Abstract
The advent of microscale technologies, such as microfluidics, has revolutionized many areas of biology yet has only recently begun to impact the field of bacterial biofilms. By enabling accurate control and manipulation of physical and chemical conditions, these new microscale approaches afford the ability to combine important features of natural and artificial microbial habitats, such as fluid flow and ephemeral nutrient sources, with an unprecedented level of flexibility and quantification. Here, we review selected case studies to exemplify this potential, discuss limitations, and suggest that this approach opens new vistas into biofilm research over traditional setups, allowing us to expand our understanding of the formation and consequences of biofilms in a broad range of environments and applications.
INTRODUCTION
Natural characteristics of microbial habitats, such as forces and substrates, are often dynamically and heterogeneously distributed at length scales relevant to the scale of biofilms (1–3). Yet, much of our understanding of biofilms comes from studies performed under highly idealized conditions, motivated by the desire to unveil the molecular and cellular mechanisms underlying biofilm formation. These idealized conditions are often not realized in practice, and the explicit consideration of additional elements of natural biofilm environments has recently begun to reveal important new features of this microbial lifestyle (4).
The ocean environment is a prime example of such dynamic heterogeneity. While seawater itself is nutrient depleted, ephemeral nutrient hot spots frequently occur in the form of microscale particles (Fig. 1), including marine snow, phytoplankton detritus, and fecal pellets (5, 6). These particles represent both the substrate and an important nutrient source for biofilms in the ocean (7), and recent work has revealed that diverse species of marine bacteria are capable of biofilm formation on these particles (7, 8). Marine particles are often formed by dead or dying phytoplankton that release dissolved organic matter (DOM) into the surrounding water, creating strong chemical gradients that attract chemotactic bacteria and promote colonization and biofilm formation. These biofilms then play a fundamental role in determining the degradation of the particles and, ultimately, the fate of carbon in the particles, with a direct impact on the flux of carbon from the upper to the deep ocean (9). Additionally, these particles represent alternative models for biofilm studies that have fundamental differences from traditional systems, both due to the shape and three-dimensional nature of the substrate, and because the substrate acts as both the surface for attachment and the source of nutrients.
FIG 1.
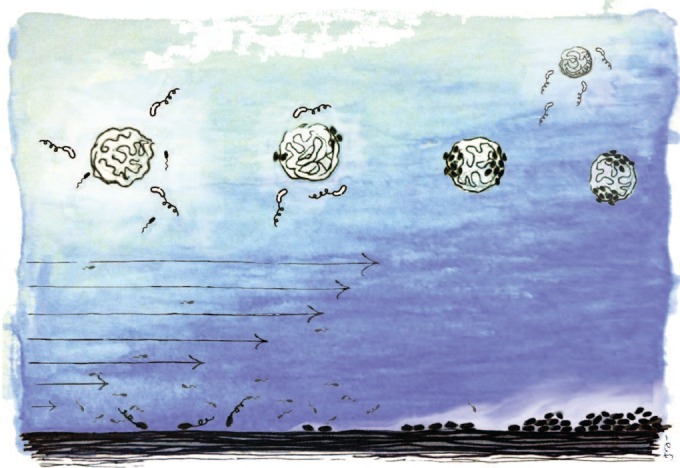
A graphic view of some environmental factors that can be fundamental for biofilm formation. Upper part, particles are prime microbial resource hot spots and biofilm substrates in the ocean. Marine bacteria often swim toward and accumulate on marine particles, yet biofilm formation on these particles is subject to a trade-off: biofilm-forming species (cells in black) can achieve stable association with the nutrient-rich particle, while non-biofilm-forming species (cells in white) are inferior competitors on the particle but are ready to migrate to fresh particles. Lower part, fluid flow can have multiple effects on biofilm formation, including the promotion of surface attachment through shear trapping near a surface and the transport of compounds, such as quorum-sensing molecules, away from producing cells and toward cells located downstream.
Biofilm research has traditionally aimed to recreate quiescent experimental conditions in topographically simple environments (often straight flat surfaces) where physical conditions are homogeneous and temporal variations in the external environment are generally suppressed by design (10). This approach has brought fundamental insights into many aspects of biofilm formation, capitalizing on the standardization of experimental conditions and absence of environmental complexities (11). Experimental tools, such as the Calgary Biofilm Device (12), significantly furthered our understanding of the genetic and physiological basis of biofilms and their antibiotic susceptibility by enabling the initiation and spontaneous dislodging of a biofilm via external chemical queues (13).
The advent of new technologies, such as microfluidics, provides unprecedented opportunities to manipulate environmental conditions over length scales relevant to bacterial motility and biofilm formation and on time scales short enough to resolve bacterial responses to rapid external stimuli (14). These approaches are enabling controlled biofilm studies that account for fundamental features of natural microbial habitats and are opening new doors to the ecological strategies underpinning biofilm formation, the role of environmental forces on biofilm development, and, ultimately, a better understanding of the physiology and consequences of biofilms. Here, we review and discuss recent efforts to directly observe the formation of bacterial biofilms under spatiotemporally heterogeneous environments and emphasize the great potential of novel technologies to expand the scope of controlled biofilm studies and to bring about new insights into this complex microbial lifestyle.
BACTERIAL ATTACHMENT UNDER FLOW: WHEN THE TRANSITION FROM THE PLANKTONIC TO THE SESSILE LIFESTYLE IS TRIGGERED BY HYDRODYNAMIC SHEAR
Understanding the physical interactions between bacteria and ambient flow (15) will benefit many scientific and industrial applications, such as the development of ecological models that account for the frequent fluid motion in microbial habitats (3), as well as the control and prevention of biofilm formation (16). Ambient flow has important ecological implications in a variety of microbial processes, including nutrient uptake (17), encounter rates (18), fertilization (19), and trophic interactions (20). In the context of biofilms, fluid flow can affect surface colonization (21), produce dislodgement (22), alter nutrient supply (23), trigger the formation of streamers (24), and wash out chemical signaling molecules (25). However, despite the ubiquity of flow in natural habitats and artificial systems, the effect of dynamic fluid environments on bacterial attachment to surfaces, the first stage in biofilm formation, and on subsequent biofilm growth has only recently received the attention necessary for a mechanistic understanding of these fundamental processes.
One feature of fluid flow that directly affects microbial interaction with surfaces is the presence of gradients in the fluid velocity, known as hydrodynamic shear (Fig. 1). Shear, which is always generated near a solid surface, where the fluid must be at rest, creates a torque on bacteria that when coupled with one or more microbial phenotypes, such as morphology, motility, or chemical sensing, can generate a rich variety of dynamics with important consequences on microbial ecology (26–28). Shear also alters the cells' swimming direction, which can strongly affect the spatial distribution of microorganisms, as recently shown by Rusconi and coworkers for Bacillus subtilis and Pseudomonas aeruginosa (29).
In those experiments, a microfluidic device and image analysis were used to determine the positions of thousands of individual bacteria within a carefully controlled flow. Without flow, the spatial distribution of cells in a dilute suspension of B. subtilis or P. aeruginosa was uniform. Conversely, in the presence of flow, a strong depletion zone in cell concentration formed at the center of the channel (the region of low shear), and an accumulation developed toward the lateral walls (the regions of high shear). The mechanism responsible for this spatial heterogeneity, elucidated by a mathematical model and termed “shear trapping,” is the competition between the shear-induced preferential alignment of the bacteria, which are highly elongated given their body shape and long nearly stiff flagella, and the reorientations due to active tumbling or passive Brownian effects. This result implies that in the presence of flow, swimming bacteria accumulate in the proximity of surfaces, thus increasing the likelihood of attaching to and colonizing those areas. This process is distinct from the association with surfaces due to hydrodynamic forces even in the absence of flow (30), as it is produced directly by fluid flow and independent of the specific type of swimming.
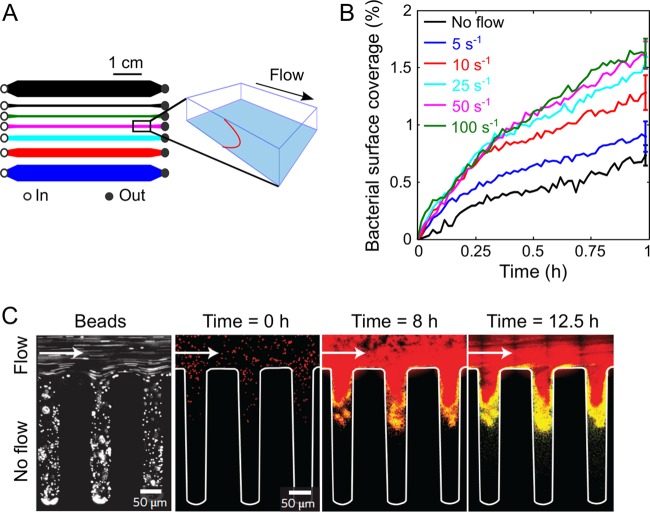
Based on these findings, Rusconi and coworkers (29) assessed the role of shear trapping in driving the transition between the planktonic and sessile lifestyles. This work again capitalized on the high level of control afforded by microfluidics and was carried out using a device consisting of seven separate microchannels, allowing a range of shear rates (0 to 100 s−1) to be assayed simultaneously with the same bacterial culture, dilute suspensions of P. aeruginosa (Fig. 2A). The progression of bacterial coverage over the bottom surface of the microchannel, made of untreated glass, consistently revealed a rapid increase in surface coverage over the first 1,000 s and then a slower increase up to 1 h (Fig. 2B).
FIG 2.
Effect of flow on bacterial attachment and biofilm formation. (A) Schematic layout of the 7-channel microfluidic device (left) used to measure the effect of shear on bacterial surface attachment at 5 different shear rates simultaneously (plus two no-shear controls) and perspective view (right) of the imaging plane (blue, lower glass surface of the channel) and the parabolic flow profile (red, vertical). (Modified from reference 29.) (B) Time series of the area coverage of P. aeruginosa PA14 on an untreated glass surface for different shear rates. Each curve represents the mean of the results from 5 replicate experiments performed under identical conditions and normalized by the mean bacterial surface coverage after 1 h of flow. (Modified from reference 29.) (C) Image of fluorescent 1-μm-diameter beads flowing into a microfluidic channel with a side groove under a flow rate of 1 μl min−1 (left) and merged images of S. aureus in the same channel (right). Red, QS-off cells; yellow, QS-on cells. (Modified from reference 25 with permission of the publisher.)
Remarkably, the magnitude of the surface coverage increased very reproducibly with increasing shear rate before saturating for shear rates of >20 s−1, where the surface coverage was double that in the quiescent case. Shear, therefore, can enhance surface attachment and thus can likely accelerate biofilm formation, a counterintuitive finding in light of the frequent association of shear with dislodging forces. The untreated glass surface used in those experiments sets this process apart from the surface-specific phenomenon of catch bonds, whereby bacteria, such as Escherichia coli, possess specific adhesins that attach to mannose-coated surfaces in a shear-dependent manner. In these species-specific cases, increasing shear rate strengthens attachment (31). Whereas catch bonds strengthen adhesion after a cell has landed on a surface, shear trapping increases the flux of cells toward the surface. Importantly, the maximum effect of flow in promoting bacterial attachment occurs at shear rates of 10 to 20 s−1, a typical range for important practical situations, such as biofilm formation on catheters (32), where P. aeruginosa is the most common pathogen in biofilm-related urinary tract infections.
Fluid flow remains a fundamental process for biofilm development after surface attachment. Recently, Kim and coworkers (25) investigated whether fluid flow represses quorum sensing (QS) in Staphylococcus aureus and Vibrio cholerae biofilms, using a microfluidic device to include geometric and topographic features. They found that QS was generally repressed by flow, which carries away signaling molecules, but in some cases it could be locally activated: at the base of a thick biofilm, in the downstream location of a long channel, and within the crevices of a groove-like surface (Fig. 2C). This work is a good illustration of the advantages of microfluidics in capturing specific features of microbial habitats, enabling not only the study of biofilms under controlled hydrodynamic conditions but also the fabrication of structures, such as grooves that mimic topographical elements of biofilm environments. As exemplified by persistent QS in a flow environment, such features significantly alter fundamental biofilm processes and, without considering them, an observation of realistic phenotypes and behaviors would otherwise be overlooked. Taken together, these studies highlight the consequences of the hydrodynamic environment on the colonization of surfaces and suggest that fluid flow should be taken into account in a more ubiquitous and controlled manner when studying biofilms.
BIOFILM FORMATION ON MARINE PARTICLES: WHEN THE DYNAMIC NATURE OF THE RESOURCE LANDSCAPE GOVERNS BIOFILM FORMATION
In the previous section, we discussed how a spatially variable flow impacts bacterial surface colonization, the first recognized step in the development of biofilms. Now, we consider how a temporally variable environment impacts the ecological role of biofilm formation, considering marine particles as a case study. Does biofilm formation confer a fitness advantage to bacteria under fluctuating environmental conditions? Past studies have highlighted the merit of biofilm formation in natural habitats, invoking the biofilm lifestyle as a strategy that allows bacteria to improve respiration and growth rates by stably localizing cells in a nutrient-rich location (5). However, cells bound to one location by biofilm formation, and in particular by the production of extracellular matrix, are conversely rather helpless to react to resource depletion in that location or to the occurrence of new and more favorable resources at nearby locations. This potential disadvantage has been pointed out in the context of sinking marine snow particles, where stable associations of cells with one particle may carry bacteria to unfavorable depths while the particle is depleted of its nutrients (9, 33). Although spontaneous dispersion has been suggested as a measure of escape from nutrient depletion (34, 35), this intrinsic trade-off makes the ecological benefit of biofilm formation on an ephemeral substrate, and more generally in a dynamic environment, difficult to predict.
In macroscale ecology, the localization of resources in patches often situates species with similar resource preferences under more direct competition (36). At the same time, ecological theory also asserts that a fluctuating resource environment allows species with similar resource preferences to coexist by modulating their ability to compete over a resource patch versus dispersing to find new resource patches, trading off their ability to access resources at a local versus global scale (36–38). In this respect, the dynamic nature of a resource landscape represents an important environmental feature for a wide range of ecological events, from competition to species coexistence (39). It is intriguing to ask if this basic tenet holds true for the microbial world, and in particular how it relates to biofilm formation in a temporally dynamic environment.
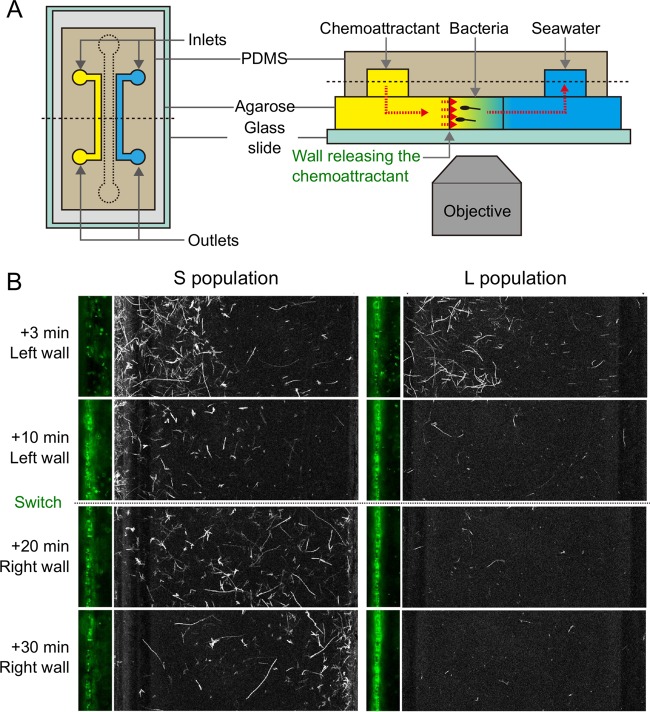
Biofilms on marine particles are assumed to confer a growth advantage to marine bacteria by affording them stable access to one of the most important sources of DOM in the ocean (40), yet the ecological consequences of this process are unclear. Recently, Yawata and coworkers (41) directly compared the behavioral strategies of two sympatric and genetically very closely related populations of marine bacteria (Vibrio cyclitrophicus) (42, 43), and they found that their differential propensities to attach to and form biofilms on particles is the key factor in determining their stable coexistence in the environment (41). Using a microfluidic setup, an experimental system that mimics the turnover of ephemeral nutrient particles (Fig. 3A) was developed to directly visualize how biofilm-forming bacteria interact with ephemeral sources of diffusing DOM. Seven isolates of V. cyclitrophicus (L population) having genes for surface adhesion (type IV pili; msh gene cluster) and biofilm formation (polysaccharide production; syp gene cluster) were compared with five isolates of V. cyclitrophicus (S population) lacking these genes. The L population was isolated from particles (>63 μm in diameter) in a size-fractionation-based field collection, whereas the S population was isolated from the free-living fraction (42) in the same field campaign.
FIG 3.
A competition-dispersal trade-off governs biofilm formation on marine particles. (A) Microfluidic system used to recreate a dynamic distribution of nutrient resources. By continuously flowing a chemoattractant solution in one of the two irrigation channels and buffer solution in the other, this device produces a steady linear chemoattractant concentration profile in the central test channel by means of diffusion in the underlying permeable layer. The direction of the chemoattractant gradient can be switched by swapping the flows in the two irrigation channels. PDMS, polydimethylsiloxane. (B) Behavioral response of the non-biofilm-forming S population and the biofilm-forming L population in a temporally varying nutrient landscape. Large panels show swimming trajectories, and small panels show the region close to the left side wall, from which serine was released for the first 10 min. Both populations swim and respond to the initial chemical gradient by accumulating in the high-serine region, but only the L population isolate attaches and forms a biofilm on the side wall, implying a benefit in the resource acquisition from the surface. In contrast, the S population isolate only hovers near the surface, without attaching, but when the direction of the chemical gradient is switched, it (and not the L isolate) is able to rapidly respond by migrating to the right side wall, implying a benefit in not forming a biofilm. (Modified from reference 41.)
Imaging in the microfluidic device afforded direct visualization of the chemotactic abilities and propensity to attach to surfaces of isolates from both the L and S populations. First, a resource gradient was generated by releasing serine from one of the two diffusion-permeable sidewalls of the device, mimicking the DOM gradient emanating from a particle. Both S and L population isolates rapidly migrated toward the resource-rich surface by chemotaxis (Fig. 3B), and no significant difference was observed in the strength of chemotaxis between isolates. However, the consequences of chemotaxis were different in the two populations: the cells from the L population attached to the surface, whereas cells from the S population only hovered near the surface without settling on it or forming a biofilm. This observation indicates that in the L population but not in the S population, chemotaxis and biofilm formation work in coordination for the rapid colonization of marine particles.
This result formed the basis for an observed trade-off in biofilm formation on particles in the L population, and ultimately for the understanding of a mechanism that enables the coexistence of the L and S populations in the field. By leveraging the flexibility of microfluidic-controlled resource conditions, the direction of the serine gradient was switched after 10 min, such that serine was at a higher concentration on the opposite side wall of the channel. This approach mimics the dynamic environment characteristic of the ocean, where particles are depleted in their resources and fresh resource-rich particles are continuously formed. Two very different responses to this microscale fluctuation emerged: the S population rapidly (within 10 min) migrated toward the new resource-rich surface (Fig. 3B), whereas the L population remained attached to the original surface (for >30 min), despite the considerable nutritional impoverishment of that surface.
This observed response to an environmental fluctuation illustrates an important ecological consequence of biofilm formation on marine particles. In the ocean, particles are nutritional hot spots in an otherwise nutrient-poor water column, which makes biofilm formation desirable per se. However, unlike archetypal setups used for biofilm studies, temporal changes in the environment are an intrinsic feature of the resource landscape, and biofilm formation thus comes at the detriment of a reduced migration ability in response to temporal changes in nutrient availability. Conversely, by not committing to particle surfaces through biofilm formation, the cells from the S population are continuously ready to migrate to new hot spots, yet at the cost of a reduced growth rate on particles compared to that of biofilm formers. Importantly, in the above-described observations, all 7 isolates of the L population behaved alike, and all 5 isolates of the S population behaved alike, denoting a clear behavioral demarcation along the boundary for horizontal gene transfer between S and L populations that reflects their genetic clustering (43). This, then, is a case in which biofilm formation is governed by a competition/dispersal trade-off, which is frequently encountered in macroecology (37, 44, 45) yet little investigated in bacteria, with one notable exception in Vibrio cholerae biofilms (46). This trade-off explains how these sympatric and genetically very closely related populations of V. cyclitrophicus can stably coexist in the ocean, ultimately because environmental fluctuations diversify the niche space pertaining to particle utilization. The biofilm-forming isolates (L population) benefit from long-lived particles, where steady attachment yields a competitive advantage, while planktonic isolates (S population) benefit from high rates of particle turnover, where frequent migration is advantageous (41).
In summary, this work exemplifies the ecological importance of a key feature in many microbial habitats, namely, the dynamic nature of resources. Beyond being a simple extension of classic biofilm studies performed under steady conditions, the explicit consideration of environmental fluctuations changes our understanding of the benefits of biofilms in environments where resources are dynamic. Thus, not only is accounting for these fluctuations made possible by microscale technology developed in recent years, it is also essential to understand the dynamics and consequences of biofilm formation in fluctuating environments.
A NEW OPPORTUNITY TO EXPLORE THE ROLE OF A DYNAMIC ENVIRONMENT IN BIOFILM FORMATION: THE MICROFLUIDIC TOOLBOX
Biofilm studies to date, and in particular over the past 3 decades, have brought about extensive knowledge of the molecular details and functions of the multiple cellular elements involved in biofilm formation, including motility (47), surface behaviors (48), quorum sensing (49), and exopolysaccharide production (50). Yet, most studies to date have rarely accounted for fundamental features of the environment, in part due to the desire to focus on the simplest case in an already-complex problem, and in part due to experimental limitations in conventional approaches. The works reviewed here illustrate how some of these environmental factors can have a substantial effect on biofilm formation. At the same time, they demonstrate that it is possible to account for selected environmental factors, be they shear, flow, or temporal variability, while still performing controlled experiments in which the ability to establish causal links and draw broad conclusions is not clouded by the complexity of real environments. The advent of microfluidic technology, coupled with advanced imaging and image analysis, has enabled exquisite control of the physical and chemical environment while allowing high-resolution observations of biofilms in space and time. In this respect, microfluidics, like classic flow cells, still represents a reductionist approach to biofilm studies, albeit one that gives better access to the study of specific features of the microbial habitat.
Microfluidics has only started to be used to investigate the effect of fluid flow and fluid shear on bacterial attachment and biofilm formation. Compared with traditional flow cell systems, which have been classically employed to study biofilms in a flowing environment, microfluidics allows for considerably greater control over flow conditions, to explore a much wider range of shear rates with great flexibility in designing flow geometries of interest, and for parallelization of experiments that can be fundamental to unravel mechanistic links in the face of the intrinsic complexity of the biofilm system. For example, the device used by Rusconi and coworkers (29) in the shear trapping study was fabricated using soft lithography so that it was much deeper than it was wide, a geometry that is not typical of flow cells. In this geometry, the velocity profile is parabolic across the width of the channel, allowing direct imaging in the plane in which shear varies, an essential feature to establish the mechanism of shear trapping. In addition, the possibility of testing multiple flow conditions at the same time, with the same culture, and on the same chip (not typical of classic approaches), has been fundamental to overcome the unavoidable biological variability among different samples and extract a robust dependence of bacterial attachment on shear rate. In this respect, parallelization is a clear advantage of a microfluidic approach for biofilm studies. In the work of Kim and coworkers (25), the advantage of microfluidics over classic flow cells also included the possibility of flexibly fabricating side grooves in which QS was studied, which would be very difficult with classic approaches. In addition, the ability to control the density of bacteria in microfluidic devices down to a few cells has given further insights into QS processes in small populations and confined environments (51, 52).
In the work of Yawata and coworkers (41), microfluidics proved essential to probe the behavior of biofilm-forming marine bacteria in a simulated ocean environment, where nutrient resources continuously turn over. Both the ability to generate gradients and, even more so, the possibility of modulating these gradients to create a temporally dynamic environment are features that go significantly beyond the capabilities of classic flow cells. In particular, the integration of a hydrogel (agarose) in the fabrication process extends the flexibility of microfluidics as an experimental approach for biofilm studies, because a hydrogel acts both as a substrate for bacterial attachment (in the case of the work of Yawata and coworkers, with properties not unlike those of real marine particles) and as a diffusion-permeable material for the supply of resources. In classic studies, the substrate for attachment is typically decoupled from the supply of nutrients, which in those approaches and indeed under many natural conditions occurs through the surrounding fluid. However, the hydrogel-based approach in the work by Yawata et al. (41) is appropriate for those situations in which the substrate is also a resource.
It is important to recognize which aspects of microfluidics represent true advantages for biofilm studies and what some limitations are. Specifically, we propose that the strength of microfluidics, as demonstrated by the studies reviewed here, resides not in attempting to fully mimic natural habitats but rather in replicating specific features of those environments and studying their effects on biofilm processes. One example is the ability to physically segregate cells belonging to single or multiple populations of bacteria while allowing them to communicate chemically (achieved through membranes, hydrogels, or nanoslits): this approach has recently started to provide insights into the role of spatial heterogeneity in competition (53), cooperation (54), and evolution (55) in microbial ecology, yet it has not been applied specifically to biofilm dynamics to date.
The use of microfluidics to study biofilms presents also some limitations. One important limitation is represented by the minute liquid volumes used in microfluidic experiments (on the order of microliters) and, usually, the difficulty of directly accessing them for endpoint sampling, for example, for chemical or genetic analysis. Alternatives have been proposed, such as integration of an electrochemical sensor for real-time analysis (56), yet they are not widespread and make fabrication and operation more complex. A second limitation is that most microfluidic devices used in microbial studies to date are essentially two-dimensional, with very few exceptions (57). To attain a closer representation of natural habitats, it will be important in the future to introduce a three-dimensional component, as was done in tissue engineering (58, 59). A third limitation, particularly for the study of biofilm in natural environments, is the very limited range of scales that can be accounted for: whereas a biofilm in a stream is subject to turbulent flow and can also develop macroscopic structures (60), a biofilm in a microfluidic device is typically exposed to laminar flow and confined spatially.
These limitations notwithstanding, we propose that microfluidics can usher in a new era of biofilm studies in which one can replicate specific features of biofilm environments and understand their interplay with physiological and behavioral processes. Having this methodological gradient spanning from simple to complex environmental conditions at our disposal can significantly expand the scope and reach of biofilm studies and ultimately bring about a deeper and more complete understanding of this important microbial lifestyle.
Funding Statement
We acknowledge support from a Gordon and Betty Moore Microbial Initiative Investigator Award (grant 3783 to R.S.).
REFERENCES
- 1.Fenchel T. 2002. Microbial behavior in a heterogeneous world. Science 296:1068–1071. doi: 10.1126/science.1070118. [DOI] [PubMed] [Google Scholar]
- 2.Stocker R. 2012. Marine microbes see a sea of gradients. Science 38:628–633. [DOI] [PubMed] [Google Scholar]
- 3.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, Wingreen NS, Bassler BL, Gitai Z, Stone HA. 2015. The mechanical world of bacteria. Cell 161:988–997. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wessel AK, Hmelo L, Parsek MR, Whiteley M. 2013. Going local: technologies for exploring bacterial microenvironments. Nat Rev Microbiol 11:337–348. doi: 10.1038/nrmicro3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azam F, Malfatti F. 2007. Microbial structuring of marine ecosystems. Nat Rev Microbiol 5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 6.Stocker R. 2015. The 100 μm length scale in the microbial ocean. Aquat Microb Ecol 76:189–194. doi: 10.3354/ame01777. [DOI] [Google Scholar]
- 7.Grossart H-P, Kiørboe T, Tang K, Ploug H. 2003. Bacterial colonization of particles: growth and interactions. Appl Environ Microbiol 69:3500–3509. doi: 10.1128/AEM.69.6.3500-3509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiørboe T, Tang K, Grossart HP, Ploug H. 2003. Dynamics of microbial communities on marine snow aggregates: colonization, growth, detachment, and grazing mortality of attached bacteria. Appl Environ Microbiol 69:3036–3047. doi: 10.1128/AEM.69.6.3036-3047.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiørboe T, Jackson GA. 2001. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol Oceanogr 46:1309–1318. doi: 10.4319/lo.2001.46.6.1309. [DOI] [Google Scholar]
- 10.McLean RJ, Bates CCL, Barnes MB, McGowin CL, Aron CM. 2004. Methods of studying biofilms, p 379–413. In Ghannoum M, O'Toole G (ed), Microbial biofilms. ASM Press, Washington, DC. [Google Scholar]
- 11.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WC365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 12.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood TK, Hong SH, Ma Q. 2011. Engineering biofilm formation and dispersal. Trends Biotechnol 29:87–94. doi: 10.1016/j.tibtech.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusconi R, Garren M, Stocker R. 2014. Microfluidics expanding the frontiers of microbial ecology. Annu Rev Biophys 43:65–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guasto JS, Rusconi R, Stocker R. 2012. Fluid mechanics of planktonic microorganisms. Annu Rev Fluid Mech 44:373–400. doi: 10.1146/annurev-fluid-120710-101156. [DOI] [Google Scholar]
- 16.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 17.Augspurger C, Küsel K. 2010. Flow velocity and primary production influences carbon utilization in nascent epilithic stream biofilms. Aquat Sci 72:237–243. doi: 10.1007/s00027-009-0126-y. [DOI] [Google Scholar]
- 18.Humphries S. 2009. Filter feeders and plankton increase particle encounter rates through flow regime control. Proc Natl Acad Sci U S A 106:7882–7887. doi: 10.1073/pnas.0809063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riffell JA, Zimmer RK. 2007. Sex and flow: the consequences of fluid shear for sperm-egg interactions. J Exp Biol 210:3644–3660. doi: 10.1242/jeb.008516. [DOI] [PubMed] [Google Scholar]
- 20.Peters F, Marrasé C, Havskum H, Rassoulzadegan F, Dolan J, Alcaraz M, Gasol JM. 2002. Turbulence and the microbial food web: effects on bacterial losses to predation and on community structure. J Plankton Res 24:321–331. doi: 10.1093/plankt/24.4.321. [DOI] [Google Scholar]
- 21.Persat A, Stone HA, Gitai Z. 2014. The curved shape of Caulobacter crescentus enhances surface colonization in flow. Nat Commun 5:3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picioreanu C, van Loosdrecht MCM, Heijnen JJ. 2001. Two-dimensional model of biofilm detachment caused by internal stress from liquid flow. Biotechnol Bioeng 72:205–218. doi:. [DOI] [PubMed] [Google Scholar]
- 23.Drescher K, Nadell CD, Stone HA, Wingreen NS, Bassler BL. 2014. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol 24:50–55. doi: 10.1016/j.cub.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rusconi R, Lecuyer S, Guglielmini L, Stone HA. 2010. Laminar flow around corners triggers the formation of biofilm streamers. J R Soc Interface 7:1293–1299. doi: 10.1098/rsif.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MK, Ingremeau F, Zhao A, Bassler BL, Stone HA. 2016. Local and global consequences of flow on bacterial quorum sensing. Nat Microbiol 1:15005. doi: 10.1038/nmicrobiol.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clay T, Grunbaum D. 2010. Morphology-flow interactions lead to stage-selective vertical transport of larval sand dollars in shear flow. J Exp Biol 213:1281–1292. doi: 10.1242/jeb.037200. [DOI] [PubMed] [Google Scholar]
- 27.Durham WM, Kessler JO, Stocker R. 2009. Disruption of vertical motility by shear triggers formation of thin phytoplankton layers. Science 323:1067–1070. doi: 10.1126/science.1167334. [DOI] [PubMed] [Google Scholar]
- 28.Taylor JR, Stocker R. 2012. Trade-offs of chemotactic foraging in turbulent water. Science 338:675–679. doi: 10.1126/science.1219417. [DOI] [PubMed] [Google Scholar]
- 29.Rusconi R, Guasto JS, Stocker R. 2014. Bacterial transport suppressed by fluid shear. Nat Phys 10:212–217. doi: 10.1038/nphys2883. [DOI] [Google Scholar]
- 30.Berke A, Turner L, Berg H, Lauga E. 2008. Hydrodynamic attraction of swimming microorganisms by surfaces. Phys Rev Lett 101:038102. doi: 10.1103/PhysRevLett.101.038102. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson LM, Thomas WE, Trintchina E, Vogel V, Sokurenko EV. 2006. Catch bond-mediated adhesion without a shear threshold: trimannose versus monomannose interactions with the FimH adhesin of Escherichia coli. J Biol Chem 281:16656–16663. doi: 10.1074/jbc.M511496200. [DOI] [PubMed] [Google Scholar]
- 32.Velraeds MMC, van de Belt-Gritter B, van der Mei HC, Reid G, Busscher HJ. 1998. Interference in initial adhesion of uropathogenic bacteria and yeasts to silicone rubber by a Lactobacillus acidophilus biosurfactant. J Med Microbiol 47:1081–1085. doi: 10.1099/00222615-47-12-1081. [DOI] [PubMed] [Google Scholar]
- 33.Azam F, Long RA. 2001. Sea snow microcosms. Nature 414:495:497–498. doi: 10.1038/35107174. [DOI] [PubMed] [Google Scholar]
- 34.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2011. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol 10:39–50. [DOI] [PubMed] [Google Scholar]
- 35.Ha D-G, O'Toole GA. 2015. c-Di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol Spectr 3(2):MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tilman D. 1994. Competition and biodiversity in spatially structured habitats. Ecology 75:2–16. doi: 10.2307/1939377. [DOI] [Google Scholar]
- 37.Lei G, Hanski I. 1998. Spatial dynamics of two competing specialist parasitoids in a host metapopulation. J Anim Ecol 67:422–433. doi: 10.1046/j.1365-2656.1998.00204.x. [DOI] [Google Scholar]
- 38.Rainey PB, Travisano M. 1998. Adaptive radiation in a heterogeneous environment. Nature 394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 39.Hastings A. 1980. Disturbance, coexistence, history, and competition for space. Theor Popul Biol 18:363–373. doi: 10.1016/0040-5809(80)90059-3. [DOI] [Google Scholar]
- 40.Ayo B, Unanue M, Azúa I, Gorsky G, Turley C, Iriberri J. 2001. Kinetics of glucose and amino acid uptake by attached and free-living marine bacteria in oligotrophic waters. Mar Biol 138:1071–1076. doi: 10.1007/s002270000518. [DOI] [Google Scholar]
- 41.Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R. 2014. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci U S A 111:5622–5627. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 43.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G, Polz MF, Alm EJ. 2012. Population genomics of early events in the ecological differentiation of bacteria. Science 336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nee S, Robert MM. 1992. Dynamics of metapopulations: habitat destruction and competitive coexistence. J Anim Ecol 61:37–40. doi: 10.2307/5506. [DOI] [Google Scholar]
- 45.Yu DW, Wilson HB, Pierce NE. 2001. An empirical model of species coexistence in a spatially structured environment. Ecology 82:1761–1771. doi: 10.1890/0012-9658(2001)082[1761:AEMOSC]2.0.CO;2. [DOI] [Google Scholar]
- 46.Nadell CD, Bassler BL. 2011. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci U S A 108:14181–14185. doi: 10.1073/pnas.1111147108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GCL, O'Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Kievit TR. 2009. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol 11:279–288. doi: 10.1111/j.1462-2920.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- 50.Wei Q, Ma LZ. 2013. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci 14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boedicker JQ, Vincent ME, Ismagilov RF. 2009. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew Chem Int Ed Engl 48:5908–5911. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeong H-H, Jin SH, Lee BJ, Kim T, Lee C-S. 2015. Microfluidic static droplet array for analyzing microbial communication on a population gradient. Lab Chip 15:889–899. doi: 10.1039/C4LC01097C. [DOI] [PubMed] [Google Scholar]
- 53.Keymer JE, Galajda P, Lambert G, Liao D, Austin RH. 2008. Computation of mutual fitness by competing bacteria. Proc Natl Acad Sci U S A 105:20269–20273. doi: 10.1073/pnas.0810792105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. 2008. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci U S A 105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung C, Pourmand N, Austin RH. 2011. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]
- 56.Toda K, Yawata Y, Setoyama E, Fukuda J, Nomura N, Suzuki H. 2011. Continuous monitoring of ammonia removal activity and observation of morphology of microbial complexes in a microdevice. Appl Environ Microbiol 77:4253–4255. doi: 10.1128/AEM.01246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 2013. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci U S A 2013:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huh D, Hamilton GA, Ingber DE. 2011. From 3D cell culture to organs-on-chips. Trends Cell Biol 21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamei K, Mashimo Y, Koyama Y, Fockenberg C, Nakashima M, Nakajima M, Li J, Chen Y. 2015. 3D printing of soft lithography mold for rapid production of polydimethylsiloxane-based microfluidic devices for cell stimulation with concentration gradients. Biomed Microdevices 17:36. doi: 10.1007/s10544-015-9928-y. [DOI] [PubMed] [Google Scholar]
- 60.Battin TJ, Besemer K, Bengtsson MM, Romani AM, Packmann AI. 2016. The ecology and biogeochemistry of stream biofilms. Nat Rev Microbiol 14:251–263. doi: 10.1038/nrmicro.2016.15. [DOI] [PubMed] [Google Scholar]