotaxane assembly, giving rise to a unique structure that is otherwise difficult to access.
Keywords: DNA nanotechnology, DNA origami, Molecular devices, Rotaxanes, Macrocycles
Graphical abstract
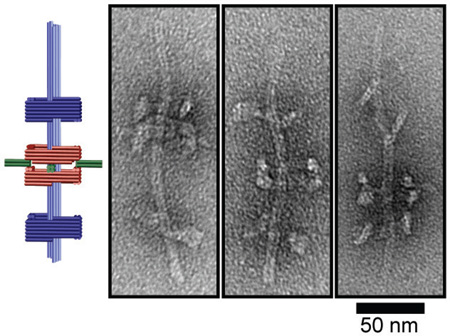
A new set of threads: Massive 200-nanometer long rotaxanes were assembled using a DNA origami approach. Threaded macrocycles can be programmably docked at either dumbbell stopper and can be reconfigured to generate novel and otherwise unfavored rotaxane topologies.
Supramolecular assemblies with mechanically interlocked components are of considerable interest to nanorobotics and nanomechanics.[1] One such mechanically bonded assembly is the rotaxane, a dumbbell-shaped molecule whose central axle is threaded through a ring- or tube-shaped “macrocycle” molecule.[2] Due to the translational and rotational freedom of the enclosed macrocycle, rotaxanes harbor potential as molecular switches and/or molecular cargo shuttles; the movement of which can be dictated by various chemical stimuli including addition of DNA strands, pH changes, and light signals.[3] Intrigued by the potential applications of rotaxanes as stimulus-responsive switches and the diverse conjugation chemistry compatible with DNA nanodevices[4], several recent investigations have focused on assembly of rotaxanes from DNA.[3b, 5]
Inspired by these antecedent DNA rotaxanes, the rapidly expanding catalog of dynamic DNA origami devices[6], and demonstrations of multimeric DNA origami assembly via shape-complementary topologies[7], we present here a strategy for assembling a rotaxane from three DNA origami structures. The large size, thermal stability, and DNA-induced macrocycle switching and reconfiguration are the most distinguishable features of this nanodevice comparing to previous rotaxanes.
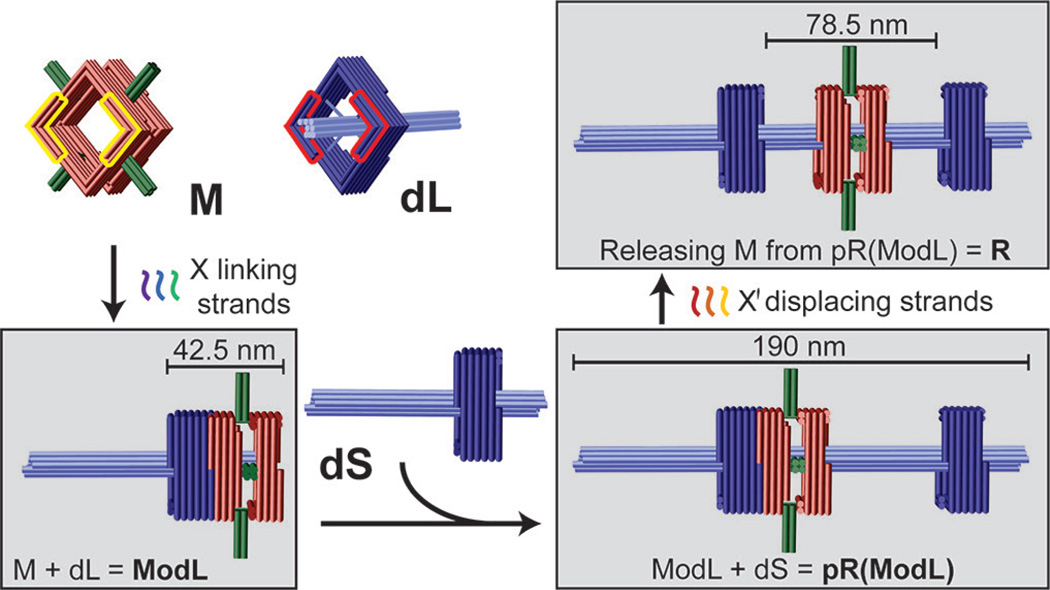
Our rotaxane (R) is composed of three scaffolded DNA origami[8] monomers: a macrocycle (M) and two half-dumbbells (dL and dS), all designed using caDNAno software (Figure S1).[9] The square-shaped macrocycle (35 nm × 35 nm × 27.5 nm, Length×Width×Height) displays four 16-nm, 4-helix bundle rods radiating outward from each side of the square (Figure 1 and Figure S2-1). The dL and dS monomers (Figure 1 and Figure S2-2) are structurally identical, each consisting of a square-shaped stopper (35 nm × 35 nm × 17.5 nm) suspended on an 8-helix bundle axle (7.5 nm × 7.5 nm × 96 nm) by four pairs of single-stranded DNA tendons (35 nt stretched over 9.8 nm), each exerting a tension force of ~6 pN as predicted by modified freely-jointed-chain models.[10] The asymmetrical half-dumbbells each feature a short axle end and a long axle end. M and each of the half-dumbbell stoppers possesses a pair of notched surfaces (highlighted by yellow and red outlines, respectively, in Figure 1 and Figure S2) that allow stacking in the presence of sequence-specific linking DNA strands. In this work we designed two sets of linking strands X and Y, which dock M to the short- or long-axle facing side of the stopper, respectively. All linking strands carry a 6-nt extension, or DNA toehold, to facilitate the releasing of M at a later stage. For example, M can be docked to the short-axle facing side of the stopper on dL by linking strand set X, giving rise to a dimeric assembly intermediate ModL (M on dL) as shown in the bottom left box of Figure 1. Additionally, one end of each half-dumbbell axle (the long end of dL and the short end of dS) is rendered inert (i.e., incapable of dimerization) while the other end is functionalized with DNA extensions to facilitate the end-to-end binding between the two half-dumbbells. Therefore, reacting dS with ModL can in principle produce a second assembly intermediate pR(ModL), which is a pseudo-rotaxane (Figure 1, lower right). This intermediate can be subsequently converted to genuine rotaxane (R) by releasing M through toehold-mediated strand displacement[11] with the addition of displacing strand set X' that binds the linking strand set X (Figure 1, top right).
Figure 1.
Schematic drawings of DNA origami rotaxane components and a viable three-step assembly route. Each cylinder represents a DNA double helix. DNA origami monomers (macrocycle M and half-dumbbell dL) are shown on the top left panel. Notches on the top faces of M and dL are highlighted in yellow and red, respectively; their bottom faces have notches of complemantary shape. Intermediate and final products are shown in gray boxes.
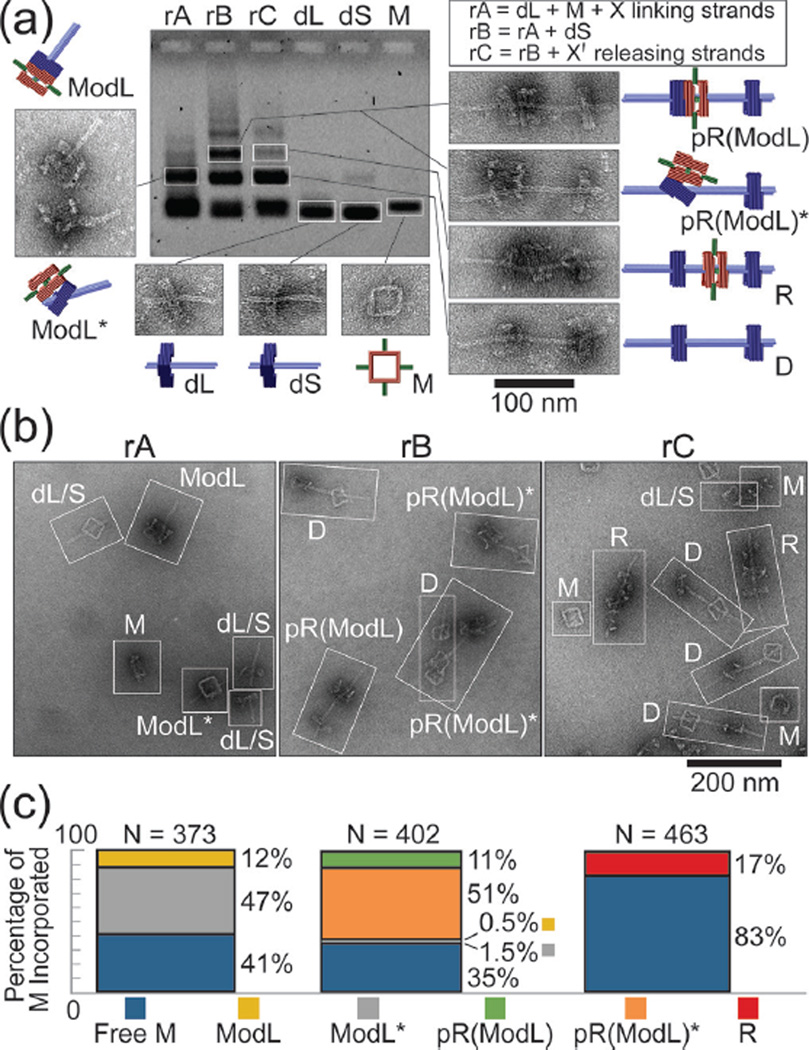
We utilized agarose gel electrophoresis, gel extraction, and negative-stain transmission electron microcopy (TEM) to identify components of the three assembly steps (Figure 2a and Figures S3&S4). The first assembly reaction (rA) consisted of M and dL in equimolar amounts and 60-fold excess of linking strand set X, which was incubated at 40 °C for 16 hours. The gel lane containing rA mixture showed only one major band besides the reactants; gel extraction and TEM revealed that this band contained the correctly formed dimer ModL and misassembled dimers that we term ModL*. The second reaction (rB) consisted of the unpurified rA mixture and dS added in equimolar proportion to dL; the reaction was incubated at 40 °C for 16 hours. Gel electrophoresis of rB mixture showed a new band in addition to the monomer and dimer bands. Analyzing the extract of this slower running band by TEM revealed the pseudo-rotaxane pR(ModL) and its misassembled counterparts pR(ModL)*. The final assembly step (rC) was to incubate rB mixture with 5-fold excess (relative to linking strand set X) of displacing strand set X' at 44 °C for 12 hours. Electrophoresis yielded a similar band pattern as with rB but with reduced intensity of the trimer band, suggesting only some trimers formed in rB remained the same molecular weight after releasing M. Despite minor structural damage during gel extraction (giving rise to a small amount of monomers and dimers), TEM analyses confirmed that all trimeric structures containing M within this band were genuine rotaxane R, in which the macrocycle sat in between the stoppers with the axle threading through all three squares. Note that the rods and the thickness of M distinguish it from the stopper of dL or dS on TEM images.
Figure 2.
DNA origami rotaxane assembly products. (a) Agarose gel electrophoresis showing individual assembly components and reaction mixtures after each assembly step. The reactants of each assembly step (rA, rB, and rC) are listed in a box on the top right corner. Representative close-up TEM images of the extracted bands of interest are shown with cartoon models. (b) Representative zoom-out TEM images of the unpurified reaction mixtures. Each distinguishable nanostructure is identified and labeled with an abbreviation. (c) Macrocycle incorporation efficiency after each reaction step presented as a bar-graph. Stacked bars from left to right correspond to rA, rB, and rC, respectively.
To quantify efficiency of each assembly step, we analyzed TEM images of the unpurified assembly products of rA, rB, and rC (Figure 2b). After categorizing and counting all resolvable nanostructures, we determined assembly efficiency as a measure of the assembly state of M at the end of each step (Figure 2c). Statistics (N=463) indicated that approximately 17% of M were incorporated in the intended product R after a 3-step assembly. We considered two possible reasons for the limited yield. One possibility is that the macrocycle of the pseudo-rotaxanes dethreaded from the axle following strand displacement due to thermal instability (i.e., two dumbbell halves temporarily detached). However, properly assembled rotaxanes stayed intact when incubated at room temperature for over a month or 44 °C for 20 hours (Figure S5), which spoke against this hypothesis. An alternative and more likely explanation is that yield of R was limited by the macrocycle threading efficiency during rA. Hence, the intermediates ModL* and pR(ModL)* were considered malformed assemblies in which M did not fully thread over the axle of dL; the release of M from pR(ModL)* necessarily yielded empty dumbbells (D) and monomeric macrocycles. Indeed, the assembly efficiency of the two properly formed intermediates (12% for ModL and 11% for pR(ModL)) roughly equaled that of rotaxane (17%); the chances of M being incorporated in misassembled intermediates in the first two assembly steps also matched well (47% ModL* and 51% for pR(ModL)*). The discrepancies were likely due to the structural distortion on the TEM grid, mainly the flattening of M-docked stopper in 24%–29% of nanostructures containing M (Figure S6), which we interpreted as misfolded intermediates, leading to underestimation of correctly formed ones.
Since our TEM images only resolve the two-dimensional projections of the three-dimensional DNA nanostructures, we designed two more experiments in order to probe the topology of the rotaxanes. In the first experiment, we added M and linking strands (set X) to the preassembled dumbbells. Although we found DNA origami trimers akin to pR(ModL)*, releasing M did not result in any rotaxane detectable by TEM (Figure S7). This confirmed that full threading of macrocycle over the axle cannot occur after dumbbell formation. In other words, dumbbells’ stoppers present unbreachable barriers to any macrocycle. In the second experiment, our attempts to assemble rotaxane by first docking M to the long-axle facing side of dS stopper (i.e., M + dS + linking strand set Y) eventually produced negligible amount of R (Figure S8). Notably, all intermediate products appeared improperly formed such that the axle did not penetrate through M. This suggested that the proper threading is critical for the formation of R, and further supported our notion that the initial docking of M on dS or dL is the yield-limiting step. We speculated that the difference between threading M over long vs short axle can be attributed to a hinging – clamping process. According to this speculation, part of the docking interface of the macrocycle can first bind with a stopper (hinging step), which does not entail threading over the axle; the rest of the interface would then engage in a clamping motion, latching M on the half-dumbbell. The latter requires a shorter axle end than macrocycle width (Figure S9). The hypothetical partial docking state can in principle lower the entropic barrier ordinarily present when randomly threading a ring over a rod (Figure S10),[12] which could explain the dismal yield of R when the assembly cannot benefit from this stepping stone (e.g., when threading M through the long axle end or adding M to a half-dumbbell without linking strands). Combining the findings described above, we established the mechanically locked state of the macrocycle on a structurally stable DNA origami rotaxane.
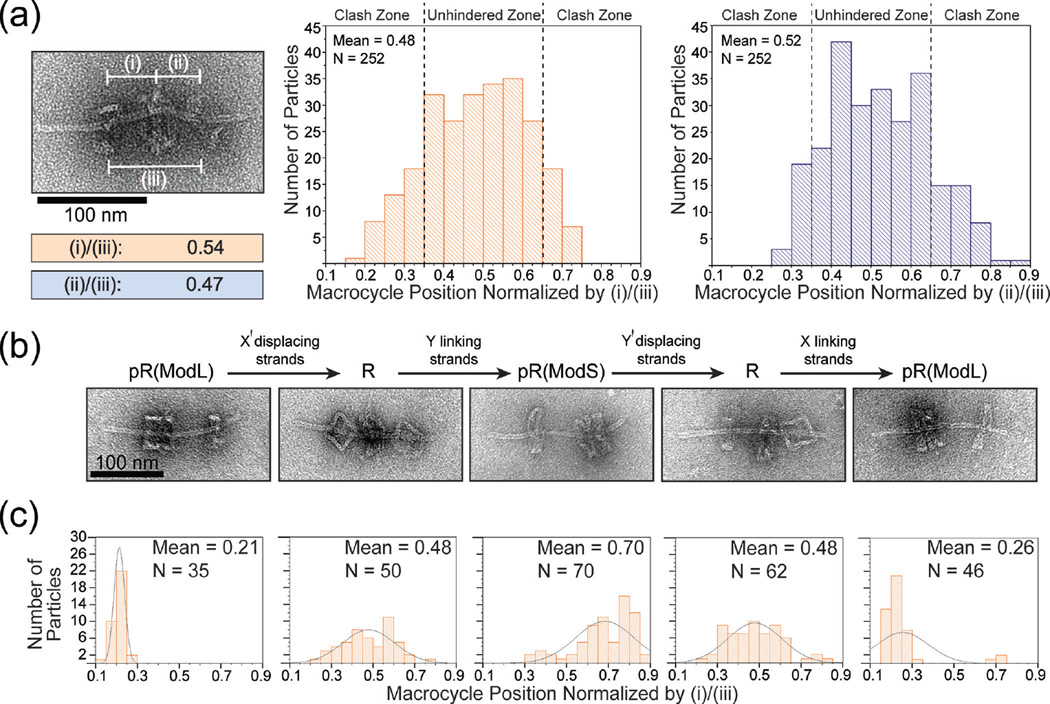
The free movement of macrocycle between stoppers is the hallmark of rotaxanes. We examined the macrocycle’s translational motion by recording the position of M in relation to either dumbbell stopper for each rotaxane identified by TEM. We measured center-to-center distances of (i) M to dL stopper, (ii) M to dS stopper, and (iii) stopper-to-stopper and defined the relative position of M as (i)/(iii) or (ii)/(iii). By plotting the relative macrocycle positions of 252 rotaxanes as histograms (Figure 3a), we found that nearly all (≥246 out of 252) normalized values fell between 0.21 and 0.79 with a mean close to 0.5, as dictated by the rotaxane’s designed dimensions (Figure S11). The data further suggested that macrocycles had equal probability to sit at any given position in the middle 1/3 of the axle (between 0.34 and 0.66); however, the frequency of M residing beyond these points tapered off rapidly towards the stoppers. In light of the tilting flexibility of the dumbbell stoppers as seen in TEM images, we reasoned that the loss of probability density in these regions (clash zone) is due to steric hindrance posed to the macrocycle by the wobbly stoppers (Figure S12). This may also explain the observation that slightly more M stayed close to the dL stopper — the dS stopper is expected to be more flexible due to its proximity to the axle end.
Figure 3.
The movement and switching of macrocycles in DNA origami rotaxanes. (a) Distribution of macrocycle positions with respect to stoppers. Left: a close-up TEM image and normalized measurements of macrocycle position of an example rotaxane. Center and right: normalized macrocycle positions plotted as histograms; note that although the two histograms are based on the same TEM images, they are not perfect mirror images due to slight bending of the rotaxane axles. (b) Top: scheme of displacing or linking strands induced rotaxane structural switching. Bottom: close-up TEM images of desired products. (c) The distributions of macrocycle position resulting from switching operations. Histograms correspond to the switching states in (b). Overlayed curves are included only to guide the reader’s eye.
After validating that the macrocycle translates freely, we examined the potential of our rotaxane as a molecular switching device. As shown in Figure 3b and 3c, the rotaxane responded correctly to the appropriate displacing and linking strands and switched reversibly between its free state R and two docked states at either stopper — pR(ModL) and pR(ModS). A caveat of the device was the incomplete switching to the pR(ModS) state such an on-axle structural switching procedure. When we (i.e., a pseudo-rotaxane with M on dS, cf. Figure 3c, middle). The reason for this inefficiency may be the orientation variability of the stopper, meaning that a portion of half-dumbbells bear a stopper facing the wrong direction. In fact, we noticed that dS sometimes docked with M on the side opposite to our design during our attempts to form pR(ModS) both through switching and through direct assembly (Figure S13), the latter case lowered the yield of R. Thus, we suspected that the portion of R incapable of assuming the pR(ModS) state likely contained such misfolded dS. Nevertheless, the macrocycle’s abilities to slide freely and change docking post in response to chemical cues are promising features for the future development of molecular switches and shuttles.
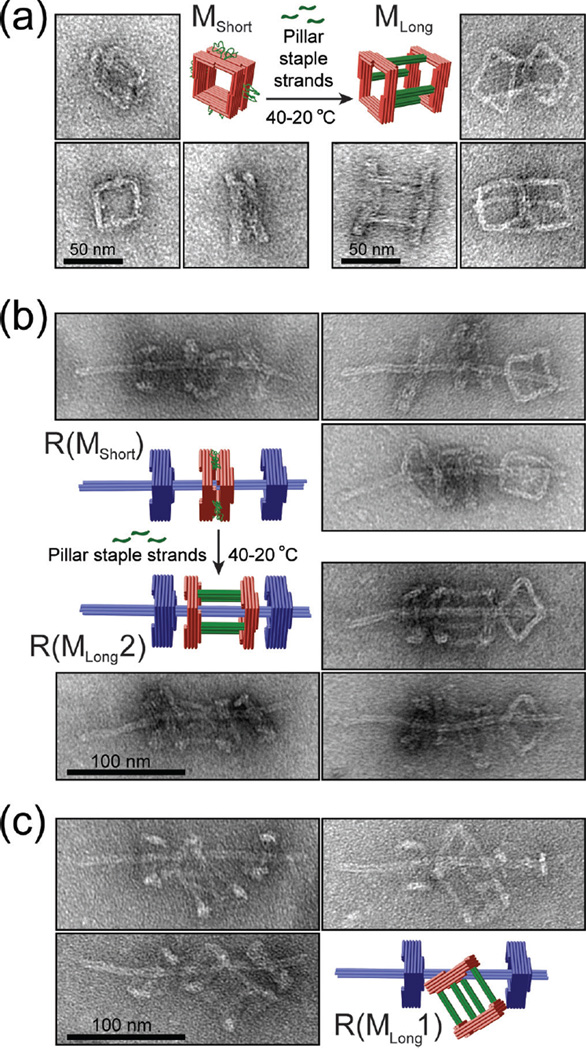
Finally, we demonstrated that we can achieve rotaxanes of different macrocycle threading topologies through varied assembly pathways. To show this, we utilized a macrocycle with single-stranded scaffold DNA loops connecting two squares. This macrocycle MShort (thickness: 30.9±11.5 nm) can be converted into a 62.5-nm long macrocycle (MLong) with the addition of an auxiliary set of 60 staple strands (Figure 4a). Rotaxanes containing the short macrocycle (termed R(MShort)) formed with efficiency similar to the original rotaxane R as assessed by negative-stain TEM (Figure 4b and Figure S14). Following this assembly, we added the auxiliary staple set to the reaction mix and annealed the entire reaction from 40 to 20 °C over an 18-hour time course. Rotaxanes generated this way contained tubular macrocycles (MLong) with both squares threaded over the axle (R(MLong2), cf. Figure 4b), which was the major product (88%) of such an on-axle structural switching procedure. When we assembled rotaxanes with premade MLong, another unique rotaxane R(MLong1) consistently emerged (96% of all rotaxanes) with only one square of macrocycle threaded and the other left on the side (Figure 4c).
Figure 4.
Assembling DNA origami rotaxanes with a tubular macrocycle. (a) Structural reconfiguration of a macrocycle by folding its unpaired scaffold loops into rigid 4-helix bundle pillars. Close-up TEM images show the typical appearances of monomeric macrocycles before and after reconfiguration. (b) Elongating a macrocycle of a complete DNA rotaxane. Close-up TEM images show the typical appearances of rotaxanes before and after reconfiguration. (c) Rotaxanes assembled using an elongated tubular macrocycle as a starting component. Note the different topologies of rotaxanes shown in (b) and (c).
In conclusion, we have constructed a class of DNA origami rotaxanes. We note that the yield and quality of these rotaxanes have much room for improvement (e.g., by rigidifying the connections between stopper and axle to limit the stopper’s tilting flexibility or by purifying desired products after each assembly step). However, the rotaxanes presented here are larger and more rigid than antecedent interlocked DNA nanodevices and harbor great potential for functionalization with a myriad of biomolecule cargos (e.g., protein) and stimulus-responsive components. Moreover, the modular design and structural switching properties of the rotaxanes open up new possibilities both for producing otherwise entropically unfavorable DNA structures and for programmably altering rotaxane geometry and dynamics. Simmel et al. recently made several multi-component rotaxanes using a different assembly scheme[13].
Supplementary Material
Acknowledgments
This work is supported by a National Institutes of Health (NIH) Director’s New Innovator Award (DP2-GM114830), an NIH grant (R21-GM109466) and a Yale University faculty startup fund to C.L. Additionally, J.T.P. and B.O.A are supported by NIH training grants T32-GM067543 and T32-GM007223, respectively.
Contributor Information
John T. Powell, Department of Cell Biology & Nanobiology Institute, Yale University, 850 West Campus Drive, West Haven, Connecticut 06516
Benjamin O. Akhuetie-Oni, Department of Molecular, Cellular, and Developmental Biology & Nanobiology Institute, Yale University, 850 West Campus Drive, West Haven, Connecticut 06516
Dr. Zhao Zhang, Department of Cell Biology & Nanobiology Institute, Yale University, 850 West Campus Drive, West Haven, Connecticut 06516
Prof. Chenxiang Lin, Email: chenxiang.lin@yale.edu, Department of Cell Biology & Nanobiology Institute, Yale University, 850 West Campus Drive, West Haven, Connecticut 06516.
References
- 1.a) Cheng C, Stoddart JF. Chemphyschem. 2016;17:1780–1793. doi: 10.1002/cphc.201501155. [DOI] [PubMed] [Google Scholar]; b) Liu X, Lu CH, Willner I. Acc. Chem. Res. 2014;47:1673–1680. doi: 10.1021/ar400316h. [DOI] [PubMed] [Google Scholar]; c) Lu CH, Cecconello A, Willner I. J Am. Chem. Soc. 2016;138:5172–5185. doi: 10.1021/jacs.6b00694. [DOI] [PubMed] [Google Scholar]; d) Schalley CA, Beizai K, Vogtle F. Acc. Chem. Res. 2001;34:465–476. doi: 10.1021/ar000179i. [DOI] [PubMed] [Google Scholar]
- 2.Harrison IT, Harrison S. J Am. Chem. Soc. 1967;89:5723–5724. [Google Scholar]
- 3.a) Lohmann F, Ackermann D, Famulok M. J Am. Chem. Soc. 2012;134:11884–11887. doi: 10.1021/ja3042096. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lohmann F, Weigandt J, Valero J, Famulok M. Angew. Chem. Int. Ed. 2014;53:10372–10376. doi: 10.1002/anie.201405447. Angew. Chem. 2014, 126, 10540–10544. [DOI] [PubMed] [Google Scholar]; c) Waeles P, Riss-Yaw B, Coutrot F. Chemistry. 2016 [Google Scholar]
- 4.a) Jones MR, Seeman NC, Mirkin CA. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]; b) Sacca B, Niemeyer CM. Chem. Soc. Rev. 2011;40:5910–5921. doi: 10.1039/c1cs15212b. [DOI] [PubMed] [Google Scholar]; c) Yang YR, Liu Y, Yan H. Bioconjug. Chem. 2015;26:1381–1395. doi: 10.1021/acs.bioconjchem.5b00194. [DOI] [PubMed] [Google Scholar]
- 5.a) Ackermann D, Jester SS, Famulok M. Angew. Chem. Int. Ed. 2012;51:6771–6775. doi: 10.1002/anie.201202816. Angew. Chem. 2012, 124, 6875–6879. [DOI] [PubMed] [Google Scholar]; b) Ackermann D, Schmidt TL, Hannam JS, Purohit CS, Heckel A, Famulok M. Nat. Nanotechnol. 2010;5:436–442. doi: 10.1038/nnano.2010.65. [DOI] [PubMed] [Google Scholar]; c) Valero J, Lohmann F, Keppner D, Famulok M. Chembiochem. 2016 doi: 10.1002/cbic.201500685. [DOI] [PubMed] [Google Scholar]; d) Weigandt J, Chung CL, Jester SS, Famulok M. Angew. Chem. Int. Ed. 2016;55:5512–5516. doi: 10.1002/anie.201601042. Angew. Chem. 2016, 128, 5602–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Andersen ES, et al. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]; b) Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]; c) Ke Y, Meyer T, Shih WM, Bellot G. Nat. Commun. 2016;7:10935. doi: 10.1038/ncomms10935. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ketterer P, Willner EM, Dietz H. Sci. Adv. 2016;2:e1501209. doi: 10.1126/sciadv.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Marras AE, Zhou L, Su HJ, Castro CE. Proc. Natl. Acad. Sci. U S A. 2015;112:713–718. doi: 10.1073/pnas.1408869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Science. 2015;347:1446–1452. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]; b) Woo S, Rothemund PW. Nat. Chem. 2011;3:620–627. doi: 10.1038/nchem.1070. [DOI] [PubMed] [Google Scholar]
- 8.a) Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rothemund PW. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 9.Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liedl T, Hogberg B, Tytell J, Ingber DE, Shih WM. Nat. Nanotechnol. 2010;5:520–524. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang DY, Seelig G. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 12.Sevick EM, Williams DR. Nano. Lett. 2016;16:671–674. doi: 10.1021/acs.nanolett.5b04440. [DOI] [PubMed] [Google Scholar]
- 13.List J, Falgenhauer E, Kopperger E, Pardatscher G, Simmel FC. Nat. Commun. 2016;7:12414. doi: 10.1038/ncomms12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.