ABSTRACT
Bacterial type IV secretion systems (T4SSs) are composed of two major subfamilies, conjugation machines dedicated to DNA transfer and effector translocators for protein transfer. We show here that the Escherichia coli pKM101-encoded conjugation system, coupled with chimeric substrate receptors, can be repurposed for transfer of heterologous effector proteins. The chimeric receptors were composed of the N-terminal transmembrane domain of pKM101-encoded TraJ fused to soluble domains of VirD4 homologs functioning in Agrobacterium tumefaciens, Anaplasma phagocytophilum, or Wolbachia pipientis. A chimeric receptor assembled from A. tumefaciens VirD4 (VirD4At) mediated transfer of a MOBQ plasmid (pML122) and A. tumefaciens effector proteins (VirE2, VirE3, and VirF) through the pKM101 transfer channel. Equivalent chimeric receptors assembled from the rickettsial VirD4 homologs similarly supported the transfer of known or candidate effectors from rickettsial species. These findings establish a proof of principle for use of the dedicated pKM101 conjugation channel, coupled with chimeric substrate receptors, to screen for translocation competency of protein effectors from recalcitrant species. Many T4SS receptors carry sequence-variable C-terminal domains (CTDs) with unknown function. While VirD4At and the TraJ/VirD4At chimera with their CTDs deleted supported pML122 transfer at wild-type levels, ΔCTD variants supported transfer of protein substrates at strongly diminished or elevated levels. We were unable to detect binding of VirD4At's CTD to the VirE2 effector, although other VirD4At domains bound this substrate in vitro. We propose that CTDs evolved to govern the dynamics of substrate presentation to the T4SS either through transient substrate contacts or by controlling substrate access to other receptor domains.
IMPORTANCE Bacterial type IV secretion systems (T4SSs) display striking versatility in their capacity to translocate DNA and protein substrates to prokaryotic and eukaryotic target cells. A hexameric ATPase, the type IV coupling protein (T4CP), functions as a substrate receptor for nearly all T4SSs. Here, we report that chimeric T4CPs mediate transfer of effector proteins through the Escherichia coli pKM101-encoded conjugation system. Studies with these repurposed conjugation systems established a role for acidic C-terminal domains of T4CPs in regulating substrate translocation. Our findings advance a mechanistic understanding of T4CP receptor activity and, further, support a model in which T4SS channels function as passive conduits for any DNA or protein substrates that successfully engage with and pass through the T4CP specificity checkpoint.
INTRODUCTION
The type IV secretion systems (T4SSs) display striking versatility among the known bacterial translocation systems in their capacity to translocate DNA and protein substrates to bacterial or eukaryotic target cells (1, 2). Members of one major T4SS subfamily, the conjugation machines, mediate transfer of mobile DNA elements and are responsible for widespread dissemination of antibiotic resistance genes and virulence determinants among pathogens in clinical settings (3, 4). A few conjugation systems have also been shown to translocate certain proteins, such as relaxases and primases, that function in recipient cells to promote establishment of the transferred mobile DNA elements (5–9). Members of a second major T4SS subfamily, the effector translocators, deliver various effector proteins to eukaryotic cells during infection processes (1, 10). Based on detailed phylogenetic analyses, it has been proposed that extant effector translocator systems arose from ancestral conjugation machines through acquisition of novel structural folds, proteins, or protein subassemblies (11). Such adaptations would have enabled the diversification of substrate repertoires, as well as delivery of secretion substrates to various prokaryotic and eukaryotic target cell types (2, 12).
In this study, we sought to define requirements for the functional repurposing of a dedicated conjugation machine as an effector translocator. One T4SS-associated ATPase, designated the type IV coupling protein (T4CP), was the focus of interest by virtue of a large body of evidence that T4CPs function as the substrate specificity checkpoints for cognate T4SSs (13–16). T4CP monomers consist minimally of three distinct domains: (i) an N-terminal transmembrane domain (NTD) implicated in establishment of critical contacts with one or more subunits of the T4SS translocation channel; (ii) a conserved nucleotide-binding domain (NBD) that is thought to provide energy through ATP hydrolysis for substrate translocation; and (iii) a sequence-variable, all-alpha domain (AAD) that participates in DNA and possibly effector protein substrate recognition (Fig. 1A) (17–19). The F plasmid-encoded TraD T4CP and many T4CPs associated with effector translocator systems possess a fourth, C-terminal domain (CTD) of variable lengths and sequence compositions. Studies of TraD's CTD established its critical role in recruitment of the F plasmid to the F-encoded T4SS (20), but the contributions of CTDs to effector protein trafficking have not been evaluated (2).
FIG 1.
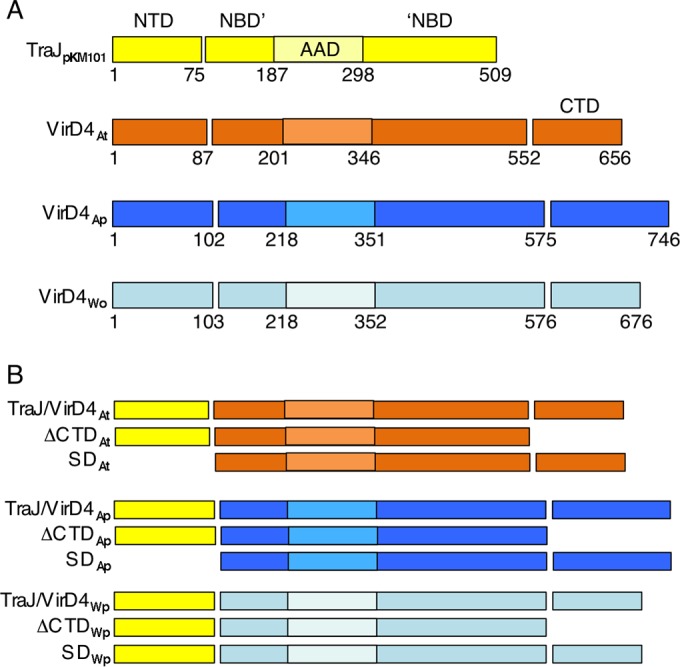
Domain architectures of type IV coupling proteins. (A) Schematic showing the domain architectures of TraJ encoded by E. coli plasmid pKM101 and VirD4 homologs from A. tumefaciens (subscript At), A. phagocytophilum (Ap), and W. pipientis (Wp). Domains: NTD, NBD (NBD′ to ′NBD), AAD, and CTD. The AAD is embedded within the NBD, as depicted by the lighter-shaded box. TraJpKM101 lacks a CTD. Domain boundaries are denoted by residue numbers relative to the N terminus. (B) Chimeric coupling proteins analyzed in this study. ΔCTD, CTD deleted; SD, soluble domain with the NTD deleted. (See Fig. S1 and S2 in the supplemental material for further information.)
T4CPs can be purified as monomers or dimers, but the hexameric form is catalytically active in vitro and is most likely the form responsible for directing substrate transfer through T4SS channels in vivo (17, 21, 22). The structural prototype for the T4CP superfamily is R388-encoded TrwB (here, the source of a protein appears in subscript, e.g., TrwBR388). A crystal structure of the soluble form of TrwBR388 with NTD deleted presents as a hexamer with 6-fold symmetry and a central channel ∼20 Å in diameter (17). In this structure, the AAD is situated at the base of the NBD hexamer, forming the entrance to the central channel. Interestingly, T4CPs structurally resemble two other hexameric ATPases, SpoIIIE and FtsK, which encircle and track unidirectionally along DNA by a mechanism involving ATP hydrolysis (23). T4CPs have thus been postulated to catalyze the unidirectional export of DNA cargoes through their central channels across the cytoplasmic membrane (24).
We report here that the Escherichia coli pKM101-encoded conjugation channel, which previously had been shown only to conjugatively transfer the pKM101 plasmid substrate to E. coli recipients, also translocates heterologous effector proteins when coupled with chimeric T4CPs. These chimeric subunits are composed of the NTD from the TraJpKM101 T4CP joined to the cytoplasmic domains of VirD4 homologs from three alphaproteobacterial species, Agrobacterium tumefaciens, Anaplasma phagocytophilum, and Wolbachia pipientis. The chimeric T4CPs supported translocation of known effectors from A. tumefaciens and A. phagocytophilum, as well as three candidate effectors from W. pipientis, through the pKM101-encoded channel. We capitalized on the functionality of these repurposed pKM101 systems to investigate the contributions of sequence-variable CTDs associated with the alphaproteobacterial T4CPs to substrate trafficking. Our findings establish, first, that a conjugation machine coupled with a reengineered T4CP receptor functions as an effector translocator and, second, that CTDs play important regulatory roles in governing the dynamics of type IV secretion.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain DH5α (Gibco-BRL) served as a host for plasmid constructions, and E. coli strain BL21(DE3) (Novagen) was used for protein production and purification. Strain HME45 [W3110 lac+ pglΔ8 gal490 λcI857Δ(cro-bioA)] has a defective λ prophage that supplies the Red recombination functions necessary for recombineering (25). Strain MS411 carrying plasmids of interest served as the donor for mating experiments (26). DH5αRifr(pUC4K) or WM1650 served as a recipient for DNA transfer experiments, and CSH26Cm::LTL(pUC4K) served as a recipient for protein transfer experiments (26, 27). E. coli strains were grown as previously described. When necessary, antibiotics were added at the following final concentrations: carbenicillin (Crb), 50 μg ml−1; chloramphenicol (Chl), 20 μg ml−1; kanamycin (Kan), 50 μg ml−1; gentamicin (Gen), 20 μg ml−1; tetracycline (Tet), 20 μg ml−1; and spectinomycin (Spc), 50 μg ml−1. A. tumefaciens A348 served as the wild-type strain, and A348Spcr served as a recipient for mating experiments (28). Other A. tumefaciens strains were LBA4404, with oncogenic transfer DNA (T-DNA) deleted (29); KA2000 (ΔvirD4) (30); At12516 (ΔvirE2) (31); Mx341 (virE3); and Mx219 (virF) (32). The conditions for growth of A. tumefaciens cells have been previously described (33), with antibiotic selection for plasmid maintenance at the following concentrations: carbenicillin, 100 μg ml−1; kanamycin, 100 μg ml−1; gentamicin, 100 μg ml−1; and spectinomycin, 400 μg ml−1. All antibiotics were obtained from Sigma Chemical Co. For induction of vir genes, cultures of A. tumefaciens strains were grown overnight with antibiotic selection, 1 ml of the culture was pelleted and diluted 5-fold in AB inducing medium (ABIM; glucose-containing minimal medium [pH 5.5], 1 mM phosphate, acetosyringone at 100 μM final concentration), and the cells were induced for 16 to 18 h with shaking at 22°C (33).
Bacterial plasmids.
The plasmids and oligonucleotides used in this study are listed in Tables 1 and 2.
TABLE 1.
Plasmids used in this study
| Plasmid(s) | Relevant characteristics | Source and/or reference |
|---|---|---|
| Vectors | ||
| pBSIISK+ and pBSIIKS+ | Crbr; cloning vectors (SacI-KpnI polylinkers in opposite orientation) | Stratagene |
| pBSIISK+.NdeI and pBSIIKS+.NdeI | Crbr; cloning vectors containing NdeI restriction site at the translational start site of lacZ | 36 |
| pBSIISK+.NcoI | Crbr; cloning vector containing NcoI restriction site at the translational start site of lacZ | 36 |
| pPC914KS+ | Crbr; pBSIIKS+ derivative with PvirB::virB1; expression vector when substituting other genes for virB1 | 36 |
| pBBR1MCS2 | Genr; broad-host-range cloning vector | 100 |
| pXZ151 | Kanr; broad-host-range IncP cloning vector | 33 |
| pML122Genr | Genr; mobilizable MOBQ derivative of RSF1010 | 31 |
| pUC4K | Kanr; source of Kanr cassette | Amersham |
| pHP45Ω | Spcr; source of Spcr cassette | 101 |
| pBAD24 | Crbr; ColE1 plasmid with arabinose-inducible PBAD promoter | 34 |
| pBAD24Kanr | Kanr Crbs; pBAD24 with nptII resistance gene from pUC4K inserted into crb resistance gene | This study |
| pBAD24Kanr-NdeI | pBAD24Kanr with NdeI site at translational start site downstream of PBAD promoter | This study |
| pBAD33 | Camr; pACYC184/p15A plasmid with arabinose-inducible PBAD promoter | 34 |
| pKM101 | Crbr; broad-host-range IncN plasmid | 102 |
| pKM101Spcr | Spcr Crbs; pKM101 with an spc resistance cassette from pHP45Ω inserted into the crb resistance gene at EcoRI | This study |
| pKM101SpcrΔtraJ | Spcr Crbs; pKM101Spcr with a ΔtraJ mutation | This study |
| pBAD33 | Camr; pACYC184/p15A plasmid with arabinose-inducible PBAD promoter | 34 |
| pMAL-c2x | Crbr; expression vector for MBP tagging | NEB |
| pET28b(+) | Kanr; expression vector for His tagging | Novagen |
| T4CP expression plasmids | ||
| pTB26 | Kanr; pBAD24Kanr with PBAD::traJpKM101 | This study |
| pNW1 | Crbr; pBSIIKS+.NcoI with Plac::traJ′ (codons 1–75 of traJpKM101 encoding NTD) | This study |
| A. tumefaciens VirD4 variants | ||
| pKA9 | Crbr; pBSIISK+ with PvirB::virD4At | 14 |
| pTB37 | Crbr; pBSIISK+ with PvirB::virD4AtΔCTD (virD4At with coding sequence for the 104-residue CTD deleted) | This study |
| pNW5 | Crbr; pBSIIKS+.NdeI with Plac::virD4At | This study |
| pNW2 | Crbr; pBSIIKS+ with Plac::traJ/virD4At (produces TraJ/VirD4At consisting of TraJ's NTD fused to the soluble domain of VirD4At spanning residues 88–656) | This study |
| pNW4 | Crbr; pBSIIKS+ with Plac::traJ/virD4ΔCTDAt (produces TraJ/VirD4At deleted of the CTD spanning residues 553–656) | This study |
| pNR1 | Crbr; pBSIISK+.NdeI with Plac::SDvirD4At (produces VirD4At's soluble domain spanning residues 88–656) | This study |
| pNR2 | Crbr; pBSIIKS+.NdeI with Plac::traJ/virD4K152QAt (produces TraJ/VirD4 with a K152Q nucleotide-binding site mutation) | This study |
| A. phagocytophilum VirD4 variants | ||
| pBT-virD4 | Crbr; cloned virD4Ap gene | Y. Rikihisa |
| pTB7 | Crbr; pPC914KS+ with PvirB::virD4Ap | This study |
| pTB48 | Kanr; pBAD24Kanr with PBAD::virD4Ap | This study |
| pTB38 | Kanr; pMK-RQ with codon-optimized traJ-virD4Ap (encodes TraJ/VirD4Ap consisting of TraJ's NTD fused to the soluble domain of VirD4Ap spanning residues 103–746) | This study, Invitrogen |
| pTB39 | Crbr; pBAD24 with PBAD::traJ/virD4Ap (produces TraJ/VirD4Ap) | This study |
| pTB50 | Crbr; pBAD24 with PBAD::traJ/virD4ΔCTDAp (produces TraJ/VirD4Ap with the CTD spanning residues 103–574 deleted) | This study |
| pNR3 | Crbr; pBSIISK+.NdeI with Plac::SDvirD4Ap (produces VirD4Ap's soluble domain spanning residues 103–746) | This study |
| W. pipientis VirD4 variants | ||
| pCR-virD4 | Crbr; cloned virD4Wp gene | K. Gentil |
| pTB12 | Crbr; pPC914KS+ with PvirB::virD4Wp | This study |
| pTB25 | Kanr; pBAD24Kanr with PBAD::virD4Wp | This study |
| pTB51 | Crbr; pBAD24 with PBAD::traJ/virD4Wp (produces TraJ/VirD4Wp consisting of TraJ's NTD fused to the soluble domain of VirD4Wp spanning residues 104–746) | This study |
| pTB52 | Crbr; pBAD24 with PBAD::traJ/virD4WpΔCTD (produces TraJ/VirD4Wp with the CTD spanning residues 103–574 deleted) | This study |
| pNR4 | Crbr; pBSIISK+.NdeI with Plac::SDvirD4Wp (produces VirD4Wp's soluble domain spanning residues 104–676) | This study |
| Constructs for protein/domain purifications | ||
| pPC2012 | pMAL-c2x with Ptac::mbp-SDvirD4At (coding sequence for VirD4At lacking the NTD) | This study |
| pPC2009 | pMAL-c2x with Ptac::mbp-NBD/CTDvirD4At (coding sequence for VirD4At lacking the NTD and AAD) | 19 |
| pNW12 | pMAL-c2x with Ptac::mbp-CTDvirD4At (coding sequence for VirD4At's CTD) | This study |
| pPC2043 | pMAL-c2x with Ptac::mbp-AADvirD4At (coding sequence for VirD4At's AAD) | This study |
| pPC2040 | pET28b(+) with pT7::His6-virE2CT100 (coding sequence for VirE2's C-terminal 100 residues) | This study |
| pPC2049 | pET28b(+) with pT7::His6-VirE1/FLAG-VirE2 | This study |
| pZZ3 | pET15b with pT7::His6-virE1 | 56 |
| Cre-effector fusions | ||
| pTB33 | Camr; pBAD33 with PBAD::cre | This study |
| pTB55 | Camr; pBAD33 with PBAD::cre-virE2 | This study |
| pTB53 | Camr; pBAD33 with PBAD::cre-virE3 | This study |
| pTB54 | Camr; pBAD33 with PBAD::cre-virF | This study |
| pTB70 | Camr; pBAD33 with PBAD::cre-virE2CT100 | This study |
| pPC56 | Camr; pBAD33 with PBAD::VirE1/Cre-VirE2 | This study |
| pPC57 | Camr; pBAD33 with PBAD::VirE1/Cre-VirE2ΔC50 | This study |
| pTB40 | Camr; pBAD33 with PBAD::cre-ats-1 | This study |
| pTB42 | Camr; pBAD33 with PBAD::cre-ats-1CT100 | This study |
| pTB43 | Camr; pBAD33 with PBAD::cre-WD0636 | This study |
| pTB44 | Camr; pBAD33 with PBAD::cre-WD0811 | This study |
| pTB45 | Camr; pBAD33 with PBAD::cre-WD0830 | This study |
TABLE 2.
Oligonucleotides used in this study
| Gene/protein/domain | Oligonucleotide | Sequencea | Plasmid |
|---|---|---|---|
| ΔtraJ constructed by recombineering | ΔTraJ_For | 5′-CTGGGAACCAAAAAAGGAGCGCTGACCATGGGTTGGGTAACGCCAGGGTTTTCC-3′ | pKM101ΔtraJ |
| ΔTraJ_Rev | 5′-TGGCGGGTAATCGTGGTTATATCAACCATGGCACACAGGAAACAGCTATGACCATGATTAC-3′ | ||
| TraJ | TraJ_For | 5′-CAGTAGCCATGGACGATAGAGAAAGA-3′ | pTB26 |
| TraJ_Rev | 5′-ACAATTGGTACCTCAGATCTCCCTCAG-3′ | ||
| TraJ′ (residues 1–75) | NW1_For | 5′-CATACCATGGACGATAGAGAAAGAGGCTTAGCATTTTTATTTG-3′ | pNW1 |
| NW1_Rev | 5′-GTATCTCGAGTATATATCATATGATAAATGATAAAAGCGATCAGACCGCCAAC-3′ | ||
| VirD4At | NW5_For | 5′-CGGTGAACATATGAATTCCAGCAA-3′ | pNW5 |
| NW5_Rev | 5′-AGTTCTCGAGCTACTTTTCAGGATCGTACGG-3′ | ||
| VirD4ΔCTDAt | VirD4AtΔ553_For | 5′-CGGTGAACATATGAATTCCAGCAA-3′ | pTB37 |
| VirD4AtΔ553_Rev | 5′-CTATTAGGTACCTCAGGGCTCAGGCAGAGA-3′ | ||
| VirD4Ap | VirD4Ap_For | 5′-AGTCGTCATATGCATAGTTCCAATCAT-3′ | pTB7 |
| VirD4Ap_Rev | 5′-TTAGTGCTCGAGCTACTTTAGTCTTCC-3′ | ||
| VirD4Wp | VirD4Wp_For | 5′-TAAGCGATCACCATGGGTCATAGC-3′ | pTB12 |
| VirD4Wp_Rev | 5′-GCTAGCTCGGGTACCTTACTTTCC-3′ | ||
| SDVirD4ΔCTDAt | NW4_For | 5′- CATATATATACATATGAATCAGAAGCATCACGGGACGG-3′ | pNW4 |
| NW4_Rev | 5′- GTTCTCGAGTCAAGGGTGCGGGCTCAGGCAG-3′ | ||
| CTDVirD4At | CTD_For | 5′-AAAAAAGAATTCCATCATCATCATCATCACAG CAGC-3′ | pNW12 |
| CTD_Rev | 5′-AAAAAAAAGCTTTCAAAAGCTGTTGACGCTTTG-3′ | ||
| TraJ/VirD4ΔCTDAp | TraJ/VirD4ApΔ574_For | 5′-TTCCGGGTCATAAGGTTCTTGG-3′ | pTB50 |
| TraJ/VirD4ApΔ574_Rev | 5′-TAAATTCGTGGTGGTGTTGAAGG-3′ | ||
| TraJ/VirD4Wp | NW1_For | 5′-CATACCATGGACGATAGAGAAAGAGGCTTAGCATTTTTATTTG-3′ | pTB51 |
| TB51_For1 | 5′-GATTATTCTTTCTCTATAAATGATAAAAGCGATCAGACCGCCAAC-3′ | ||
| TB51Rev1 | 5′-GCTTTTATCATTTATAGAGAAAGAATAATCGAGTGGCGGCC-3′ | ||
| TB51_Rev | 5′-GTATCTCGAGTTACTTTCCATTACTTTTTGGTTTATCACCATCATCTTCATC-3′ | ||
| TraJ/VirD4ΔCTDWp | NW1_For | 5′-CATACCATGGACGATAGAGAAAGAGGCTTAGCATTTTTATTTG-3′ | pTB52 |
| TB51_For1 | 5′-GATTATTCTTTCTCTATAAATGATAAAAGCGATCAGACCGCCAAC-3′ | ||
| TB51Rev1 | 5′-GCTTTTATCATTTATAGAGAAAGAATAATCGAGTGGCGGCC-3′ | ||
| TB52_Rev5′ | 5′-CTATCTCGAGTTATGGGTCATATGGCTCCTGTGTAGGTACATAAGTC-3′ | ||
| His6-VirE1/FLAG-VirE2 | VirE1_For | 5′-GGAGAGAACATATGGCCATCATC-3′ | pPC2049 |
| VirE1_RevFLAG | 5′-TTTGTCGTCGTCGTCTTTGTAGTCCATCGTCTCACTGGTTGTGAC-3′ | ||
| VirE2_ForFLAG | 5′-ATGGACTACAAAGACGACGACGACAAAATGGATCTTTCTGGCAAT-3′ | ||
| VirE2_Rev | 5′-TATCCTCGAGTCAAAAGCTGTTGA-3′ | ||
| VirE2CT100 | E2CT100_For | 5′ TAACTGCAGGCATATGTTGCGTGACATCCATGAC-3′ | pPC2040 |
| VirE2_Rev | 5′-TATCCTCGAGTCAAAAGCTGTTGA-3′ | pTB70 | |
| Cre | Cre_For | 5′-GATAGAGCTCAGGAGGTATTCACCATGTCCAATTTACTGACCGTACACCAAAATTTGC-3′ | pTB33 |
| Cre_Rev | 5′-CATGGTACCTATATATCATATGATCGCCATCTTCCAGCAGGC-3′ | ||
| VirE3 | VirE3_For | 5′-GATGCATATGGTGAGCACTACGAAG | pTB53 |
| VirE3_Rev | 5′-GATCGGTACCTTAGAAACCTCTGGAGG | ||
| VirF | VirF_For | 5′-GCACCATATGAGAAATTCGAGTTTGCG | pTB54 |
| VirF_Rev | 5′-GATATTGGTACCTCATAGACCGCGCGTTG | ||
| Ats-1 | Ats-1_For | 5′-GTGCTCCATATGCTAATAAGAAGAATTCTG-3′ | pTB40 |
| Ats-1_Rev | 5′-GTAATTGGTACCCTCGAGTTACCTCGTACCTTTACC-3′ | ||
| Ats-1 CT100 | AtsCT_For | 5′-GTACTTCATATGGAACGCATTTTCTCATTG-3′ | pTB42 |
| AtsCT_Rev | 5′-GTAATTGGTACCCTCGAGTTACCTCGTACCTTTACC-3′ | ||
| WD0636 | WD0636_For | 5′-GCCGAGCATATGAGTAAAAAAGAAAAAGAG-3′ | pTB43 |
| WD0636_Rev | 5′-CACGGTACCTCATAATTTCTCAAATAACTTTTC-3′ | ||
| WD0811 | WD0811_For | 5′-GTCCATATGATGATATCCAATAATTCT-3′ | pTB44 |
| WD0811_Rev | 5′-GATGGTACCTCAATTCATTTGTAA-3′ | ||
| WD0830 | WD0830_For | 5′-TCGTAGCATATGAAACAAGGAGATAAG-3′ | pTB45 |
| WD0830_Rev | 5′-GTAGGTACCTTACACTGTTCCTGGAGT |
Restriction sites for cloning are underlined.
(i) Plasmid vectors.
Plasmid pBAD24Kanr was made by isolation of the nptII gene conferring kanamycin resistance as a HincII fragment from pUC4K (Amersham) and its insertion into the single ScaI restriction site within pBAD24 (34). pBAD24Kanr-NdeI was constructed by inverse PCR to replace the NcoI site with an NdeI site using primers PBAD24Nde_For and PBAD24Nde_Rev. The conjugative plasmid pKM101Spcr was made by isolation of an Spcr gene as an EcoRI fragment from pHP45Ω and insertion into similarly digested pKM101. The resulting plasmid (Spcr Crbs) retains all of its conjugative abilities. pKM101SpcrΔtraJ with traJ precisely deleted was generated by recombineering using E. coli strain HME45(pKM101Spcr) according to the method of Thomason et al. (25). Briefly, pKM101Spcr was introduced by conjugation into HME45. A PCR product with an nptII gene flanked by NcoI restriction sites and 25 bp of DNA corresponding to sequences located upstream and downstream of pKM101 traJ was amplified with primers ΔTraJ_For and ΔTraJ_Rev using pUC4K as a template. HME45(pKM101Spcr) was temperature induced for expression of the red-gam genes and then transformed with the purified PCR product by electroporation. pKM101Spcr is a multicopy plasmid requiring enrichment of strains carrying only the ΔtraJ mutant plasmid by sequential subculturing of Kanr strains for 4 days in LB broth with Kan (200 μg/ml). Plasmid DNA was isolated from Kanr colonies, and the nptII gene was deleted by NcoI digestion and religation, followed by transformation into strain DH5α. Plasmids from Spcr Kans transformants were screened for the ΔtraJ mutation by PCR amplification and sequence analysis across the deletion junction with primers TraJ_For and ΔTraJ_Rev.
(ii) E. coli expression plasmids.
Plasmid pPC2012 produces the maltose binding protein (MBP) fused to a soluble domain of VirD4At lacking its N-terminal transmembrane domain (here designated MBP-SDVirD4). It was constructed by introduction of an XbaI/XhoI fragment from pKA28 (35) into XbaI/SalI-digested pMAL-c2x. Plasmid pPC2009 produces MBP fused to VirD4 lacking its NTD and AAD (MBP-NBD/CTDVirD4) (19). Plasmid pPC2043 produces MBP fused to VirD4At's AAD (MBP-AADVirD4; residues 201 to 346). The AAD gene fragment was obtained by digestion of pPC2013 (19) with BamHI/XhoI, and the product was introduced into BamHI/SalI-digested pMAL-c2x (New England BioLabs). Plasmid pNW12 produces MBP fused to VirD4's CTD (MBP-CTDVirD4; residues 552 to 656). It was constructed by PCR amplification of the CTD coding sequence with pKA28 as a template and CTD_For and CTD_Rev primers, digestion of the PCR product with EcoRI and HindIII, and introduction of the gene fragment into EcoRI/HindIII-digested pMAL-c2x. Plasmid pPC2049 produces His6-VirE1 and FLAG-VirE2. The VirE1/FLAG-VirE2 coding fragment was obtained by overlapping PCR by amplification of the upstream virE1 fragment using primers VirE1_For and VirE1_RevFLAG and the downstream virE2-FLAG fragment using primers VirE2_ForFLAG and VirE2_Rev, with pXZ237 (33) as a template. The PCR products were joined by overlapping PCR amplification with the outside primers VirE1_For and VirE2_Rev and digested with NdeI and XhoI, and the resulting fragment was introduced into similarly digested pET28b(+) (Novagen). Plasmid pPC2040 producing His6-VirE2CT100 (the C-terminal 100 residues of VirE2) was constructed by PCR amplification of the CT100 fragment using primers E2CT100_For and VirE2_Rev with pPC731 (33) as a template. The PCR product was digested with Nde/Xho, and the resulting fragment was introduced into similarly digested pET28b(+).
(iii) A. tumefaciens T4CP expression plasmids.
Plasmid pTB37 produces VirD4ΔCTDAt (VirD4At with its C-terminal 104 residues deleted) from the PvirB promoter. virD4AtΔCTD was PCR amplified with primers VirD4AtΔ553_For and VirD4AtΔ553_Rev with plasmid pKA9 (14) as a template. The PCR product was digested with NdeI and KpnI, and the resulting fragment was introduced into similarly digested pXZ27 (33), replacing the virE2 gene. Plasmid pTB7 produces the A. phagocytophilum VirD4 homolog (VirD4Ap) from the PvirB promoter. It was constructed by amplification of virD4Ap with the primers VirD4Ap_For and VirD4Ap_Rev with pBT-virD4 as a template (kindly provided by Y. Rikihisa), digestion of the PCR fragment with NdeI and XhoI, and introduction of the digested product into the similarly digested ColE1 plasmid pPC914KS+. Plasmid pTB12 produces the W. pipientis VirD4 homolog (VirD4Wp) from the PvirB promoter. It was constructed by PCR amplification of virD4Wp with primers VirD4Wp_For and VirD4Wp_Rev using pCR-VirD4 as a template (kindly provided by K. Gentil). The PCR product was digested with NcoI and KpnI, and the resulting fragment was introduced into similarly digested pPC914KS+. All the ColE1-based expression plasmids were ligated to the broad-host-range IncP plasmid pXZ151 or pBBR1MCS for introduction into A. tumefaciens (33).
(iv) E. coli T4CP expression plasmids.
Plasmid pTB26 produces TraJpKM101 from the PBAD promoter. It was constructed by PCR amplification of traJ with primers TraJ_For and TraJ_Rev using pKM101 as a template, digestion of the PCR fragment with NcoI and KpnI, and introduction of the resulting fragment into similarly digested pBAD24Kanr. Plasmid pNW5 produces VirD4At from the Plac promoter. It was generated by amplifying full-length virD4 using primers NW5_For and NW5_Rev with pKA9 as a template, digestion of the PCR fragment with NdeI/XhoI, and introduction of the resulting fragment into similarly digested pBSIIKS+.NdeI (36). Plasmid pTB48 producing A. phagocytophilum VirD4 (VirD4Ap) from the PBAD promoter was constructed by introduction of the virD4Ap gene as an NdeI/XhoI fragment from pTB7 into pBAD24Kanr-NdeI digested with NdeI and SalI. Plasmid pTB25 producing W. pipientis VirD4 (VirD4Wp) from the PBAD promoter was constructed by introducing the virD4Wp gene as an NcoI/KpnI fragment from pTB12 into similarly digested pBAD24Kanr-NdeI.
(v) E. coli T4CP chimera expression plasmids.
Plasmid pNW1 is pBSIIKS+NcoI (37) with the first 75 codons of TraJpKM101 encoding the NTD. It was constructed by amplification of the corresponding traJ fragment using primers NW1_For and NW1_Rev with pKM101 as a template, digestion with NcoI and XhoI, and introduction into similarly digested pBSIIKS+NcoI; an NdeI site was also located immediately 5′ of the XhoI site. Plasmid pNW2 produces the TraJ/VirD4At fusion protein composed of TraJ's NTD joined to a truncated, soluble form of VirD4At (lacking its NTD) from the Plac promoter. It was generated by inserting a fragment of virD4 encoding residues 88 to 656 of VirD4 as an NdeI/XhoI fragment from plasmid pKA38 (14) into similarly digested pNW1. Plasmid pNW4 produces TraJ/VirD4ΔCTDAt from the Plac promoter. It was generated by amplifying the virD4 sequence encoding residues 88 to 552 using primers NW4_For and NW4_Rev with pKM101 as a template, digestion with NdeI/XhoI, and introduction of the digested fragment into similarly digested pNW1. Plasmid pNR1 producing the soluble fragment of virD4 from the Plac promoter was generated by insertion of an NdeI/XhoI fragment from plasmid pKA38 into pBSIISK+.NdeI. Plasmid pNR2 produces TraJ/VirD4K152QAt from the Plac promoter generated by inverse PCR using pNW2 as a template. Plasmid pTB38 carries a codon-optimized traJ-virD4Ap chimeric gene within the vector plasmid pMK-RQ (Invitrogen). This chimeric gene codes for the TraJpKM101 NTD fused to the soluble fragment of VirD4Ap (residues 103 to 746). Plasmid pTB39 produces TraJ/VirD4Ap from the PBAD promoter. The chimeric gene was obtained as an NcoI/KpnI fragment from pTB38, and inserted into similarly digested pBAD24. Plasmid NR3 producing the soluble fragment of VirD4Ap from Plac was generated by cloning residues 103 to 746 as an NdeI/KpnI fragment into pBSIISK+.NdeI. Plasmid pTB50 produces TraJ/VirD4ΔCTDAp from the PBAD promoter. This plasmid was generated by inserting an amber mutation at codon 574 of pTB39 by inverse PCR with primers TraJ/VirD4ApΔ574_For and TraJ/VirD4ApΔ574_Rev. Plasmid pTB51 produces TraJ/VirD4Wp from the PBAD promoter. It was constructed by overlapping PCR with the gene fragments corresponding to the TraJ NTD (residues 1 to 75) and the soluble domain (residues 104 to 676) of VirD4Wp. PCR products were digested with NcoI and KpnI, and the digested product was ligated into similarly digested pBAD24. Plasmid NR4 producing the soluble domain of VirD4Wp from Plac was generated by insertion of the corresponding restriction fragment from pTB51 into pBSIISK+. Plasmid pTB52 produces TraJ/VirD4ΔCTDWp from the PBAD promoter. It was constructed by overlapping PCR with the gene fragments corresponding to the TraJ NTD (residues 1 to 75) and the soluble domain with the CTD (residues 104 to 574) of VirD4Wp deleted. The PCR products were joined by amplification with outside primers digested with NcoI and KpnI, and the resulting fragments were ligated into similarly digested pBAD24.
(vi) Effector expression plasmids.
Plasmid pTB33 producing Cre recombinase from the PBAD promoter was constructed by PCR amplification of the cre fragment using pCreTraIF (8) as a template and the primers Cre_For and Cre_Rev. The PCR products incorporated a SacI site followed by the Shine-Dalgarno sequence (AGG AGG) at the 5′ end and NdeI and KpnI sites at the 3′ end. The two restriction sites at the 3′ end allow the creation of Cre-effector fusion proteins. Plasmid pTB55 producing Cre-VirE2 from PBAD was constructed by introducing the NdeI/XhoI fragment carrying virE2 from pXZ27 (33) into similarly digested pTB33. Plasmid pPC56 producing VirE1 and Cre-VirE2 from PBAD was constructed by separate PCR amplification of an upstream Shine-Dalgarno–virE1 fragment and cre-virE2, followed by overlapping PCR to join the two fragments. The PCR product was digested with SacI and XhoI for introduction into similarly digested pBAD33. Plasmid pPC57 producing VirE1/Cre-VirE2ΔC50 was constructed by introduction of an amber codon at codon 484 by inverse PCR using pPC56 as a template. Plasmid pTB40 producing Cre–Ats-1 from PBAD was constructed by PCR amplification of the ats-1 gene using primers Ats-1_For and Ats-1_Rev with A. phagocytophilum genomic DNA as the template. The PCR fragment was digested with NdeI/KpnI, and the resulting product was introduced into similarly digested pTB33. The following plasmids producing Cre-effector fusion proteins were constructed using the same strategy with primers listed in Table 2: pTB43 (Cre-WD0636) using pENTR-D/TOPO-WD0636 as the template, pTB44 (Cre-WD0811) using pENTR-D/TOPO-WD0811 as the template, pTB45 (Cre-WD0830) using pENTR-D/TOPO-WD0830 as the template, pTB53 (Cre-VirE3) using pTiA6NC as the template, pTB54 (Cre-VirF) using pTiA6NC as the template, pTB42 (Cre–Ats-1CT100 (the C-terminal 100 residues of Ats-1) with pTB40 as the template, and pTB70 (Cre-VirE2CT100) using pXZ27 (33) as the template. All the plasmid constructs were verified by PCR amplification, restriction digestion analysis, and sequencing.
Protein enrichment.
Cells engineered to produce His6-tagged proteins were lysed by sonication in lysis buffer 1 (0.1% Triton X-100, 10 mM imidazole, 100 mM NaCl, 100 mM Tris, pH 7.5). The cell lysates were centrifuged at 22,700 × g for 15 min, and the resulting supernatant was applied to a Talon affinity resin (Clontech) column. The column was then washed with 10 column volumes of wash buffer 1 (10 mM imidazole, 1 M NaCl, 10 mM phenylmethylsulfonyl fluoride [PMSF[, 20 μg ml−1 leupeptin, 100 μg ml−1 pepstatin, 100 mM Tris, pH 7.5). Bound proteins were eluted with elution buffer 1 (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 250 mM imidazole, pH 7.4). Cells engineered to produce MBP-tagged proteins were lysed by sonication in lysis buffer 2 (10 mM EDTA, 1 mM KCl, 10 mM PMSF, 20 μg ml−1 leupeptin, 100 μg ml−1 pepstatin, 100 mM Tris, pH 7.5). The cell lysates were centrifuged at 22,700 × g for 15 min, and the resulting supernatant was applied to an amylose resin (New England BioLabs) column. The column was washed with 10 column volumes of wash buffer 2 (1 mM KCl, 20 mM NaCl, 1 mM EDTA, 20 mM Tris, pH 7.4), and bound proteins were eluted with elution buffer 2 (10 mM maltose, 1 mM KCl, 20 mM NaCl, 1 mM EDTA, and 20 mM Tris, pH 7.4). For isothermal calorimetry (ITC) (see below), enriched protein domains were further purified by gel filtration chromatography with a Superose 6 10/300 GL column and an AKTA/FPLC (fast protein liquid chromatography) system (GE Healthcare).
In vitro binding assays.
E. coli total extracts of cells engineered to produce MBP alone or fused to VirD4 domains of interest were prepared by growth of 10-ml cultures as described above. Cells were harvested by centrifugation at 22,700 × g for 10 min, and the cell pellets were resuspended in 200 μl of lysis buffer 2 and lysed by sonication. Insoluble material was removed by centrifugation at 22,700 × g for 15 min. Purified His6-VirE1/FLAG-VirE2 complex (100 μM) or the His6-VirE2CT100 fragment (150 μM) was added, along with amylose resin (100 μl; New England BioLabs) to the soluble cell extracts, and the resulting mixture was incubated at 4°C for 10 min with gentle shaking. The mixture was then applied to an amylose column, and the resin was washed with 10 column volumes of wash buffer 2 by gravity flow. Bound proteins were eluted with 100 μl of elution buffer 2 and resolved by SDS-PAGE.
Protein detection.
Proteins recovered in the pulldown assays were detected by SDS-PAGE and immunostaining of proteins transferred to polyvinylidene fluoride (PVDF) membranes with primary antibodies against the His6 (Sigma-Aldrich), MBP (New England BioLabs), or FLAG (Sigma-Aldrich) epitope tag, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies and visualization by chemiluminescence (38). To assay for the accumulation of Cre-effector fusion proteins, cells were induced with arabinose (0.2%), harvested at mid-log phase, and normalized to equivalent optical densities at 600 nm (OD600). Total protein extracts were subjected to SDS-PAGE, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes, and the blots were developed with anti-Cre antibodies (Novus Biologicals) and HRP-conjugated secondary antibodies.
ITC binding studies.
Binding of purified VirE2CT100 to MBP-AADVirD4At or MBP-NBD/CTDVirD4At was observed using a VP-ITC calorimeter (MicroCal, LLC, Northampton, MA, USA) at 25°C in 1× phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4). Bound ligands and coupling protein domains were extensively dialyzed in the same buffer, centrifuged to remove precipitates, and thoroughly degassed before use. Samples (5 μl) were injected at 2-min intervals to allow the peaks to reach baseline levels.
Conjugation assays.
A. tumefaciens and E. coli donor strains were assayed by DNA transfer through the VirB/VirD4 and pKM101-encoded transfer (Tra) T4SSs. In A. tumefaciens, wild-type (WT) A348 or the Mx355 (virD4) mutant carrying plasmids of interest served as DNA donors and A348-Spcr as the bacterial recipient. The mobilizable MOBQ plasmid pML122 (Genr) served as a DNA substrate for interbacterial matings carried out as described previously (28). For E. coli matings, strains DH5α and WM1650 carrying plasmids of interest served as the donor and recipient, respectively. The strains were grown overnight with antibiotic selection at 37°C, diluted 1:10 in fresh LB medium with the addition of 0.2% arabinose or 200 μM IPTG (isopropyl-β-d-thiogalactopyranoside) as appropriate for induction of gene expression, and incubated for 1 h at 37°C. Donors and recipients (100 μl each), alone or mixed in a 1:1 ratio, were pelleted by centrifugation, and the cells were suspended in 10 μl of the supernatant and spotted onto sterile filters on LB induction plates. The filters were incubated for 2 h or overnight at 37°C; the cells were resuspended in 1× PBS; and serial dilutions were plated on LB plates with antibiotics selective for donors, recipients, or transconjugants. The frequency of transfer was calculated as the number of transconjugants (Tcs) per donor (D). Experiments were performed at least three times in duplicate or triplicate, and the results are reported as the mean frequency of transfer.
Virulence assays.
A. tumefaciens strains were assayed for T-DNA and effector protein transfer using a tumor formation assay with Kalanchoe daigremontiana as previously described (36). The wild-type strain A348 and avirulent strains, e.g., KA2000 (virD4), served as positive and negative controls, respectively. Briefly, bacterial strains were freshly streaked onto LB agar plates supplemented with mannitol and glutamate (MG/L agar plates) with the appropriate antibiotics and grown at room temperature for 2 days (36). Plant leaves were wounded by scratching with a sterile toothpick and immediately inoculated with cells from the 2-day plates. Mixed infections were performed by coinoculation of two strains of interest on the same wound (29). All strains were inoculated onto at least 5 different leaves, and virulence was assessed after 6 to 8 weeks. Tumor production was scored as follows: +++, WT virulence; ++, tumors reproducibly smaller than those formed by WT strain A348; +, detectable tumor production but small and delayed in appearance by 2 or more weeks relative to WT A348; −, avirulent.
CRAfT.
The Cre reporter was used to assay for translocation of known or putative effector proteins from E. coli MS411 donor cells into the E. coli recipient strain CHS26Cm::LTL, which contains a loxP-Tetr-loxP cassette interrupting a Chlr gene on the bacterial chromosome (26). Cre-mediated excision of the loxP cassette restores the integrity of the Chlr gene cells, conferring a Chlr Tets phenotype. The recipient strain was also mutated to rifampin resistant (Rifr) by growth on rifampin-containing (50 μg/ml) plates and carried a pUC4K plasmid (conferring Crbr and Kanr) for additional antibiotic selection of loxP excisants. For Cre recombinase reporter assay for translocation (CRAfT), strains were grown overnight in LB broth with antibiotic selection at 30°C. Cells were diluted 1:10 in fresh LB broth containing 0.2% arabinose or 200 μM IPTG as appropriate for plasmid induction and grown at 30°C for 1 h. The cell suspensions were normalized by OD600. Donor and recipient cells (1 ml each) alone were mixed in a 1:1 ratio and centrifuged for 1 min. Cell pellets were resuspended in 20 μl of the supernatant and spotted onto sterile nitrocellulose filters on LB induction plates. The plates were incubated overnight at 30°C; the cells were resuspended in 1× PBS and serially diluted; and donors, recipients, and loxP excisants were selected on LB agar plates containing the appropriate antibiotics. The frequency of Cre recombination was calculated as the number of recombinants (Rcs) per donor (D). All CRAfT experiments were performed at least three times in duplicate, and the results are reported as the mean frequency of recombination with standard deviations.
RESULTS
A chimeric TraJ/VirD4At receptor mediates transfer of a MOBQ plasmid through the pKM101-encoded T4SS channel.
The T4CP nomenclature arose from early evidence that these subunits physically couple DNA substrates with conjugation channels (13, 39, 40). In these early studies, in some cases, a T4CP from one conjugation system was shown to functionally substitute for that of a second system in mobilizing the transfer of a non-self-transmissible MOBQ plasmid, RSF1010 (13). These findings prompted us to test whether T4CPs associated with effector translocator systems could functionally replace a T4CP associated with a conjugation system to support protein trafficking through the conjugation channel. Our model conjugation system was that encoded by plasmid pKM101, which to date has only been shown to transfer pKM101 in E. coli and related enterobacterial species. The pKM101-encoded TraJ T4CP was swapped with A. tumefaciens VirD4 (here called VirD4At), a versatile receptor that recruits and mediates transfer of oncogenic T-DNA, plasmid pML122 (an RSF1010 derivative), and effector proteins through the VirB channel (15, 30, 41). We also replaced TraJpKM101 with VirD4 homologs from the obligate intracellular species A. phagocytophilum and W. pipientis (Fig. 1A). These rickettsial species are phylogenetically closely related to A. tumefaciens and carry VirB/VirD4-like T4SSs that are thought to function as effector translocators during establishment of pathogenic or symbiotic relationships with eukaryotic hosts (10, 42, 43).
We showed that a pKM101ΔtraJ plasmid was transfer minus (Tra−) and that trans expression of traJ from a multicopy plasmid efficiently complemented the ΔtraJ mutation in mediating transfer of pKM101 and pML122 (Table 3). In fact, the traJ-complemented donor delivered these plasmid substrates at elevated frequencies compared with a donor expressing traJ from native pKM101, in agreement with evidence that the T4CP is rate limiting for transfer of cognate DNA substrates (18, 26). Complementation of the ΔtraJ mutation with virD4At, virD4Ap, or virD4Wp, however, did not restore transfer of pKM101 or pML122 (Table 3). Similar negative results were obtained when we attempted to complement an A. tumefaciens virD4At null mutation by trans-expression of virD4Ap or virD4Wp, as monitored by pML122 transfer (data not shown).
TABLE 3.
Complementation of the pKM101ΔtraJ mutation with genes encoding native and chimeric T4CPs
| Donor transmissible plasmid(s) | Donor complementing plasmid | Transfer frequency for: |
|
|---|---|---|---|
| pKM101 | pML122 | ||
| pKM101 | 2.2 × 10−1 | ||
| pKM101, pML122 | 2.5 × 10−1 | 7.6 × 10−4 | |
| pKM101ΔtraJ | pTB26 (PBAD::traJ) | 2.5 × 101 | 9.9 × 10−3 |
| pKM101ΔtraJ, pML122 | pTB26 (PBAD::traJ) | 2.3 × 102 | 4.3 × 10−3 |
| pKM101ΔtraJ, pML122 | pTB47 (PBAD::virD4At) | ||
| pKM101ΔtraJ, pML122 | pTB48 (PBAD::virD4Ap) | ||
| pKM101ΔtraJ, pML122 | pTB25 (PBAD::virD4Wp) | ||
| pKM101ΔtraJ, pML122 | pNW2 (Plac::traJ/virD4At) | 3.1 × 10−3 | |
| pKM101ΔtraJ, pML122 | pNW (Plac::traJ/virD4K152QAt) | ||
| pKM101ΔtraJ, pML122 | pNW13 (Plac::SDvirD4At) | ||
| pKM101ΔtraJ, pML122 | pTB39 (PBAD::traJ/virD4Ap) | ||
| pKM101ΔtraJ, pML122 | pTB51 (PBAD::traJ/virD4Wp) | ||
NTDs are thought to mediate the coupling of T4CPs with cognate T4SS channels (14, 44–47). We therefore engineered T4CPs consisting of the NTD from TraJpKM101 joined to ΔNTD variants of VirD4At, VirD4Ap, and VirD4Ap (Fig. 1B). The fusion junctions for each chimeric T4CP were located after the second transmembrane helix of the NTD and prior to the conserved NBD motifs (Fig. 1B), with junction sites selected on the basis of primary sequence alignments, hydrophobicity profiles, and structural modeling (data not shown; see Fig. S1 and S2 in the supplemental material). Strikingly, TraJ/VirD4At supported pML122 transfer through the pKM101-encoded T4SS at a frequency comparable to that of native TraJ (Table 3). TraJ's NTD or the soluble fragment of VirD4At (SDVirD4At) alone did not support plasmid transfer (data not shown), nor did a TraJ/VirD4At chimera bearing a K152Q mutation in the Walker A motif shown previously to abolish VirD4At function in A. tumefaciens (Table 3) (30). The TraJ/VirD4Ap and TraJ/VirD4Wp chimeras did not support pML122 transfer in E. coli (Table 3). We show below that these chimeras do support protein transfer, suggesting that the VirD4Ap and VirD4Wp receptor domains might fail to productively engage the pML122 substrate. In this context, although some rickettsial species carry possible mobile DNA elements (48, 49), the sequenced genomes of A. phagocytophilum and W. pipientis lack such elements or genes encoding processing factors, e.g., relaxases, required for DNA transfer. The T4CPs from these species therefore might have completely lost the capacity to recognize mobile DNA elements during evolution.
The TraJ/VirD4At chimera mediates transfer of A. tumefaciens effectors through the pKM101 channel.
To determine if the chimeric T4CPs support transfer of effector proteins through the pKM101 T4SS, we fused known or candidate effectors at their N termini to Cre recombinase, whose intercellular translocation is monitored by CRAfT (50). We used a version of the assay in which donors are mated with strain CHS26Cm::LTL and assayed for Cre-mediated excision of a loxP-Tetr-loxP cassette from the recipient cell chromosome (26). Excision of the loxP cassette restores the integrity of a Chlr gene, resulting in a Chlr Tets phenotype. In the initial experiments, the loxP cassette did not excise at detectable frequencies, as shown previously (22). In contrast, transformation of the recipient strain with plasmids encoding Cre or the Cre-effector fusion proteins under investigation resulted in loxP excision, as evidenced by the appearance of Chlr Tets colonies and by restoration of Chlr gene integrity, as monitored by PCR amplification and sequence analyses (data not shown).
Strikingly, the TraJ/VirD4At chimera supported translocation of Cre when fused to two A. tumefaciens effector proteins, VirE3 (51) and VirF (50). Donors delivered these Cre-effector fusion proteins at frequencies approaching or exceeding 10−6 Rcs/D, as determined by CRAfT (Fig. 2A). These transfer frequencies were comparable to frequencies reported for the transfer of Cre-effector fusion proteins through the Legionella pneumophila Dot/Icm T4SS (52). In control experiments, we confirmed that donors lacking pKM101ΔtraJ or the TraJ/VirD4At-producing plasmid failed to translocate the Cre-effector fusions and also that donors did not conjugatively transfer plasmids encoding the Cre-effector fusion proteins at detectable frequencies, in agreement with a lack of discernible oriT sequences on these plasmids. In many repetitions of the mating assays using donors carrying either native pKM101 or pKM101ΔtraJ and the TraJ/VirD4At-producing plasmid, we occasionally detected the appearance of a few Chlr Tets colonies (Fig. 2A). Since such colonies did not arise in the above-mentioned control experiments, Cre apparently is capable of delivery through the pKM101 channel at a low frequency (≤10−8 Rcs/D). Similar results have been reported for the F-like and R751 conjugation systems (26, 53), suggesting that Cre promiscuously transfers at low but detectable levels through different T4SSs. Donors carrying native pKM101 also were capable of transferring Cre-VirE3 and Cre-VirF, although at frequencies that were not statistically different than those observed for Cre-only transfer. In contrast, donors harboring the chimeric TraJ/VirD4At translocated Cre-VirE3 and Cre-VirF at levels significantly higher than those for Cre only (P < 0.01; Student's t test), in line with previous findings that A. tumefaciens effectors carry translocation signals specifying interactions with the VirD4 T4CP (19, 50). As expected, the TraJ/VirD4K152QAt variant did not support transfer of the A. tumefaciens effectors through the E. coli pKM101 T4SS (Fig. 2A).
FIG 2.
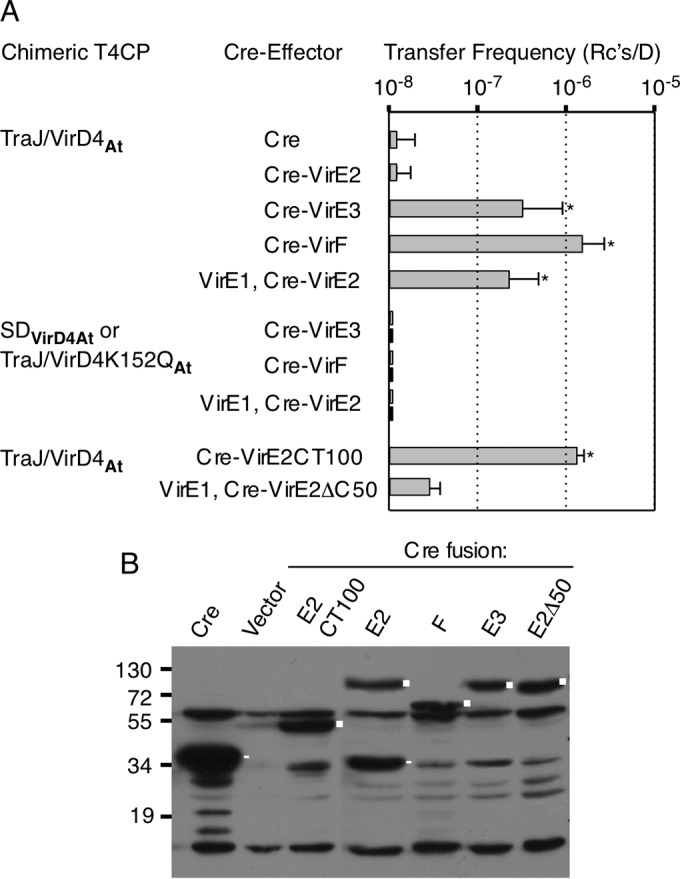
Transfer of A. tumefaciens effectors through the E. coli pKM101 conjugation channel. (A) E. coli donors produced the TraJ/VirD4At chimera. As controls, donors produced either the VirD4At soluble domain (SDVirD4At) (upper bars) or the TraJ/VirD4K152QAt variant bearing a nucleotide-binding site mutation (lower bars). Transfer of Cre alone; Cre fused to the VirE2, VirE3, or VirF effector; or Cre fused to VirE2's C-terminal 100 residues (CT100) or VirE2 with its last 50 residues deleted (VirE2ΔC50) was monitored by CRAfT (50). The values represent the means of at least three experiments with standard deviations. *, P < 0.01 versus Cre-only transfer; statistical analyses using Student's t test. (B) Steady-state levels of Cre or Cre-effector fusion proteins in E. coli donors, detected by immunostaining with anti-Cre antibodies. Lane Vector, donors carrying the vector plasmid only. Cre fusion proteins are marked by squares; Cre and prominent Cre breakdown products are marked by rectangles. Cre antibodies also cross-reacted with unknown proteins.
In these initial experiments, we failed to detect transfer of a Cre-VirE2 fusion protein (Fig. 2A), despite evidence that E. coli donors accumulated Cre-VirE2 at levels comparable to those of the Cre-VirE3 and Cre-VirF fusion proteins (Fig. 2B). VirE2 is a single-stranded DNA binding protein (SSB) that, when delivered to plants, associates with the T-DNA substrate and protects it from degradation (50, 54, 55). However, prior to export through the A. tumefaciens VirB/VirD4 channel, VirE2 must interact with its secretion chaperone, VirE1, to prevent self-aggregation and premature binding to DNA (56, 57). When E. coli donors were engineered to coproduce VirE1 and Cre-VirE2, the latter was translocated to recipients (Fig. 2A). Although the frequency of Cre-VirE2 transfer was slightly lower than those observed for the Cre-VirE3 and Cre-VirF fusion proteins, it was significantly higher than the frequency of Cre-only transfer (P < 0.01). Prior formation of the VirE1 chaperone/VirE2 effector complex in E. coli thus enabled docking with the TraJ/VirD4At chimera and VirE2 transfer through the pKM101 channel.
The C termini of the A. tumefaciens effectors VirE2, VirE3, and VirF are unstructured and have a high proportion of positively charged Arg residues (see Fig. S3 in the supplemental material) (58). A systematic mutational analysis established the importance of a putative translocation signal (TS) consisting of an Arg(7X)Arg(X)Arg(2X)Arg motif (where X represents any residue) at the C terminus of VirF for transfer through the A. tumefaciens VirB/VirD4 T4SS (58). While not as extensively characterized, VirE2's charged C terminus was shown to be necessary for a detectable VirE2-VirD4 interaction in A. tumefaciens (14) and its export to plants (33, 59, 60). Furthermore, VirE2 and VirE1 complex formation appears to maintain the accessibility VirE2's C terminus for potential binding to the VirD4 T4CP, as deduced from the X-ray crystal structure of the VirE1/VirE2 complex (57). To examine the requirement for VirE2's C-terminal domain for transfer through the pKM101 T4SS, we fused Cre to VirE2's C-terminal 100 residues (VirE2CT100) or VirE2 lacking its C terminus (VirE2ΔC50). Both Cre fusion proteins accumulated in donors at detectable levels (Fig. 2B). E. coli donors delivered Cre-VirE2CT100 at frequencies exceeding those of Cre-VirE2 and significantly higher than those of Cre-only transfer. In contrast, donors translocated Cre-VirE2ΔC50 at frequencies no different than those of Cre-only transfer, even upon coproduction of the VirE1 chaperone (Fig. 2A). VirE2's C-terminal domain bearing a putative TS thus appears to be necessary and sufficient for transfer via the TraJ/VirD4At chimera through the E. coli pKM101 T4SS.
Finally, A. tumefaciens also delivers oncogenic T-DNA through the VirB/VirD4 T4SS to plant cells. To assay for T-DNA transfer through the pKM101 T4SS, we engineered donors to carry the T-DNA border-containing plasmid pCIT20 or pBIN19 (61, 62) and a plasmid encoding the VirD1 and VirD2 processing factors, which are known to catalyze T-DNA border cleavage in both A. tumefaciens and E. coli (63). We further engineered donors to additionally produce VirC1 and VirC2, which were shown to enhance the T-DNA border cleavage efficiency and participate in the spatial coupling of the T-DNA substrate with VirD4 in A. tumefaciens (35). Donors producing these factors, however, failed to transfer the T-DNA vectors to recipient cells at detectable levels (data not shown). We suspect additional factors produced in A. tumefaciens, e.g., VirD2-binding proteins (64) or specific physiological conditions associated with the infection process are necessary for T-DNA transfer in E. coli, and we are currently examining these possibilities.
Chimeric T4CPs mediate transfer of known or candidate effectors from rickettsial species in E. coli.
We expanded our studies by testing the functionality of chimeric T4CPs composed of TraJ's NTD joined to the soluble domains of VirD4Ap and VirD4Wp from A. phagocytophilum and W. pipientis, respectively. Rickettsial species carry T4SS gene clusters, and a growing body of evidence indicates that type IV secretion is a general requirement for intracellular survival and proliferation (10, 43, 65, 66). Identification of T4SS effectors, however, has been hampered by the inability to grow rickettsial species axenically and by lack of robust genetic systems. The list of candidate effectors remains small, and of these, only a few are confirmed T4SS substrates by virtue of demonstrations of translocation through surrogate T4SSs, such as the A. tumefaciens VirB/VirD4 and L. pneumophila Dot/Icm systems (67, 68).
One of the best-characterized effectors of A. phagocytophilum, Ats-1, was identified in a two-hybrid screen using VirD4Ap as bait (69). Ats-1 abundantly accumulates in the mammalian cell cytosol and mitochondria during infection and is implicated in inhibition of apoptosis, among other activities (10, 69). We engineered E. coli donors to produce the TraJ/VirD4Ap chimera and assayed for Cre–Ats-1 transfer through the pKM101 channel (Fig. 3). Donors accumulated Cre–Ats-1 at detectable levels and also delivered the substrate through the pKM101 channel at frequencies nearly an order of magnitude higher than those observed for the A. tumefaciens effectors (Fig. 3). As shown for the A. tumefaciens effectors, E. coli donors producing only TraJ's NTD or only the soluble domain of VirD4Ap (SDVirD4Ap) failed to translocate Cre–Ats-1 (Fig. 3A and data not shown).
FIG 3.
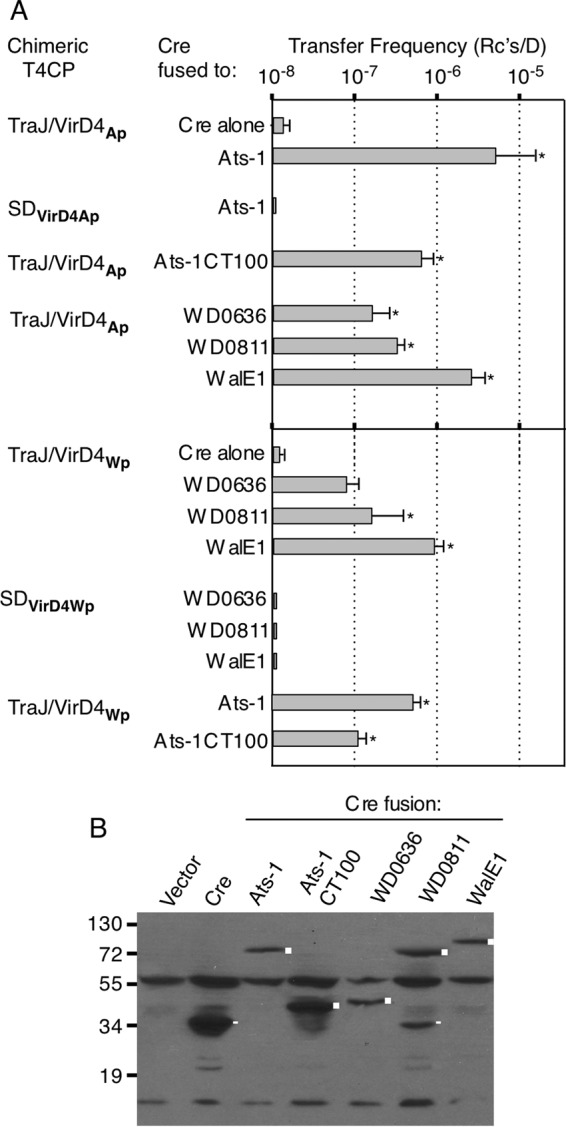
Transfer of known or candidate effectors from A. phagocytophilum and W. pipientis through the E. coli pKM101 conjugation channel. (A) E. coli donors produced the TraJ/VirD4Ap or TraJ/VirD4Wp chimera or the respective soluble domains of the VirD4 T4CPs. Transfer of Cre alone, Cre fused to the effector proteins listed, or Cre fused to the C-terminal 100 residues of Ats-1 (CT100) was monitored by CRAfT. The values represent the means of at least three experiments with standard deviations. *, P < 0.01 versus Cre transfer. (B) Steady-state accumulation of Cre or Cre-effector fusion proteins in E. coli donors, detected by immunostaining with anti-Cre antibodies. Lane Vector, donors carrying the vector plasmid. Cre fusion proteins are marked by squares; Cre or prominent Cre breakdown products are marked by rectangles. Cre antibodies also cross-reacted with unknown proteins.
Ats-1 has an unstructured, positively charged C terminus reminiscent of the A. tumefaciens effectors (see Fig. S3 in the supplemental material) (10). A TraJ/VirD4Ap-producing donor strain was engineered to produce Cre fused to the C-terminal 100 residues of Ats-1 (Cre–Ats-1CT100) (Fig. 3B). Interestingly, this donor translocated Cre–Ats-1CT100 less efficiently than a donor producing Cre–Ats-1, although still at levels significantly higher than Cre-only transfer (Fig. 3A). These finding suggest that Ats-1's C terminus suffices to mediate transfer of the Cre reporter but that additional internal motifs might contribute to optimal transfer. Synergistic contributions of C-terminal TSs and internal motifs have been reported for other T4SS effectors (70–72).
Presently, no T4SS effectors have been confirmed for any Wolbachia species, although these species encode many potential effectors bearing eukaryotic protein motifs (73). Here, we tested for translocation of three such candidates: (i) WD0636 (Ank domain), (ii) WD0811 (WASP-homology 2 actin-binding module [WH2]), and (iii) WD0830 (α-synuclein actin-binding domain). As discussed further below, WD0830 was recently shown to be secreted by Wolbachia and has been renamed WalE1 (for Wolbachia actin localizing effector) (see Discussion).
E. coli donors were engineered to produce TraJ/VirD4Wp and Cre-WD0636, Cre-WD0811, or Cre-WalE1. The respective donor strains accumulated detectable levels of the Cre-effector proteins, indicating that the fusion proteins were stably produced (Fig. 3B). Intriguingly, donors translocated each of the three Cre-effector fusion proteins through the pKM101 channel. Transfer frequencies were lower than those observed for the A. tumefaciens and A. phagocytophilum effectors but for WD0811 and WalE1 were significantly higher than Cre-only transfer frequency (compare Fig. 2A and 3A). Donors lacking the TraJ/VirD4Wp chimera or producing TraJ's NTD or the VirD4Wp soluble domain (SDVirD4Wp) did not translocate the Cre fusion proteins at detectable levels (Fig. 3A and data not shown).
A. phagocytophilum VirD4Ap and W. pipientis VirDWp exhibit extensive sequence similarities, especially throughout their soluble domains (see Fig. S2 in the supplemental material). We therefore asked whether the two chimeric T4CPs, TraJ/VirD4Ap and TraJ/VirD4Wp, reciprocally supported translocation of each other's substrates. Interestingly, donors producing TraJ/VirD4Ap transferred the three W. pipientis candidate effectors at frequencies higher than those for the corresponding TraJ/VirDWp-producing donors and significantly higher than that for the Cre-only control (Fig. 3A). Conversely, TraJ/VirD4Wp-producing donors translocated Ats-1 and Ats-1CT100, but at frequencies slightly lower than those for the TraJ/VirD4Ap-producing donors. We suspect that reduced substrate transfer by the TraJ/VirD4Wp-producing donors might be attributable to less efficient expression of this chimeric T4CP than of TraJ/VirDAp, because during construction the latter but not the former had been codon optimized for expression in E. coli (see Materials and Methods). Regardless, the present findings support a conclusion that the TraJ/VirD4Ap and TraJ/VirD4Wp chimeras productively engage with and support translocation of the tested effectors from both rickettsial species at levels significantly higher than that for the Cre-only control through the pKM101 conjugation system.
The CTDs of VirD4 T4CPs contribute to substrate discrimination in vivo.
Many T4CPs carry C-terminal domains that are missing from the structurally characterized TrwBR388 or pKM101-encoded TraJ (Fig. 1A). The CTDs of the VirD4 homologs under study here range in length from 105 to 169 residues, are almost entirely α-helical in their predicted secondary structures, and, most intriguingly, are highly enriched in Glu and Asp residues (see Fig. S1 in the supplemental material). Such features were of interest in view of previous work showing that nucleic acid binding proteins, e.g., SSBs and helicases, often carry acidic tails important for coordinating nucleic acid-protein or protein-protein interactions (74–78).
To evaluate the functional importance of the alphaproteobacterial VirD4 CTDs, we first characterized the effects of a VirD4ΔCTDAt mutation on substrate transfer through the A. tumefaciens VirB/VirD4 T4SS. A virD4At null mutant strain is defective for substrate transfer and displays wild-type transfer functions when expressing virD4At in trans (Fig. 4A) (30). Interestingly, a virD4At mutant donor complemented with a virD4ΔCTD-expressing plasmid also transferred pML122 to A. tumefaciens recipients at wild-type levels, establishing that VirD4At's CTD is dispensable for MOBQ plasmid transfer (Fig. 4A).
FIG 4.
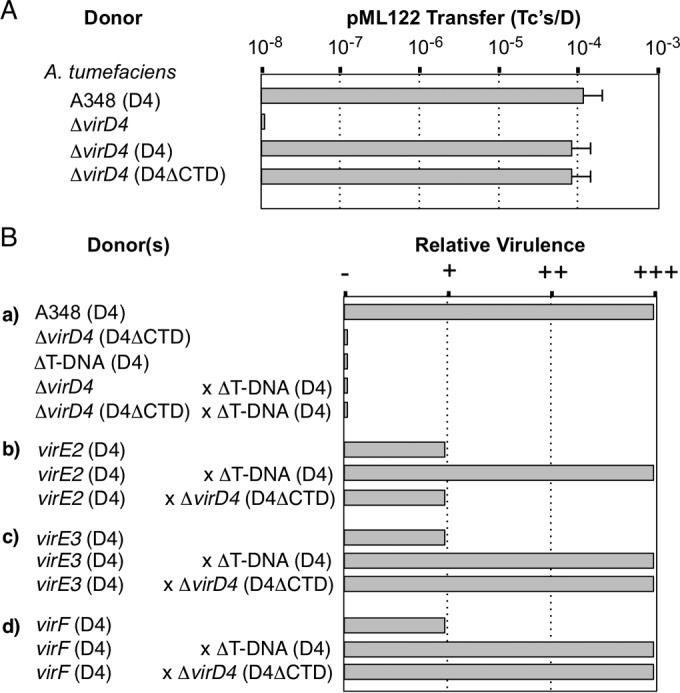
Contribution of VirD4At's CTD to function. (A) Transfer of the MOBQ plasmid pML122 by A. tumefaciens. A. tumefaciens strains: A348, wild-type strain producing native VirD4At (D4); ΔvirD4, strain KA2000 lacking or producing native VirD4At or VirD4ΔCTDAt (D4ΔCTD) from a broad-host-range plasmid. The histogram presents pML122 transfer frequencies determined for A. tumefaciens with 3-day solid-surface matings. (B) Transfer of T-DNA and effector proteins through the A. tumefaciens VirB channel as monitored by tumor production on plant leaves. (a) A348 is fully virulent (+++); ΔvirD4 strain KA2000 lacking or producing VirD4ΔCTDAt and the ΔT-DNA mutant strain LBA4404 are avirulent (−). Coinoculation of plant tissues with the ΔT-DNA mutant (which is capable of exporting effector proteins) and the ΔvirD4 mutant engineered to produce VirD4ΔCTDAt failed to incite virulence, showing that the latter strain does not export T-DNA. (b) The virE2 mutant (strain At12516) produces VirD4At and is highly attenuated for virulence (it occasionally produces small tumors in multiple repetitions of virulence assays [+]). Coinoculation of plant tissues with the virE2 mutant (which exports T-DNA) and the ΔT-DNA mutant (which exports VirE2) restores full virulence. Coinoculation of the virE2 mutant and the ΔvirD4 mutant engineered to produce VirD4ΔCTDAt did not restore virulence, indicating that the latter strain is incapable of exporting VirE2. (c and d) Coinoculation assays performed as for graph b showing that the VirD4ΔCTDAt-producing strain is capable of delivering VirE3 and VirF to plant cells. ++, tumors reproducibly smaller than those formed by WT strain A348.
The VirD4ΔCTDAt-producing strain, however, was avirulent in plant assays, suggestive of a block in transfer of oncogenic T-DNA, effector proteins, or both types of substrates through the VirB channel (Fig. 4B). To distinguish whether the ΔCTD mutation selectively blocks T-DNA or effector protein substrates, we employed a mixed-infection assay (29). In this assay, when an avirulent strain with T-DNA deleted or highly attenuated strains with an effector gene deleted are coinoculated on the same plant wound site, the mixture incites robust tumor development (Fig. 4B). This is thought to result from the transfer of effector proteins by the ΔT-DNA mutant and T-DNA by the effector mutant into the same plant cell, enabling formation of T-DNA–effector protein complexes necessary for efficient substrate translocation to the plant nucleus (29, 79).
In mixed infections, we coinoculated the VirD4AtΔCTD-producing strain with a ΔT-DNA mutant to evaluate the importance of the CTD for T-DNA transfer (Fig. 4B, a). This coinfection failed to incite tumor production, indicating a requirement for the CTD for T-DNA transfer through the A. tumefaciens VirB channel. We then coinoculated the VirD4ΔCTDAt-producing strain with a virE2, virE3, or virF mutant strain to assess the importance of the CTD for effector protein transfer. Consistent with previous findings (54), the effector mutant strains exhibited a highly attenuated virulence phenotype, as evidenced by the capacity to incite formation of small tumors after prolonged incubation periods of several months (Fig. 4B, b to d). For each mutant, mixed infection with the ΔT-DNA strain restored virulence to wild-type levels (Fig. 4B, b to d). For the virE2 mutant, however, a mixed infection with the VirD4ΔCTDAt-producing strain failed to restore virulence, suggesting that the CTD also strongly contributes to VirE2 transfer (Fig. 4B, b). In striking contrast, coinfection of the virE3 or virF mutant with the VirD4ΔCTDAt-producing strain resulted in robust tumor development, suggesting that VirD4At's CTD is dispensable for transfer of these effectors to plant cells (Fig. 4B, c and d).
We next characterized the effects of the CTD deletion on TraJ/VirD4At function in E. coli. Overall, the results paralleled our findings with the A. tumefaciens infection assays. The TraJ/VirD4ΔCTDAt-producing donor transferred pML122 to E. coli recipients at a frequency comparable to that of the TraJ/VirD4At-producing donor (Fig. 5A) and also showed a reduction in levels of transfer of Cre-VirE2 to levels that were not significantly different than that of Cre-only transfer (Fig. 5B). Intriguingly, however, the ΔCTD mutant donors reproducibly transferred Cre-VirE3 at elevated frequencies and Cre-VirF at significantly (P < 0.05) higher frequencies than the isogenic TraJ/VirD4At-producing donors (Fig. 5B). The ΔCTD mutant donor translocated Cre-VirE2CT100 at frequencies comparable to or higher than those observed for the TraJ/VirD4At-producing donor (Fig. 5B). These findings suggest that VirD4At's CTD not only is dispensable for transfer of VirE3, VirF, and VirE2CT100, but its presence antagonizes transfer of these effectors.
FIG 5.
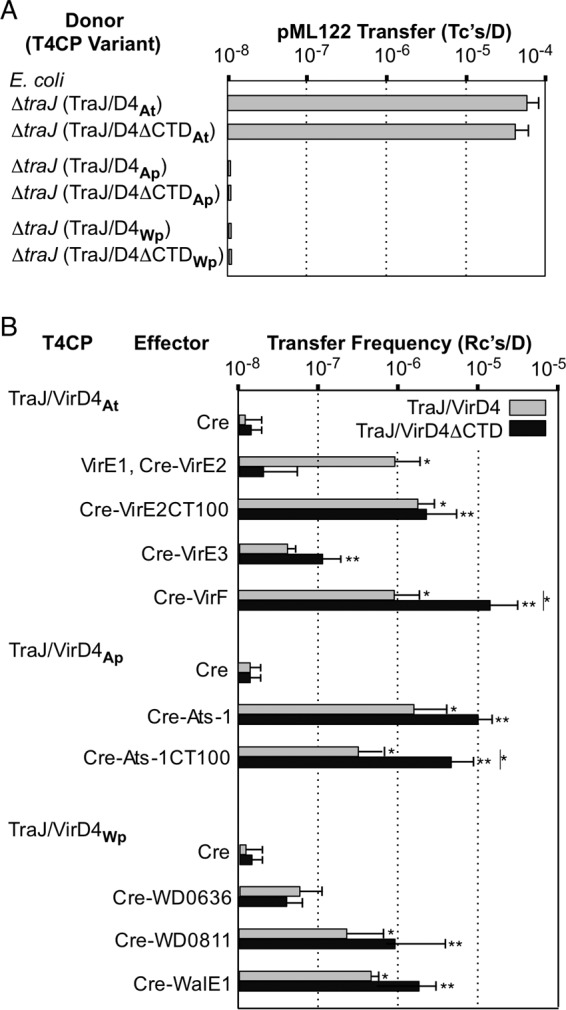
Effects of ΔCTD mutations on functions of VirD4 homologs in E. coli. (A) E. coli donors carrying pKM101ΔtraJ (ΔtraJ) and producing the TraJ/VirD4 or TraJ/VirD4ΔCTD chimeras listed were assayed for pML122 transfer. Transfer frequencies were measured in 2-h solid-surface matings. (B) The E. coli donors producing full-length TraJ/VirD4 chimeras or ΔCTD variants listed were assayed for transfer of Cre or Cre-effector fusion proteins, as monitored by CRAfT. The values represent the means of at least three experiments with standard deviations. *, P < 0.01 versus Cre-only transfer mediated by the TraJ/VirD4 chimeras; **, P < 0.01 versus Cre-only transfer via the TraJ/VirD4ΔCTD chimeras; |*, P < 0.01, transfer via a TraJ/VirD4ΔCTD chimera versus the isogenic TraJ/VirD4 chimera.
Donors producing the TraJ/VirD4ΔCTDAp and TraJ/VirD4ΔCTDWp variants were also assayed for the capacity to translocate rickettsial effectors through the pKM101 channel (Fig. 5B). Donors producing TraJ/VirD4ΔCTDAp also reproducibly transferred Cre–Ats-1 at elevated frequencies and Cre–Ats-1CT100 at significantly higher frequencies (P < 0.01) than the TraJ/VirD4Ap-producing donor. Similarly, donors producing TraJ/VirD4ΔCTDWp also transferred the W. pipientis candidate effectors WD0811 and WalE1 at slightly elevated frequencies, although the differences were not statistically significant. In contrast, the ΔCTDWp mutant delivered Cre-WD0636 at lower levels than the isogenic TraJ/VirD4Wp-producing donor (Fig. 5B).
Finally, we speculated that the failure of donors producing TraJ/VirDAp or TraJ/VirD4Wp to translocate the MOBQ plasmid pML122 might be due to the presence of the acidic CTDs acting as DNA mimics. However, donors producing the ΔCTD variants of TraJ/VirD4Ap or TraJ/VirD4Wp still failed to transfer pML122, arguing against the notion that the acidic CTDs occlude access of negatively charged DNA substrates to T4CP receptor domains (Fig. 5A).
VirD4's AAD and NBD, but not the CTD, bind the VirE2 effector in vitro.
In view of the demonstrated importance of VirD4At's CTD for VirE2 translocation, we assayed for binding of VirD4At's CTD with the VirE1/VirE2 complex or the VirE2CT100 fragment in vitro. Despite repeated attempts, in pulldown assays we were unable to detect stable interactions between purified forms of MBP-tagged CTD and either a His6-VirE1/FLAG-VirE2 complex or the His6-VirE2CT100 fragment (Fig. 6). These findings, coupled with the results of our recent studies exploring VirD4 receptor domain-DNA substrate interactions (19), raised the possibility that other VirD4 domains directly bind the VirE2 effector. To test this possibility, we purified MBP-tagged (i) VirD4At with its NTD deleted (SDD4), (ii) SDD4 with the AAD deleted (NBD/CTDD4), and (iii) AADD4 (Fig. 6A). Interestingly, all three VirD4 variants bound the His6-VirE1/FLAG-VirE2 complex (Fig. 6B, left). These domains did not detectably bind VirE1, nor did MBP alone interact with VirE1 or VirE2, confirming that the VirD4At domains were responsible for binding the VirE2 component of the chaperone/effector complex (Fig. 6B and data not shown). The MBP-tagged SDD4, NBD/CTDD4, and AADD4 variants also bound the His6-VirE2CT100 fragment bearing the putative C-terminal TS (Fig. 6B, right).
FIG 6.
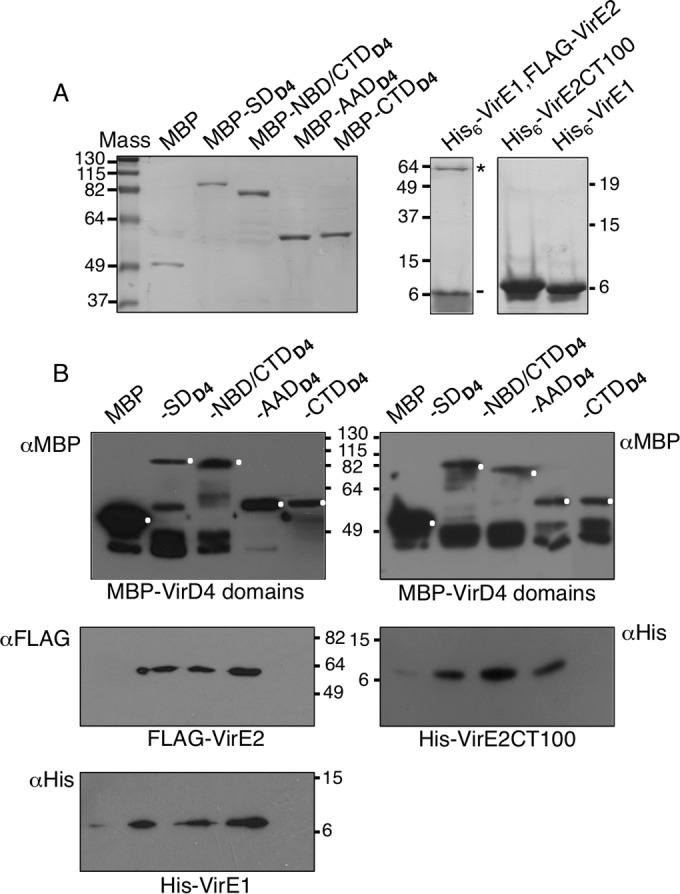
VirD4At domain interactions with the VirE1/VirE2 complex and the VirE2CT100 fragment bearing a putative translocation signal. (A) Purified forms of proteins/domains subjected to pulldown assays. (Left) MBP-tagged VirD4At domains: SDD4, soluble VirD4At with its N-terminal transmembrane domain deleted; NBD/CTDD4, SDD4 with the AAD deleted; AADD4, AADD4 only; CTDD4, VirD4At's C-terminal 104 residues. Mass, molecular mass markers (in kilodaltons); MBP, purified MBP tag. (Right) His6-VirE1/FLAG-VirE2 complex with positions of VirE2 (*) and VirE1 (-) indicated; His6-VirE2CT100, His-tagged C-terminal 100 residues of VirE2; His6-VirE1, His-tagged VirE1 chaperone. Proteins were identified by staining with Coomassie blue G-250 (Bio-Rad). (B) MBP-tagged VirD4 domains were bound to the amylose resin, and the protein-bead complexes were mixed with His6-VirE1/FLAG-VirE2 complex (left) or His-VirE2CT100 (right). Following extensive washes, the bound proteins were eluted from the beads with maltose and identified by immunostaining with anti-MBP, -FLAG, or -His antibodies, as indicated. Full-length proteins detected by anti-MBP immunostaining are marked with white dots; other immunoreactive species are putative breakdown products. The MBP-tagged domains did not detectably interact with purified His6-VirE1 (data not shown).
We attempted to purify MBP-NBD/CTDD4, MBP-AADD4, and His6-VirE2CT100 in sufficient amounts to evaluate the relative binding affinities of the VirD4 domains to the VirE2 substrate by ITC. While we were able to gain further evidence for binding of VirD4At's AAD and NBD/CTD to VirE2CT100, the binding affinities were not high enough to produce quantitative data (see Fig. S4 in the supplemental material). We suspect that this was due either to insufficient concentrations of soluble forms of the MBP-tagged domains, which tended to aggregate, or to weak affinity or transient binding. Regardless, the findings support the results from the pulldown assays suggesting that more than one of VirD4At's domains engage with VirE2. This could reflect a complex docking reaction in which multiple domains of the T4CP simultaneously engage with the substrate or a sequence of domain-substrate binding reactions corresponding to distinct substeps of the translocation pathway.
DISCUSSION
The T4CPs are a unique receptor superfamily associated with nearly all T4SSs (2, 16). T4CPs fulfill the complex tasks of serving as docking sites for cognate DNA or protein substrates, energizing substrate transfer, and productively engaging with translocation channels (17, 19, 22, 30, 47, 80, 81). In this study, we showed that chimeric T4CPs composed of the NTD of TraJpKM101 and soluble domains of VirD4 homologs from alphaproteobacterial species support transfer of heterologous protein substrates through the pKM101 conjugation system. We further advanced a mechanistic understanding of T4CP function by determining that acidic C-terminal tails, when present, positively or negatively modulate substrate transfer efficiencies. Finally, our studies established a proof of principle for the use of conjugation systems in genetically tractable E. coli for monitoring translocation of known or candidate effectors of T4SSs operating in recalcitrant species.
T4CP coupling activities.
To fulfill their coupling functions, T4CPs must interact with secretion substrates and cognate T4SS channels. The latter contacts were not a major focus of our study, but prior work has shown that T4CPs interact with VirB10-like subunits, as well as the VirB4- and VirB11-like ATPases (15, 18, 30, 44, 47, 82). This interaction network is required not only for early-stage reactions mediating substrate transfer across the inner membrane but also for transduction of intracellular signals across the T4SS channel, enabling substrate translocation to the cell exterior. Earlier studies showing that the transmembrane domains (NTDs) of T4CPs are necessary for the VirB10 interaction (18, 44, 47, 83, 84) formed a basis for our design of chimeric T4CP receptors. The fact that these chimeras were functional confirmed the necessity for the NTD and further suggested that this domain suffices to couple receptors to cognate translocation channels. The nature of NTD contacts with VirB10 or other components of the translocation channel, however, remains a particularly intriguing area for further investigation in view of stoichiometries reported for the T4SS subunits. For example, T4CPs assemble as hexamers (17), whereas subunits of a large complex of proteins at the inner membrane have copy numbers of 12 or 24 and subunits of an outer membrane subassembly (that includes VirB10) have copy numbers of 14 (85, 86).
In the A. tumefaciens VirB/VirD4 T4SS, the VirB4 and VirB11 ATPases coordinate with the VirD4 receptor to mediate translocation of DNA substrates across the cell envelope (15, 30, 47). The nature of these or other T4SS ATPase interactions is not fully defined but is predicted to involve contacts between their cytoplasmic domains (37, 82, 87, 88). Accordingly, the VirD4 moieties of the chimeric T4CPs must productively engage with the pKM101-encoded VirB4-like TraB and VirB11-like TraG ATPases for protein transfer through the pKM101 channel. It is interesting that these VirD4 homologs exhibit only weak sequence similarities with the TraJpKM101 T4CP (see Fig. S2 in the supplemental material), which raises the possibility that the ATPase subunit contacts are transient and possibly stimulated by activating signals, such as substrate docking, ATP binding, or hydrolysis. Such a proposal is consistent with a recent model, based on genetic and structural studies of the R388-encoded ATPases, positing that transient contacts between the TrwBR388 substrate receptor and the VirB11-like TrwD ATPase act as a switch to convert the R388 channel from a pilus assembly system to a translocation channel (82).
With respect to the coupling of T4CPs with cognate substrates, our findings add to earlier evidence that the AAD functions as a substrate specificity determinant and further suggest that the CTD, when present, also plays a role in modulating the efficiency of the docking reaction. Mutational analyses of the AADs of E. coli TraDF and TrwBR388 (18, 89) and, more recently, A. tumefaciens VirD4At and Enterococcus faecalis PcfCpCF10 (19) provided genetic evidence for their contributions to substrate discrimination. The AADs of VirD4At and PcfCpCF10 also bind the VirD2At and PcfGpCF10 relaxases (19) that are responsible for processing and then piloting DNA cargoes through the A. tumefaciens VirB/VirD4 and E. faecalis Prg/Pcf T4SSs, respectively (70, 90). VirD2 carries a charged C terminus, the deletion of which abolishes translocation of VirD2 and T-DNA substrates to plant cells (70). VirD2's C-terminal TS is also essential for docking of the T-DNA substrate with VirD4At in vivo (47) and for a detectable VirD2-AADVirD4 interaction in vitro (19). Together, these findings indicate that the AAD mediates binding of cognate DNA substrates at least partly through recognition of relaxase translocation signals (19). A recent study identified a second, internal motif that is also important for VirD2 transfer, and it will now be of interest to determine if this motif also contributes to the relaxase-VirD4 docking reaction (70). Other relaxases also carry internal TSs whose contributions to T4CP docking are presently undefined (7, 9, 53, 91).
VirD4At's AAD is also required for VirE2 translocation to plant cells (19). Here, we gained further evidence that the AAD is involved in binding of this effector through pulldown assays (Fig. 6B) and by ITC (see Fig. S4 in the supplemental material). However, we also showed that VirD4 lacking the AAD also bound these VirE2 substrates (Fig. 6B) and, acknowledging the limitations of the ITC data, we were unable to distinguish affinity differences in binding of the VirD4 AAD or ΔAAD variant with VirE2's C-terminal TS (see Fig. S4 in the supplemental material). These findings are consistent with a role for VirD4's AAD in recruitment of VirE2 but do not exclude the possible involvement of other VirD4 domains in effector docking or later-stage reactions.
We were particularly interested in evaluating the contribution of VirD4At's CTD to substrate docking or translocation. T4CPs associated with effector translocators often carry such domains, which vary considerably in length but typically consist of a high proportion of acidic residues (2). The latter feature is most strikingly illustrated with the A. phagocytophilum VirD4 CTD, nearly half of which is composed of Glu and Asp residues (see Fig. S1 in the supplemental material). Underscoring the potential importance of these acidic termini for substrate binding, DNA binding proteins often carry acidic C-terminal tails important for binding to nucleic acid or protein partners or for stimulating NTP hydrolysis activities of protein partners (74–78). In fact, in the one T4CP-substrate contact structurally solved to date, acidic C-terminal residues were shown to specify substrate binding. In the E. coli F plasmid transfer system, the TraDF receptor recruits the F plasmid in part through binding of the relaxosome subunit TraM. An X-ray structure of the TraD-TraM interaction revealed specific contacts between TraDF's C-terminal acidic residues and TraM (20, 92). Genetic studies confirmed that the TraDF-TraMF interaction promotes efficient F plasmid transfer and also that this interaction strongly inhibits transfer of the MOBQ plasmid RSF1010 through the F T4SS (20, 93). TraD's C terminus thus confers selective binding of the cognate F plasmid substrate while simultaneously blocking receptor access by a promiscuous MOBQ plasmid substrate.
VirD4At's CTD similarly was required for translocation of the cognate T-DNA substrate in A. tumefaciens (Fig. 4B). In contrast to the F transfer system, however, VirD4At's CTD is completely dispensable for and also does not interfere with transfer of a MOBQ substrate (Fig. 4), arguing against a competitive-interference model as invoked for the F plasmid system. Of further interest, VirD4At's CTD exerted opposing effects on translocation of different effector proteins, as evidenced by its requirement for VirE2 transfer and its antagonistic role in the transfer of VirE3 and VirF (Fig. 4B and 5B). The CTDs of the VirD4Ap and VirD4Wp homologs also generally negatively impacted translocation of the known or candidate rickettsial effectors (Fig. 5B). VirD4At's CTD might play a critical role in VirE2 transfer through direct binding of the effector; however, thus far, we have been unable to gain evidence for direct contacts with either the VirE1/VirE2 complex or the VirE2CT100 fragment (Fig. 6). It remains possible that the CTD weakly or transiently interacts with VirE2 or other effectors to regulate their access to other T4CP domains. Alternatively, the CTD might form intrasubunit contacts with other T4CP domains to expose or occlude access to effectors as a means of regulating docking reactions. Deciphering the mechanism of action of CTDs in modulating effector docking or translocation reactions remains an intriguing area for further investigation.
In sum, our studies have further defined contributions of the all-alpha and C-terminal domains of T4CPs to T4SS substrate-docking reactions. VirD4At's AAD is essential for translocation of all the tested substrates in vivo and binds DNA and protein substrates in vitro. In contrast, VirD4At's CTD exerts differential effects on substrate translocation but is completely dispensable for translocation of a promiscuous MOBQ plasmid. The CTDs of VirD4Ap and VirD4Wp also exerted differential effects on substrate transfer, supporting a general proposal that T4CPs acquired these domains over evolutionary time as a means of exerting spatiotemporal control over the translocation of different effector cargoes during infection processes.
Chimeric T4CPs for T4SS effector identification and other applications.
Thus far, candidate T4SS effectors from obligate intracellular rickettsial species have been identified through two-hybrid screens for VirD4-interacting proteins, bioinformatics screens for proteins with eukaryote-like motifs, and iTRAQ or immunofluorescence methods for detection of bacterial proteins within the eukaryotic host cell (67–69, 73, 94–96). Only a few such effectors, however, were confirmed as T4SS substrates by demonstrations of trafficking through surrogate A. tumefaciens VirB/VirD4 or L. pneumophila or Coxiella burnetii Dot/Icm T4SSs (67, 68, 96). Translocation of rickettsial effectors through the L. pneumophila and C. burnetii Dot/Icm systems is in fact surprising in view of the fact that DotL and rickettsial VirD4 T4CPs are only weakly phylogenetically related (see Fig. S2 in the supplemental material). The results of our studies suggest that E. coli conjugation systems configured with chimeric T4CPs offer distinct advantages over other surrogate systems in ease of use, genetic manipulation, and, most importantly, fidelity with respect to recruitment of cognate substrates. One possible limitation, common to all surrogate systems (97, 98) and evidenced here with A. tumefaciens VirE2 (Fig. 2A), is that effectors requiring a chaperone or adaptor for translocation are not efficiently delivered via chimeric T4CPs through E. coli conjugation channels.
We confirmed that A. phagocytophilum Ats-1 is a bona fide T4SS substrate and provided the first evidence that the charged C terminus of Ats-1 (10) suffices to mediate transfer through a T4SS. Both TraJ/VirD4Ap and TraJ/VirD4Wp supported Ats-1 transfer, which at the outset established the utility of both chimeras for monitoring transfer of candidate effectors from W. pipientis. We also demonstrated transmissibility of three candidate effectors bearing eukaryote-like domains, WD0636 (Ank domain), WD0811 (WH2 domain), and WD0830 (WalE1; synuclein domain), via the TraJ/VirD4Wp and TraJ/VirD4Ap chimera T4CPs. A demonstration of WalE1 translocation was of particular interest in view of recent work showing that WalE1 production in Saccharomyces cerevisiae induces a growth defect with formation of aberrant, filamentous structures that colocalize with actin (99). In vitro studies confirmed that purified WalE1 bundles actins and cosediments with actin filaments in vitro. WalE1 transcripts also were shown to be upregulated during critical stages of Drosophila development and, when overexpressed, WalE1 localizes to the developing oocyte. Taken together, these data led to a proposal that Wolbachia secretes WalE1 during development to manipulate host actin and facilitate replication in and infection of important niches in the fly host (99). Our present findings add to this intriguing story by providing strong evidence that WalE1 is secreted via the Wolbachia T4SS. This is the first confirmed T4SS effector for a Wolbachia sp., and our results suggest that phenotypic profiling of two more candidate effectors, WD0636 and WD0811, in model yeast and natural fly hosts is now warranted.
We attribute the ability of the TraJ/VirD4Ap and TraJ/VirD4Wp chimeras to translocate effectors from both A. phagocytophilum and W. pipientis to the fact that VirD4Ap and VirD4Wp exhibit >90% sequence identities across their NBDs and AADs (see Fig. S2 in the supplemental material). Nevertheless, it is reasonable to predict that A. phagocytophilum and W. pipientis secrete distinct sets of effectors during infection of mammalian, fly, or nematode hosts. This is supported by the results of bioinformatics screens, which identified as many as 50 candidate effectors in A. phagocytophilum, only 4 of which possess Ank domains, whereas Wolbachia genomes encode as many as 60 Ank proteins (68, 69, 73, 96). It is conceivable that the CTDs of these T4CPs, which differ extensively in their primary sequences (see Fig. S1 and S2 in the supplemental material), play critically important roles in specifying effector repertoires of rickettsial T4SSs during infection processes. Indeed, even with Wolbachia, species-specific variations exist in the lengths and sequence compositions of the encoded VirD4 T4CPs (see Fig. S2E in the supplemental material). It is interesting that of the characterized rickettsial effectors, only Ats-1 and WD0811 possess positively charged C termini (see Fig. S3 in the supplemental material). These findings raise intriguing questions about the locations and properties of TSs in specifying effector repertoires among the different Rickettsia species.
The results of our studies confirmed that the T4CPs are critical substrate specificity checkpoints for T4SSs and advanced a mechanistic understanding of the T4CP domain requirements for substrate docking. The data support a general model in which functional diversification of T4SSs over evolutionary time was achieved through acquisition of novel structural features, e.g., CTDs, by T4CP receptors (11). Future work devoted to the engineering of chimeric T4CPs in different species might prove useful for targeted delivery of therapeutic protein or DNA substrates via repurposed T4SSs into specific eukaryotic host cells of interest.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Christie, Carlyon, and Newton laboratories for helpful discussions. We thank Yasuko Rikihisa for plasmid pBT-virD4 and Katrin Gentil for plasmid pCR-virD4.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00378-16.
REFERENCES
- 1.Cascales E, Christie PJ. 2003. The versatile bacterial type IV secretion systems. Nat Rev Microbiol 1:137–150. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev 73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Cruz F, Frost LS, Meyer RJ, Zechner EL. 2010. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev 34:18–40. doi: 10.1111/j.1574-6976.2009.00195.x. [DOI] [PubMed] [Google Scholar]
- 4.Juhas M. 2015. Horizontal gene transfer in human pathogens. Crit Rev Microbiol 41:101–108. doi: 10.3109/1040841X.2013.804031. [DOI] [PubMed] [Google Scholar]
- 5.Rees CE, Wilkins BM. 1989. Transfer of Tra proteins into the recipient cell during bacterial conjugation mediated by plasmid ColIb-P9. J Bacteriol 171:3152–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rees CED, Wilkins BM. 1990. Protein transfer into the recipient cell during bacterial conjugation: studies with F and RP4. Mol Microbiol 4:1199–1205. doi: 10.1111/j.1365-2958.1990.tb00695.x. [DOI] [PubMed] [Google Scholar]
- 7.Parker C, Meyer RJ. 2007. The R1162 relaxase/primase contains two, type IV transport signals that require the small plasmid protein MobB. Mol Microbiol 66:252–261. doi: 10.1111/j.1365-2958.2007.05925.x. [DOI] [PubMed] [Google Scholar]
- 8.Lang S, Zechner EL. 2012. General requirements for protein secretion by the F-like conjugation system R1. Plasmid 67:128–138. doi: 10.1016/j.plasmid.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alperi A, Larrea D, Fernandez-Gonzalez E, Dehio C, Zechner EL, Llosa M. 2013. A translocation motif in relaxase TrwC specifically affects recruitment by its conjugative type IV secretion system. J Bacteriol 195:4999–5006. doi: 10.1128/JB.00367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rikihisa Y, Lin M, Niu H. 2010. Type IV secretion in the obligatory intracellular bacterium Anaplasma phagocytophilum. Cell Microbiol 12:1213–1221. doi: 10.1111/j.1462-5822.2010.01500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guglielmini J, de la Cruz F, Rocha EP. 2013. Evolution of conjugation and type IV secretion systems. Mol Biol Evol 30:314–331. doi: 10.1093/molbev/mss221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christie PJ, Whitaker N, Gonzalez-Rivera C. 2014. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta 1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabezon E, Sastre JI, de la Cruz F. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol Gen Genet 254:400–406. doi: 10.1007/s004380050432. [DOI] [PubMed] [Google Scholar]
- 14.Atmakuri K, Ding Z, Christie PJ. 2003. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol 49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cascales E, Christie PJ. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170–1173. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabezon E, Ripoll-Rozada J, Pena A, de la Cruz F, Arechaga I. 2015. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 17.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, de la Cruz F, Coll M. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637–641. doi: 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 18.de Paz HD, Larrea D, Zunzunegui S, Dehio C, de la Cruz F, Llosa M. 2010. Functional dissection of the conjugative coupling protein TrwB. J Bacteriol 192:2655–2669. doi: 10.1128/JB.01692-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitaker N, Chen Y, Jakubowski SJ, Sarkar MK, Li F, Christie PJ. 2015. The all-alpha domains of coupling proteins from the Agrobacterium tumefaciens VirB/VirD4 and Enterococcus faecalis pCF10-encoded type IV secretion systems confer specificity to binding of cognate DNA substrates. J Bacteriol 197:2335–2349. doi: 10.1128/JB.00189-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu J, Wong JJ, Edwards RA, Manchak J, Frost LS, Glover JN. 2008. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol Microbiol 70:89–99. doi: 10.1111/j.1365-2958.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- 21.Schroder G, Lanka E. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J Bacteriol 185:4371–4381. doi: 10.1128/JB.185.15.4371-4381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tato I, Zunzunegui S, de la Cruz F, Cabezon E. 2005. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA dependent ATPase. Proc Natl Acad Sci U S A 102:8156–8161. doi: 10.1073/pnas.0503402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. 2004. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des 10:1551–1565. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- 24.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol 45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 25.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 106:1.16.1–1.16.39. doi: 10.1002/0471142727.mb0116s106. [DOI] [PubMed] [Google Scholar]
- 26.Lang S, Gruber K, Mihajlovic S, Arnold R, Gruber CJ, Steinlechner S, Jehl MA, Rattei T, Frohlich KU, Zechner EL. 2010. Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol Microbiol 78:1539–1555. doi: 10.1111/j.1365-2958.2010.07423.x. [DOI] [PubMed] [Google Scholar]
- 27.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sagulenko E, Sagulenko V, Chen J, Christie PJ. 2001. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol 183:5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otten L, De Greve H, Leemans J, Hain R, Hooykaas P, Schell J. 1984. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol Gen Genet 195:159–163. doi: 10.1007/BF00332739. [DOI] [Google Scholar]
- 30.Atmakuri K, Cascales E, Christie PJ. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54:1199–1211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fullner KJ. 1998. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J Bacteriol 180:430–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stachel SE, Nester EW. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J 5:1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X-R, Christie PJ. 1999. Mutagenesis of Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J Bacteriol 181:4342–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atmakuri K, Cascales E, Burton OT, Banta LM, Christie PJ. 2007. Agrobacterium ParA/MinD-like VirC1 spatially coordinates early conjugative DNA transfer reactions. EMBO J 26:2540–2551. doi: 10.1038/sj.emboj.7601696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger BR, Christie PJ. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol 176:3646–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashkova S, Spudich GM, Christie PJ. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J Bacteriol 179:583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harlow E, Lane D. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 39.Cabezon E, Lanka E, de la Cruz F. 1994. Requirements for mobilization of plasmids RSF1010 and ColE1 by the IncW plasmid R388: trwB and RP4 traG are interchangeable. J Bacteriol 176:4455–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamilton CM, Lee H, Li PL, Cook DM, Piper KR, von Bodman SB, Lanka E, Ream W, Farrand SK. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol 182:1541–1548. doi: 10.1128/JB.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cascales E, Atmakuri K, Liu Z, Binns AN, Christie PJ. 2005. Agrobacterium tumefaciens oncogenic suppressors inhibit T-DNA and VirE2 protein substrate binding to the VirD4 coupling protein. Mol Microbiol 58:565–579. doi: 10.1111/j.1365-2958.2005.04852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masui S, Sasaki T, Ishikawa H. 2000. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J Bacteriol 182:6529–6531. doi: 10.1128/JB.182.22.6529-6531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pichon S, Bouchon D, Cordaux R, Chen L, Garrett RA, Greve P. 2009. Conservation of the type IV secretion system throughout Wolbachia evolution. Biochem Biophys Res Commun 385:557–562. doi: 10.1016/j.bbrc.2009.05.118. [DOI] [PubMed] [Google Scholar]
- 44.Llosa M, Zunzunegui S, de la Cruz F. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci U S A 100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tato I, Matilla I, Arechaga I, Zunzunegui S, de la Cruz F, Cabezon E. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J Biol Chem 282:25569–25576. doi: 10.1074/jbc.M703464200. [DOI] [PubMed] [Google Scholar]
- 46.Mihajlovic S, Lang S, Sut MV, Strohmaier H, Gruber CJ, Koraimann G, Cabezon E, Moncalian G, de la Cruz F, Zechner EL. 2009. Plasmid R1 conjugative DNA processing is regulated at the coupling protein interface. J Bacteriol 191:6877–6887. doi: 10.1128/JB.00918-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cascales E, Atmakuri K, Sarkar MK, Christie PJ. 2013. DNA substrate-induced activation of the Agrobacterium VirB/VirD4 type IV secretion system. J Bacteriol 195:2691–2704. doi: 10.1128/JB.00114-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillespie JJ, Beier MS, Rahman MS, Ammerman NC, Shallom JM, Purkayastha A, Sobral BS, Azad AF. 2007. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One 2:e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heu CC, Kurtti TJ, Nelson CM, Munderloh UG. 2015. Transcriptional analysis of the conjugal transfer genes of Rickettsia bellii RML 369-C. PLoS One 10:e0137214. doi: 10.1371/journal.pone.0137214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290:979–982. doi: 10.1126/science.290.5493.979. [DOI] [PubMed] [Google Scholar]
- 51.Schrammeijer B, den Dulk-Ras AA, Vergunst AC, Jurado Jacome E, Hooykaas PJ. 2003. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res 31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo ZQ, Isberg RR. 2004. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A 101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer R. 2015. Mapping type IV secretion signals on the primase encoded by the broad-host-range plasmid R1162 (RSF1010). J Bacteriol 197:3245–3254. doi: 10.1128/JB.00443-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christie PJ, Ward JE, Winans SC, Nester EW. 1988. The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J Bacteriol 170:2659–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward DV, Zambryski PC. 2001. The six functions of Agrobacterium VirE2. Proc Natl Acad Sci U S A 98:385–386. doi: 10.1073/pnas.98.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao Z, Sagulenko E, Ding Z, Christie PJ. 2001. Activities of virE1 and the VirE1 secretion chaperone in export of the multifunctional VirE2 effector via an Agrobacterium type IV secretion pathway. J Bacteriol 183:3855–3865. doi: 10.1128/JB.183.13.3855-3865.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dym O, Albeck S, Unger T, Jacobovitch J, Branzburg A, Michael Y, Frenkiel-Krispin D, Wolf SG, Elbaum M. 2008. Crystal structure of the Agrobacterium virulence complex VirE1-VirE2 reveals a flexible protein that can accommodate different partners. Proc Natl Acad Sci U S A 105:11170–11175. doi: 10.1073/pnas.0801525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vergunst AC, van Lier MC, den Dulk-Ras A, Grosse Stuve TA, Ouwehand A, Hooykaas PJ. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A 102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simone M, McCullen CA, Stahl LE, Binns AN. 2001. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol Microbiol 41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 60.Vergunst AC, Van Lier MC, Den Dulk-Ras A, Hooykaas PJ. 2003. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol 133:978–988. doi: 10.1104/pp.103.029223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bevan M. 1984. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma H, Yanofsky MF, Klee HJ, Bowman JL, Meyerowitz EM. 1992. Vectors for plant transformation and cosmid libraries. Gene 117:161–167. doi: 10.1016/0378-1119(92)90725-5. [DOI] [PubMed] [Google Scholar]
- 63.Yanofsky MF, Porter SG, Young C, Albright LM, Gordon MP, Nester EW. 1986. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 47:471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- 64.Guo M, Jin S, Sun D, Hew CL, Pan SQ. 2007. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc Natl Acad Sci U S A 104:20019–20024. doi: 10.1073/pnas.0701738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Khedery B, Lundgren AM, Stuen S, Granquist EG, Munderloh UG, Nelson CM, Alleman AR, Mahan SM, Barbet AF. 2012. Structure of the type IV secretion system in different strains of Anaplasma phagocytophilum. BMC Genomics 13:678. doi: 10.1186/1471-2164-13-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gillespie JJ, Brayton KA, Williams KP, Diaz MA, Brown WC, Azad AF, Sobral BW. 2010. Phylogenomics reveals a diverse Rickettsiales type IV secretion system. Infect Immun 78:1809–1823. doi: 10.1128/IAI.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, Broschat SL. 2011. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS One 6:e27724. doi: 10.1371/journal.pone.0027724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol 9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- 69.Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. 2010. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog 6:e1000774. doi: 10.1371/journal.ppat.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Kregten M, Lindhout BI, Hooykaas PJ, van der Zaal BJ. 2009. Agrobacterium-mediated T-DNA transfer and integration by minimal VirD2 consisting of the relaxase domain and a type IV secretion system translocation signal. Mol Plant-Microbe Interact 22:1356–1365. doi: 10.1094/MPMI-22-11-1356. [DOI] [PubMed] [Google Scholar]
- 71.Jeong KC, Sutherland MC, Vogel JP. 2015. Novel export control of a Legionella Dot/Icm substrate is mediated by dual, independent signal sequences. Mol Microbiol 96:175–188. doi: 10.1111/mmi.12928. [DOI] [PubMed] [Google Scholar]
- 72.Schindele F, Weiss E, Haas R, Fischer W. 2016. Quantitative analysis of CagA type IV secretion by Helicobacter pylori reveals substrate recognition and translocation requirements. Mol Microbiol 100:188–203. doi: 10.1111/mmi.13309. [DOI] [PubMed] [Google Scholar]
- 73.Walker T, Klasson L, Sebaihia M, Sanders MJ, Thomson NR, Parkhill J, Sinkins SP. 2007. Ankyrin repeat domain-encoding genes in the wPip strain of Wolbachia from the Culex pipiens group. BMC Biol 5:39. doi: 10.1186/1741-7007-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haberland J, Becker J, Gerke V. 1997. The acidic C-terminal domain of rna1p is required for the binding of Ran GTP and for RanGAP activity. J Biol Chem 272:24717–24726. doi: 10.1074/jbc.272.39.24717. [DOI] [PubMed] [Google Scholar]
- 75.Kong D, Richardson CC. 1998. Role of the acidic carboxyl-terminal domain of the single-stranded DNA-binding protein of bacteriophage T7 in specific protein-protein interactions. J Biol Chem 273:6556–6564. doi: 10.1074/jbc.273.11.6556. [DOI] [PubMed] [Google Scholar]
- 76.Marintcheva B, Marintchev A, Wagner G, Richardson CC. 2008. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc Natl Acad Sci U S A 105:1855–1860. doi: 10.1073/pnas.0711919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stott K, Watson M, Howe FS, Grossmann JG, Thomas JO. 2010. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J Mol Biol 403:706–722. doi: 10.1016/j.jmb.2010.07.045. [DOI] [PubMed] [Google Scholar]
- 78.Curtis FA, Reed P, Wilson LA, Bowers LY, Yeo RP, Sanderson JM, Walmsley AR, Sharples GJ. 2011. The C-terminus of the phage lambda Orf recombinase is involved in DNA binding. J Mol Recognit 24:333–340. doi: 10.1002/jmr.1079. [DOI] [PubMed] [Google Scholar]
- 79.Gelvin SB. 2012. Traversing the cell: Agrobacterium T-DNA's journey to the host genome. Front Plant Sci 3:52. doi: 10.3389/fpls.2012.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol 184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lang S, Kirchberger PC, Gruber CJ, Redzej A, Raffl S, Zellnig G, Zangger K, Zechner EL. 2011. An activation domain of plasmid R1 TraI protein delineates stages of gene transfer initiation. Mol Microbiol 82:1071–1085. doi: 10.1111/j.1365-2958.2011.07872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezon E. 2013. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol 195:4195–4201. doi: 10.1128/JB.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cascales E, Christie PJ. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci U S A 101:17228–17233. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Paz HD, Sangari FJ, Bolland S, Garcia-Lobo JM, Dehio C, de la Cruz F, Llosa M. 2005. Functional interactions between type IV secretion systems involved in DNA transfer and virulence. Microbiology 151:3505–3516. doi: 10.1099/mic.0.28410-0. [DOI] [PubMed] [Google Scholar]
- 85.Chandran V, Fronzes R, Duquerroy S, Cronin N, Navaza J, Waksman G. 2009. Structure of the outer membrane complex of a type IV secretion system. Nature 462:1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, Lu F, Redzej A, Fronzes R, Orlova EV, Waksman G. 2014. Structure of a type IV secretion system. Nature 508:550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J 22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pena A, Matilla I, Martin-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, Cabezon E, Arechaga I. 2012. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem 287:39925–39932. doi: 10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee MH, Kosuk N, Bailey J, Traxler B, Manoil C. 1999. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J Bacteriol 181:6108–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Y, Staddon JH, Dunny GM. 2007. Specificity determinants of conjugative DNA processing in the Enterococcus faecalis plasmid pCF10 and the Lactococcus lactis plasmid pRS01. Mol Microbiol 63:1549–1564. doi: 10.1111/j.1365-2958.2007.05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Redzej A, Ilangovan A, Lang S, Gruber CJ, Topf M, Zangger K, Zechner EL, Waksman G. 2013. Structure of a translocation signal domain mediating conjugative transfer by type IV secretion systems. Mol Microbiol 89:324–333. doi: 10.1111/mmi.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu J, Frost LS. 2005. Mutations in the C-terminal region of TraM provide evidence for in vivo TraM-TraD interactions during F-plasmid conjugation. J Bacteriol 187:4767–4773. doi: 10.1128/JB.187.14.4767-4773.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G. 2004. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J Bacteriol 186:6999–7006. doi: 10.1128/JB.186.20.6999-7006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rikihisa Y, Lin M. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol 13:59–66. doi: 10.1016/j.mib.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. 2015. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev 39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sinclair SH, Garcia-Garcia JC, Dumler JS. 2015. Bioinformatic and mass spectrometry identification of Anaplasma phagocytophilum proteins translocated into host cell nuclei. Front Microbiol 6:55. doi: 10.3389/fmicb.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sutherland MC, Nguyen TL, Tseng V, Vogel JP. 2012. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog 8:e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP. 2012. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol 85:378–391. doi: 10.1111/j.1365-2958.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sheehan KB, Martin M, Lesser CF, Isberg RR, Newton IL. 2016. Identification and characterization of a candidate Wolbachia pipientis type IV effector that interacts with the actin cytoskeleton. mBio 7:e00622–16. doi: 10.1128/mBio.00622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kovach ME, Phillips RW, Elzer PH, Roop RM II, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802. [PubMed] [Google Scholar]
- 101.Prentki P, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 102.Winans SC, Walker GC. 1985. Conjugal transfer system of the IncN plasmid pKM101. J Bacteriol 161:402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


