ABSTRACT
Bacterial colonization of animal epithelial tissue is a dynamic process that relies on precise molecular communication. Colonization of Euprymna scolopes bobtail squid by Vibrio fischeri bacteria requires bacterial aggregation in host mucus as the symbiont transitions from a planktonic lifestyle in seawater to a biofilm-associated state in the host. We have identified a gene, binK (biofilm inhibitor kinase; VF_A0360), which encodes an orphan hybrid histidine kinase that negatively regulates the V. fischeri symbiotic biofilm (Syp) in vivo and in vitro. We identified binK mutants as exhibiting a colonization advantage in a global genetic screen, a phenotype that we confirmed in controlled competition experiments. Bacterial biofilm aggregates in the host are larger in strains lacking BinK, whereas overexpression of BinK suppresses biofilm formation and squid colonization. Signaling through BinK is required for temperature modulation of biofilm formation at 28°C. Furthermore, we present evidence that BinK acts upstream of SypG, the σ54-dependent transcriptional regulator of the syp biofilm locus. The BinK effects are dependent on intact signaling in the RscS-Syp biofilm pathway. Therefore, we propose that BinK antagonizes the signal from RscS and serves as an integral component in V. fischeri biofilm regulation.
IMPORTANCE Bacterial lifestyle transitions underlie the colonization of animal hosts from environmental reservoirs. Formation of matrix-enclosed, surface-associated aggregates (biofilms) is common in beneficial and pathogenic associations, but investigating the genetic basis of biofilm development in live animal hosts remains a significant challenge. Using the bobtail squid light organ as a model, we analyzed putative colonization factors and identified a histidine kinase that negatively regulates biofilm formation at the host interface. This work reveals a novel in vivo biofilm regulator that influences the transition of bacteria from their planktonic state in seawater to tight aggregates of cells in the host. The study enriches our understanding of biofilm regulation and beneficial colonization by an animal's microbiome.
INTRODUCTION
During colonization of animal tissue, communication between colonizing bacteria and the host regulates processes in both organisms that influence the outcome of the interaction. The host innate immune system senses bacterial products and plays a major role in delimiting the species and strains of bacteria that can colonize specific niches. In turn, bacteria release products that modulate host responses and produce signaling proteins that respond at the host interface (1). To understand the impact of the molecular dialogue that occurs between colonizing bacteria and their animal hosts, it is useful to examine a reduced system in which individual steps can be isolated. Colonization of the Hawaiian bobtail squid Euprymna scolopes by the luminous Gram-negative bacterium Vibrio fischeri provides such a system (2, 3). V. fischeri is the only species that colonizes the squid light organ despite the large abundance of bacteria in seawater (approximately 106 bacteria per milliliter) and the proportionally low representation of V. fischeri in the seawater population (0.02% as a high estimate) (4). A powerful combination of bacterial genetics and direct imaging at the host-microbe interface has allowed for the mapping of specific stages of symbiotic colonization with high spatial and temporal resolution (5, 6). During initiation of the symbiosis, bacterial peptidoglycan stimulates host mucus secretion, which harvests bacteria from the seawater (7). The bacteria adhere in the mucus, and V. fischeri bind host cilia and aggregate in a process that requires biofilm development through production of the symbiosis polysaccharide (Syp) (8). Biofilm development is required for robust colonization of the animal and for host colonization specificity (9, 10). After the biofilm aggregate is formed, the bacteria migrate through the host mucus and predominate over other bacteria through a process that does not require flagellar motility or chemotaxis (11–13). The bacteria proceed to use flagellar motility and chemotaxis to migrate through one of the six pores into an internal crypt of the light organ, completing initiation of the symbiosis (5, 12, 14). Bacterial products, including the peptidoglycan fragment tracheal cytotoxin and the lipopolysaccharide (LPS), are shed by the colonizing V. fischeri and lead to apoptosis and regression of the host tissue that recruits the symbiont (7, 15, 16). Overall, this highly selective process of colonization initiation requires proper temporal and spatial regulation of bacterial behaviors necessary for efficient colonization of the squid host.
Many bacterial behaviors required for robust colonization of the squid host—biofilm formation, flagellar motility, and chemotaxis—are governed by two-component signaling (TCS) (17). TCS systems are prevalent throughout bacteria and enable the coupling of environmental stimulus perception with appropriate behavioral outputs (18–20). TCS systems include a sensor histidine kinase (HK) and a response regulator (RR) that effects an output, and often variations of TCS are connected in more complicated circuitry termed a phosphorelay (21–23). Usually cognate HK-RR pairs are encoded adjacent to each other in the genome; in other cases an “orphan” HK or RR does not have a known partner.
TCS systems play important roles during squid colonization by V. fischeri (17). Light organ pore entry requires biogenesis of its polar flagella and chemotaxis, which rely on the HK-RR pairs CheA-CheY, CheA-CheB, and FlrB-FlrC (5, 12). Biofilm development involves the HKs RscS and SypF. RscS, a hybrid histidine kinase, is proposed to autophosphorylate within its dimerization and histidine phosphotransferase (DHp) domain and transfer the phosphate to the conserved aspartate residue within the receiver (REC) domain. Although RscS contains a histidine phosphotransferase (HPt) domain, there is convincing evidence that the phosphorylation signal from RscS instead uses the HPt domain of SypF (24). SypF then phosphorylates two RRs, SypG and SypE, which act to positively and negatively regulate biofilm formation, respectively (24–26).
In a previous report we conducted a high-throughput, global analysis of V. fischeri factors required for squid colonization (6). Given the importance of HKs in regulating bacterial behaviors in the host environment, we examined the HKs in the global data set. HKs known to be important for robust colonization of the animal exhibited significant competitive defects, validating this analysis. An unstudied HK, VF_A0360, exhibited a dramatic competitive advantage upon gene interruption. We report here that VF_A0360 (i) negatively regulates aggregation behavior in the squid host environment, (ii) inhibits Syp polysaccharide production and biofilm phenotypes, and (iii) reduces syp transcriptional regulation. Together these data identify VF_A0360 as a negative regulator of Syp polysaccharide production. Following the nomenclature in Bassis and Visick (27) we have named VF_A0360 as binK, for biofilm inhibitor-kinase, and we use the binK nomenclature for the remainder of this report.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
V. fischeri and Escherichia coli strains used in this study are listed in Table 1. V. fischeri strains were grown at 25°C in Luria-Bertani salt (LBS) medium (per liter, 10 g Bacto Tryptone, 5 g yeast extract and 20 g NaCl, 50 ml 1 M Tris buffer [pH 7.5], in distilled water). E. coli strains, as used for cloning and conjugation, were grown at 37°C in Luria-Bertani (LB) medium (28). When necessary, antibiotics were added to the media at the following concentrations: tetracycline, 5 μg/ml for V. fischeri; erythromycin, 5 μg/ml for V. fischeri; kanamycin, 100 μg/ml for V. fischeri and 50 μg/ml for E. coli; and chloramphenicol, 5 μg/ml for V. fischeri and 25 μg/ml for E. coli. Growth media were solidified with 1.5% agar as needed. Standard microbial techniques were used to mobilize plasmids into V. fischeri strains (28). In brief, the binK alleles used in this report were generated by PCR amplification and cloned into either the mobilizable vector pVSV104 or the mini-Tn7 delivery vector pEVS107. Constructs generated using the pVSV104 vector backbone were introduced into V. fischeri by triparental mating (100-μl aliquots of each pEVS104-containing helper, pVSV104-type-containing donor, and V. fischeri recipient were mixed), and constructs generated using the pEVS107 vector backbone were introduced into V. fischeri by tetraparental mating (100-μl aliquots of each pEVS104-containing helper, pUX-BF13-containing transposase, pEVS107-type plasmid-containing donor, and V. fischeri recipient were mixed) (29).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or descriptiona | Reference(s)b |
|---|---|---|
| Vibrio fischeri strains | ||
| MJM1100 | ES114 | 54, 55 |
| MJM1107 | MJM1100/pVSV102 | 6 |
| MJM1198 | MJM1100 rscS* | 37 |
| MJM1575 | MJM1100/pVSV103 | |
| MJM1778 | MJM1100 rscS*/pVSV104 | |
| MJM1782 | MJM1100/pVSV104 | |
| MJM1946 | MJM1100 rscS* sypB::Tnerm | |
| MJM1952 | MJM1100 rscS* sypQ::Tnerm | |
| MJM1954 | MJM1100 rscS* sypG::Tnerm | |
| MJM2251 | MJM1100 ΔbinK | |
| MJM2255 | MJM1100 rscS* ΔbinK | |
| MJM2256 | MJM1100 ΔbinK/pVSV104 | |
| MJM2265 | MJM1100 ΔbinK/pVSV102 | |
| MJM2385 | MJM1100 rscS*/pBinK | |
| MJM2386 | MJM1100/pBinK | |
| MJM2387 | MJM1100 ΔbinK/pBinK | |
| MJM2388 | MJM1100 rscS* ΔbinK/pBinK | |
| MJM2389 | MJM1100 rscS* ΔbinK/pVSV104 | |
| MJM2476 | MJM1100 rscS* ΔbinK attTn7::binK+ | |
| MJM2478 | MJM1100 ΔbinK attTn7::binK+ | |
| MJM2479 | MJM1100 rscS* attTn7::erm | |
| MJM2480 | MJM1100 rscS* ΔbinK attTn7::erm | |
| MJM2487 | MJM1100 rscS* ΔbinK/pLostfoX-Kan | |
| MJM2488 | MJM1100 rscS* ΔbinK sypG::Tnerm | |
| MJM2489 | MJM1100 rscS* attTn7::erm/pM1422 | |
| MJM2490 | MJM1100 rscS* ΔbinK attTn7::erm/pM1422 | |
| MJM2491 | MJM1100 rscS* ΔbinK attTn7::binK+/pM1422 | |
| MJM2492 | MJM1100 rscS* sypG::Tnerm/pM1422 | |
| MJM2495 | MJM1100 ΔbinK attTn7::binK+/pVSV102 | |
| KV3299 | ES114 ΔsypE | 46 |
| MJM2546 | KV3299/pEAH73, pVSV104 | |
| MJM2547 | KV3299/pEAH73, pBinK | |
| KV5479 | ES114 ΔsypA attTn7:: sypA+ | 45 |
| MJM2569 | KV5479/pEAH73, pVSV104 | |
| MJM2570 | KV5479/pEAH73, pBinK | |
| KV5481 | ES114 ΔsypA attTn7:: sypAS56A | 45 |
| MJM2573 | KV5481/pEAH73, pVSV104 | |
| MJM2574 | KV5481/pEAH73, pBinK | |
| MJM2647 | MJM1100/pKV69 | |
| MJM2648 | MJM1100/pEAH73 | |
| MJM2649 | MJM2251/pKV69 | |
| MJM2650 | MJM2251/pEAH73 | |
| MJM2656 | KV3299/pKG11, pVSV104 | |
| MJM2658 | KV3299/pKG11, pBinK | |
| MJM2660 | KV5479/pKG11, pVSV104 | |
| MJM2662 | KV5479/pKG11, pBinK | |
| MJM2664 | KV5481/pKG11, pVSV104 | |
| MJM2666 | KV5481/pKG11, pBinK | |
| Plasmids | ||
| pEVS122 | V. fischeri suicide vector (Ermr) | 56 |
| pUC19 | Cloning vector (Ampr) | Lab stock |
| pVSV102 | Constitutive GFP (Kanr) | 34 |
| pVSV103 | Constitutive LacZ (Kanr) | 34 |
| pVSV104 | Complementation vector (Kanr) | 34 |
| pEVS104 | Conjugal helper plasmid (Kanr) | 28 |
| pEVS107 | Mini-Tn7 mobilizable vector (Ermr Kanr) | 29 |
| pUX-BF13 | Tn7 tranposition helper (Ampr) | 57 |
| pKV69 | Complementation vector (Camr Tetr) | 41 |
| pEAH73 | pKV69 carrying wild-type sypG (Camr Tetr) | 46 |
| pKG11 | pKV69 carrying rscS* (Camr Tetr) | 25 |
| pM1422 | pTM267 sypA′-gfp+ (Camr) | |
| pBinK | pVSV104 carrying wild-type binK (Kanr) | |
| pTn7BinK | pEVS107 carrying wild-type binK (Ermr) | |
| pLostfoX-Kan | Arabinose-inducible TfoX for transformation (Kanr) | 6 |
Erm, erythromycin; Amp, ampicillin; Kan, kanamycin; Cam, chloramphenicol; Tet, tetracycline.
Strains and plasmids with no reference listed were constructed for this study.
DNA synthesis and sequencing.
Each of the primers listed in Table 2 was synthesized by Integrated DNA Technologies (Coralville, IA). Full inserts from all cloned constructs were verified by Sanger DNA sequencing at Northwestern University Feinberg School of Medicine Center for Genetic Medicine.
TABLE 2.
Primer list
| Name | Sequencea (5′ to 3′) | Construct(s) (purpose[s])b |
|---|---|---|
| JFB_275 | GAGCCTTTTAAATCCCCTAACATT | ΔbinK (P) |
| JFB_276 | GCCATCGATTAATGACATATTATTATTCAT | ΔbinK (P) |
| JFB_277 | GCCATCGATGCGTATACATAAATAATGATTC | ΔbinK (P) |
| JFB_278 | TTTCAATACTGTGTTTTTATGCTGT | ΔbinK (P) |
| MJM-675 | GCTTTCGAGCCTTTTAAA | ΔbinK (S) |
| MJM-676 | GTTTTTGTATTCAACACG | ΔbinK (S) |
| MJM-677 | CCAACAGCAAGACTTACT | ΔbinK (S) |
| MJM-678 | AGAGTTTATTGAATTCGG | ΔbinK (S) |
| MJM-679 | CAAAACGCTTATCCAAAA | ΔbinK (S) |
| MJM-680 | GAGGGTAAGATCAAACTT | ΔbinK (S) |
| MJM-681 | AGGGTGTAGATATTTGGC | ΔbinK (S) |
| MJM-682 | GTTGATGTAGGCTAAATG | ΔbinK (S) |
| MJM-683 | ACCATCAACGGCTTTGAT | ΔbinK (S) |
| MJM-684 | CGTTTTCAATCTTAATGC | ΔbinK (S) |
| MJM-685 | GCGTGGTGAGACTTCAGA | ΔbinK (S) |
| JFB_287 | ATGGAGTTTCTACGTCAACCAGAA | ΔbinK (V) |
| JFB_288 | TGTTATAACGATTACATGGCAGCG | ΔbinK (V) |
| MJM-713 | CTAATGACAGATGTGTATGTCAG | pBinK (P, V) |
| MJM-714 | TTATAACGATTACATGGCAGCG | pBinK (P, V) |
| M13 Rev | AGCGGATAACAATTTCACACAGG | Multiple constructs (V, S) |
| M13 For | CGCCAGGGTTTTCCCAGTCACGAC | Multiple constructs (V, S) |
| JFB_355 | CTATGCGGCATCAGAGCA | pTn7BinK, pBinK (S) |
| JFB_356 | CGACGTTTTATAACGATT | pTn7BinK, pBinK (S) |
| JFB_357 | ATTTATGTATACGCTTCC | pTn7BinK, pBinK (S) |
| JFB_358 | CGCAAAATCCGGCCTTTT | pTn7BinK, pBinK (S) |
| JFB_359 | AAATGATAATCGCTGGTC | pTn7BinK, pBinK (S) |
| JFB_360 | ACCCTTTTTCTGAATCAA | pTn7BinK, pBinK (S) |
| JFB_361 | GATGTTCATCAAGCATTA | pTn7BinK, pBinK (S) |
| JFB_362 | GAGGTGTTCGAATTTCGT | pTn7BinK, pBinK (S) |
| JFB_363 | GAGCGAAAGTCTCATCAG | pTn7BinK, pBinK (S) |
| JFB_364 | AAACCTCAGACCATGAAA | pTn7BinK, pBinK (S) |
| JFB_365 | GGAAAGAGAATGATTAAG | pTn7BinK, pBinK (S) |
| JFB_366 | ATTCAAAGAATATGGTGC | pTn7BinK, pBinK (S) |
| JFB_367 | ATGACCATGATTACGCCA | pTn7BinK, pBinK (S) |
| JFB_368 | TACGACAAAGTACTTAAG | pTn7BinK, pBinK (S) |
| JFB_369 | GTTACTCTATCGATGTTC | pTn7BinK, pBinK (S) |
| JFB_370 | TCACCGCTTCACAACCTT | pTn7BinK, pBinK (S) |
| JFB_371 | CTATTTTATTGGCTTGTG | pTn7BinK, pBinK (S) |
| JFB_372 | AACTGAAACCGATTTAAC | pTn7BinK, pBinK (S) |
| JFB_373 | ATGCCGTTAAATTTACTC | pTn7BinK, pBinK (S) |
| JFB_374 | TTGAGGTGATTGAGCCAA | pTn7BinK, pBinK (S) |
| JFB_375 | TTGAACGTACAATTGAAG | pTn7BinK, pBinK (S) |
| JFB_376 | TAGATATGGTGATGAGTA | pTn7BinK, pBinK (S) |
| JFB_377 | ACTGAATTACGTTTAACG | pTn7BinK, pBinK (S) |
| JFB_378 | AGTGAGTCGTATTACAAT | pTn7BinK, pBinK (S) |
| JFB_424 | GGCGCGCCTAGGGCCCTC | pTn7BinK (P) |
| JFB_425 | TCGAGGTACCTGGCCACTAGTAGATCTCTG | pTn7BinK (P) |
| JFB_426 | GGCCCTAGGCGCGCCGGTACCTTATAACGATTACATGGCAGC | pTn7BinK (P) |
| JFB_427 | GGCCAGGTACCTCGAGGTACCCTAATGACAGATGTGTATGTC | pTn7BinK (P) |
| Tn7 Site F | TGTTGATGATACCATTGAAGCTAAA | attTn7::binK+ (V) |
| Tn7 Site R | CTTGCTGTATGTATTTGCTGATGA | attTn7::binK+ (V) |
| pEVS107 F | ACCTATCAAGGTGTACTGCCTTCC | pTn7BinK (V) |
| pEVS107 R | GTCGTTAAATGCCCTTTACCTGT | pTn7BinK (V) |
| JFB_379 | GATAGCATTTTGAATGACTTCACG | sypG::Tnerm (V) |
| MJM-477 | TTCCATAACTTCTTTTACGTTTCC | Transposon-specific primer (V) |
| MJM-475F | GCGCATGCCCGGGGCCTTACTTGGACACGAATCA | sypA′-gfp+ (P, S) |
| MJM-476R | GCACTAGTCTAGATTAGTCCATATCACCTTGAACTGATAGC | sypA′-gfp+ (P, S) |
ClaI restriction site is underlined.
Primers were used for PCR during construction (P), PCR to verify insertion/orientation (V), and/or sequencing (S) to confirm the construct.
Construction of ΔbinK strain.
A strain lacking the binK gene was constructed using allelic exchange, resulting in an in-frame deletion (30, 31). Briefly, the ΔbinK allele was generated by PCR amplifying two fragments, each approximately 1.6 kb of DNA, flanking the binK coding sequence. The gene fragments were amplified using 1 U Pfx50 polymerase (Invitrogen) and the reaction buffer provided, 300 μM (each) dNTP, 0.3 μM (each) primer, and 250 ng MJM1100 genomic DNA. Primers for the upstream fragment include JFB_275 and JFB_276, and primers for the downstream fragment include JFB_277 and JFB_278. The upstream and downstream fragments were blunt cloned into the vectors pUC19 and pEVS122, respectively, after digestion with SmaI. The resulting plasmid constructs were fused, creating a “megaplasmid,” using the engineered ClaI restriction enzyme sites (designated in the primers JFB_276 and JFB_277). Proper orientation of the fragments was screened with PCR using the primer pair JFB_275 and JFB_278. The megaplasmid was sequenced using the primers MJM-675 through MJM-685. Once confirmed, the megaplasmid was conjugated into V. fischeri ES114, and transconjugants were screened for double recombinants using the primer pair JFB_287 and JFB_288.
Construction of pTn7BinK.
A gene fragment containing the binK allele was integrated into the attTn7 site using the mini-Tn7 delivery vector pEVS107. PCR amplification generated a 3,196-bp fragment that included the binK open reading frame (2,595 kb) and 300 bp upstream and 301 bp downstream of the open reading frame. PCR was conducted using Pfx50 as described above with primers JFB_426 and JFB_427 and MJM1100 genomic DNA. The vector backbone (pEVS107) was amplified using the primer pair JFB_424 and JFB_425. The gene fragment was introduced into pEVS107 with Gibson Assembly using the Gibson Assembly master mix (New England BioLabs). The plasmid construct was sequenced using the primers JFB_355 through JFB_378 and pEVS107 F and pEVS107 R. Integration of the binK allele into the attTn7 site was verified using the primers Tn7 Site F and Tn7 Site R.
Construction of pBinK.
A gene fragment containing the wild-type binK open reading frame, along with 300 bp upstream and 300 bp downstream, was PCR amplified using Pfx50 as described above with primers MJM-713 and MJM-714 and MJM1100 genomic DNA. The gene fragment was blunt cloned into the HpaI-digested vector pVSV104. The plasmid construct was sequenced using the primers JFB_355 through JFB_378.
Construction of pM1422.
The sypA′-gfp+ transcriptional reporter was cloned by amplifying the ES114 sypA promoter with Pfx50 as described above using primers MJM-475F and MJM-476F and MJM1100 genomic DNA. The resulting PCR product was cut with XmaI and XbaI, and the XmaI-XbaI-cut product was introduced into the XmaI and XbaI sites of pTM267, respectively (32). The resulting plasmid, termed pM1422, was shown to have green fluorescent protein (GFP) activity regulated by RscS and to contain a wild-type ES114 sequence across the length of the insert.
Construction of rscS* ΔbinK strain.
A strain containing an in-frame deletion in binK and harboring the constitutive rscS allele was constructed using TfoX-based transformation as previously described (6, 33). The plasmid pLostfoX-Kan was introduced into the ΔbinK (recipient) strain by conjugation. The recipient was grown overnight in LBS containing kanamycin and subcultured 1:100 into 3 ml of Tris minimal N-acetylglucosamine containing kanamycin and grown overnight with aeration. The recipient was subcultured again at 1:50 into 20 ml of Tris minimal N-acetylglucosamine containing kanamycin and grown with aeration to an optical density at 600 nm (OD600) of 0.25 to 0.30. DNA was isolated from the strain MJM1198, which harbors the marked rscS* allele, with the Qiagen DNeasy kit; 2.4 μg of donor genomic DNA was incubated with 500 μl of prepared recipient. Following a brief vortex, the samples were incubated statically at room temperature for 30 min. LBS (1 ml) was added, and the samples were allowed to recover overnight. Cells (50 μl) were plated onto LBS containing chloramphenicol to select for transformants. After transformation, loss of the pLostfoX-Kan plasmid was confirmed by ensuring that the resulting strain was sensitive to kanamycin.
Construction of rscS* ΔbinK sypG::Tnerm strain.
The marked sypG allele was transformed into MJM2255/pLostfoX-Kan (MJM2488) as described above with the following modifications: donor genomic DNA was prepared from strain MJM1954, which harbors the marked sypG::Tnerm allele, and transformants were selected on LBS containing erythromycin (6, 33).
Competition assays in vitro.
The ΔbinK mutant and the wild-type strain MJM1100 (ES114) carrying plasmid pVSV103 that constitutively expresses LacZ (β-galactosidase) were grown overnight with shaking at 25°C in LBS and LBS containing kanamycin, respectively. Both of the overnight cultures were diluted 1:80 in LBS and grown at 25°C with shaking to an OD600 of 0.2 to 0.3. The two strains were normalized by optical density and mixed at a 1:1 ratio (34). The mixed inoculum was diluted 181-fold in LBS in triplicate, allowed to grow at 25°C with shaking for 7.5 generations, diluted 181-fold again and grown for another 7.5 generations, for a total of 15 generations, and plated. The blue/white ratios were used to score these competitions as done previously.
Competition assays in vivo.
The ΔbinK mutant and the wild-type strain MJM1100 (ES114) carrying plasmid pVSV103 that constitutively expresses LacZ (β-galactosidase) were grown overnight with shaking at 25°C in LBS and LBS containing kanamycin, respectively. Both of the overnight cultures were diluted 1:80 in LBS and grown at 25°C with shaking to an OD600 of 0.2 to 0.3. The two strains were normalized by optical density and mixed at a 1:1 ratio (34). E. scolopes hatchlings were exposed to mixed inoculum concentrations of 2 × 103 CFU/ml for 3 h. Then, squid were transferred to 40 ml of uninoculated filter-sterilized Instant Ocean (FSIO) for an additional 45 h (water was changed at 24 h postinoculation), at which point they were euthanized and surface sterilized by storage at −80°C. Each squid was homogenized and plated, and the blue/white colony ratios were used to score these competitions as done previously (6, 35). In these experiments, all squid were colonized by both strains in the competition.
Squid colonizations for CFU counts.
E. scolopes hatchlings were colonized by exposure to approximately 2 × 103 CFU/ml of each strain in a total volume of 40 ml of FSIO for 3 h. Squid were then transferred to 40 ml of uninoculated FSIO for an additional 45 h (water was changed at 24 h postinoculation), at which point they were euthanized and surface sterilized by storage at −80°C according to standard procedures (35). For determination of CFU per light organ, each squid was thawed and homogenized, and 50 μl of each homogenate was plated onto LBS plates. Bacterial colonies from each plate were counted and recorded.
Aggregate assessment.
E. scolopes hatchlings were exposed to inoculum concentrations of 2 × 106 CFU/ml for 3 to 5 h prior to dissection. The juvenile squid were anesthetized in FSIO containing 2% ethanol. Each squid was placed ventral side up on a depression well slide and dissected to expose the light organ. GFP-labeled (green) bacteria were viewed using a Nikon Eclipse 90i fluorescence microscope with appropriate filter sets. Aggregate sizes were assessed using the NIS-Elements Advanced Research version 3.2 autodetection tool, which calculates area measurements in square micrometers based on views with increased fluorescent intensity.
Wrinkled colony assay.
Cultures were grown overnight in LBS with shaking at 25°C. Eight-microliter aliquots were spotted onto LBS, LBS containing tetracycline, or LBS containing tetracycline and kanamycin, as required for plasmid maintenance. Plates were incubated at 25°C, 28°C, and 30°C for 48 h. Colonies were imaged on the Leica Firecam microscope at 48 h postspotting. For the time course assay, plates were incubated at 25°C, and colonies were imaged at the indicated time points postspotting.
Syp exopolysaccharide immunoblotting analysis.
Exopolysaccharide (EPS) from V. fischeri cells was isolated. Antiserum raised against Syp EPS was first blocked with non-EPS-producing V. fischeri ES114 and then used for immunoblotting against the isolated EPS, as described in detail previously (36, 37).
syp expression analysis.
Bacterial strains harboring the pM1422 plasmid were grown overnight with shaking at 25°C, 28°C, and 30°C. A 400-μl aliquot of each overnight culture was spun down in a centrifuge at 6,000 × g for 10 min. The supernatant was decanted, and each pellet was resuspended in 100 μl of FSIO. Each resuspension was arrayed in a 96-well flat-bottom Costar plate. The fluorescence of each well was read using 485 nm excitation/535 nm emission for the GFP and 535 nm excitation/612 nm emission for the mCherry sequentially on the Synergy H1 hybrid multi-mode microplate reader (BioTek).
Growth curves.
Bacterial microplate growth curves were obtained as described previously (6). Strains were grown overnight with shaking at 25°C and arrayed in a 96-well plate. The master plate, containing glycerol as a cryoprotectant, was frozen at −80°C. For growth curves, the master plate was thawed on ice, and 1 μl was pin replicated into a new 96-well plate containing 100 μl of fresh LBS per well; cultures in the new plate were grown overnight at 25°C with aeration. A new 96-well plate was prepared with 99 μl of LBS per well, into which 1-μl amounts of the overnight cultures were pin replicated. The inoculated plate was incubated in a Synergy H1 hybrid multimode microplate reader at 25°C, 28°C, or 30°C. The OD600 of each well was recorded every 10 min for 30 h with a 5-min period of shaking every 10 min. To prevent condensation on the lids, each lid was precoated for 30 min with 10 ml of 0.05% Triton X-100 in 20% ethanol and then air dried.
RESULTS
The hybrid histidine kinase BinK is a negative regulator of squid colonization.
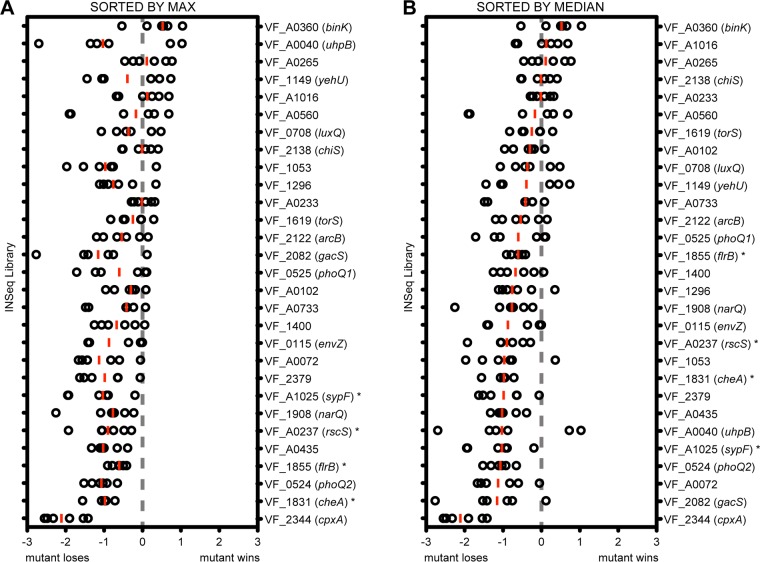
Two-component signaling systems are important mediators of bacterial interaction with their environment, including animal host niches. A previous study in V. fischeri identified response regulators that affect squid colonization, but there has not been a comparable examination of V. fischeri histidine kinases (HKs) (17). We recently completed a global investigation of bacterial mutant behavior during squid colonization using insertion sequencing (INSeq) technology, and here we have proceeded to examine the distribution of histidine kinases across the colonization data set (6). Gene products for which the corresponding mutants exhibited a substantial colonization defect included sensors in known colonization pathways. These included the Syp biofilm kinases RscS and SypF and the flagellar motility and chemotaxis kinases FlrB and CheA (Fig. 1). There was one HK, CpxA, for which the mutant yielded a more severe colonization defect, but follow-up analysis revealed it to have a growth defect in vitro, and, therefore, we did not pursue it in this analysis (6).
FIG 1.
Histidine kinase mutant dynamics during colonization in an INSeq experiment. Colonization data for the 29 histidine kinases that were sampled in vivo in our previous INSeq study (6). Circles represent INSeq analysis of 250 squid each, and the log ratio is computed as in the previous study. Red bars represent the median replicate for each mutant. Genes indicated by an asterisk are ones for which there was prior evidence for a role in squid colonization. The same data are sorted by their maximum (A) or median (B) as described in the text.
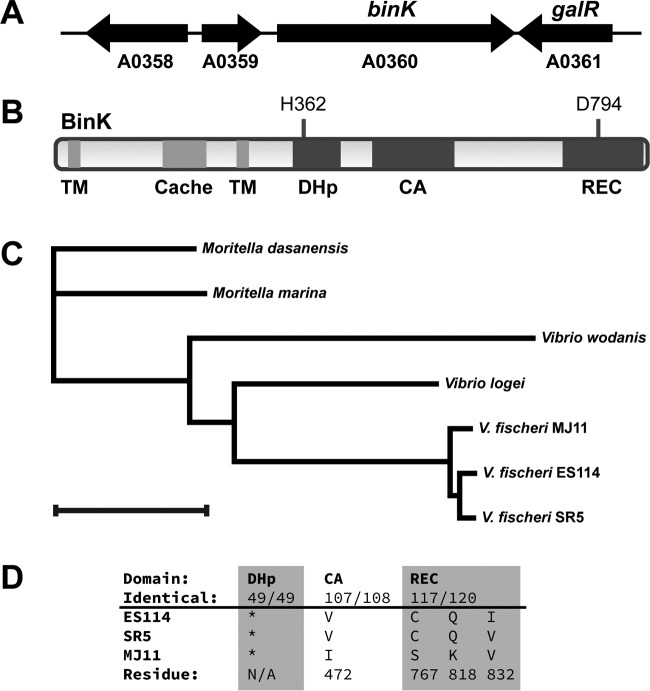
At the other end of the distribution, there appeared to be genes for which disruption was predicted to enhance competitive squid colonization. In contrast to the large number of studies on mutants with colonization defects in V. fischeri, there are only a small number that describe mutants that exhibit enhanced colonization (38, 39). As a result, we did not have a strong data set on which to train our methods for identification of new negative regulators of colonization. To determine whether the INSeq screen identified mutants with bona fide competitive advantages in the squid host environment, we selected the gene with the largest such phenotype for examination and characterization in this report, an orphan hybrid histidine kinase encoded by binK (Fig. 1). Mutations in binK were not predicted to be polar on a downstream gene, and BinK encoded domains typical of a hybrid histidine kinase that signals through an HPt domain-containing protein and a response regulator (Fig. 2). In fact, BinK is encoded in all three sequenced V. fischeri genomes; the predicted phosphorylation sites are conserved, and the annotated domains have at least 97% identity (Fig. 2C and D). The binK mutant was predicted to have a dramatic colonization advantage in the INSeq study: among INSeq library-colonized replicates of 250 squid, binK mutants exhibited a median 4-fold enrichment compared to their abundance in the library prior to colonization. To test whether this phenotype would be observed in a more controlled assay, we constructed a ΔbinK deletion strain and performed competitive colonization of the mutant against that of the wild-type strain ES114. In this assay, the mutant exhibited a similar phenotype as predicted from the global analysis, and the median competitive advantage for the mutant was 4-fold (Fig. 3). The ΔbinK strain did not exhibit a competitive advantage in culture, providing evidence that the observed advantage in vivo was specific to the host environment (Fig. 3). Introduction of a wild-type allele of binK at the Tn7 site complemented the mutant, supporting the fact that the observed phenotypes are attributable to BinK and that the presence of BinK reduces competitive colonization fitness. Therefore, BinK is a putative negative regulator of squid colonization. A major goal of this study was to identify a negative regulator from the INSeq data set and examine how it performed in controlled competition assays. The competition data obtained here confirmed that the binK mutant reproducibly outcompeted the isogenic wild-type strain. We therefore proceeded to identify the developmental stage at which BinK influences colonization.
FIG 2.
binK locus and BinK domain structure. (A) The binK locus. Locus tag “VF_” suffixes (e.g., VF_A0360) are shown, along with gene names where assigned. VF_A0358 encodes a predicted acriflavin resistance periplasmic protein, VF_A0359 encodes a predicted TetR family transcriptional regulator, and VF_A0361 is galR, which encodes a predicted galactose-responsive DNA-binding transcriptional repressor. (B) Predicted domain structure of BinK, including the transmembrane (TM) domains, the periplasmic loop that includes a Cache domain, and the cytoplasmic two-component protein signaling domains: dimerization and histidine phosphotransferase (DHp), catalytic and ATPase (CA), and receiver (REC). (C) Neighbor-joining tree of BinK orthologs. Bar, 0.1 distance metric. Proteins are from V. fischeri ES114 (GenBank accession AAW87430.1), V. fischeri SR5 (EHN69059.1), V. fischeri MJ11 (ACH63581.1), Vibrio logei (RefSeq accession WP_035470791.1), Vibrio wodanis (WP_045104527.1), Moritella dasanensis (WP_026006314.1), and Moritella marina (WP_019439447.1). (D) DHp domains among the V. fischeri orthologs are identical (*), and CA and REC domains are identical except for the residues listed.
FIG 3.
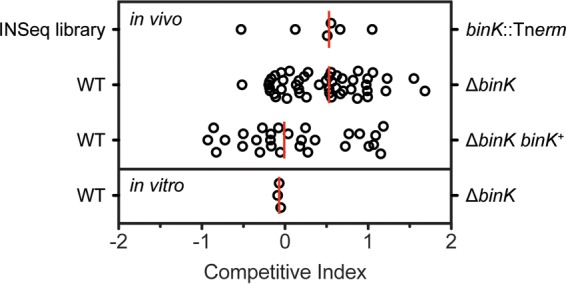
Mutants lacking BinK exhibit an advantage when competing directly against the wild-type parent strain. The top row (INSeq library) shows the same data as Fig. 1 for reference. The other rows represent competition experiments in which two strains were coinoculated and permitted to colonize squid (in vivo) or were coinoculated into culture medium and grown for a corresponding number of generations (n = 15). Individual samples (squid or culture tubes) are plotted as circles, and medians from at least three biological replicates are plotted as red lines. The competitive index is equal to the log-transformed value of the mutant/wild-type ratio after competition normalized to its measured ratio at the beginning of the competition. All of the animals were colonized by both strains. Competition of wild-type ES114 with the isogenic ΔbinK strain shows the 4-fold advantage of the mutant. This advantage is not observed upon complementation with binK+ at the attTn7 site or in vitro.
BinK is a negative regulator of in vivo aggregation.
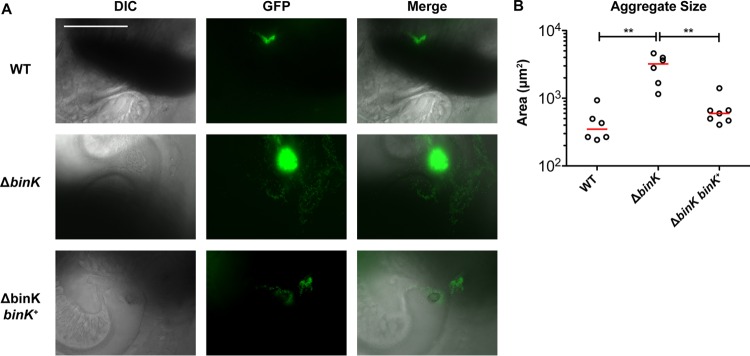
The earliest stages of bacterial colonization can be directly imaged in E. scolopes squid juveniles. Newly hatched squid acquire bacteria from the seawater, and the colonizing V. fischeri aggregate on cilia within host-produced mucus (8, 40). Furthermore, aggregation is known to be critical for proper colonization (9, 10). Using constitutive green fluorescent protein (GFP) expression to mark colonizing bacteria, we examined aggregation behavior in wild-type V. fischeri and in strains lacking BinK. GFP-expressing wild-type cells exhibited consistent aggregation 3 to 5 h postinoculation (hpi) against the ciliated appendages of the host light organ (Fig. 4). Aggregation of ΔbinK occurred in a comparable time frame but with many more bacterial cells, resulting in a larger biofilm-like aggregate. The complemented attTn7::binK+ strain exhibited aggregation comparable to that of the wild-type strain. Quantification of the area bounded by bacterial fluorescence in aggregates in at least n = 6 replicates per sample was plotted (Fig. 4B), which demonstrated that across animals BinK negatively regulates aggregate size in the squid host during the critical stage of colonization initiation.
FIG 4.
BinK negatively regulates aggregation in the squid mucus. Imaging of bacterial aggregates in host mucus by differential interference contrast (DIC) and epifluorescence microscopy. V. fischeri cells (4 × 105 to 6 × 105 CFU/ml) constitutively expressing GFP from the pVSV102 plasmid were inoculated into filter-sterilized Instant Ocean containing juvenile squid and imaged at 3 to 5 h postinoculation. (A) Bar, 100 μm; all panels are at the same scale. (B) Quantification of the area of the aggregates shows a significant BinK-dependent effect. WT, wild type.
BinK is a negative regulator of Syp biofilm phenotypes in culture.
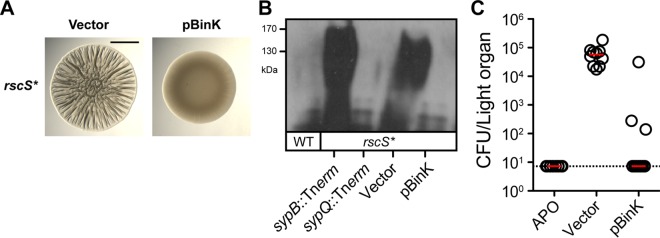
V. fischeri aggregation in the host is dependent on the hybrid histidine kinase RscS and its 18-gene target locus sypABCDEFGHIJKLMNOPQR (25, 26, 41). There is a strong correlation between phenotypes observed in the host and those in the colony biofilm assay, in which a biofilm-induced strain forms polysaccharide-rich rugose colonies on rich medium agar plates (37, 42). Unless noted otherwise, for colony biofilm formation, we typically employ the rscS-overexpressing strain MJM1198, which we denote rscS* (6, 37). We proceeded to ask whether overexpression of BinK alone was sufficient to interfere with biofilm formation in such an assay. Expression of BinK from a medium-copy vector (pVSV104 backbone [34]) led to complete inhibition of wrinkled colony phenotypes when assayed 48 h after plating (Fig. 5A). When polysaccharide production was assessed with an antibody against the Syp polysaccharide, overexpression of BinK was sufficient to reduce the levels of Syp polysaccharide below detection, even in this otherwise biofilm-induced background (Fig. 5B). For squid colonization of strains overexpressing BinK, we observed a substantial deficit (more than 3 orders of magnitude) in the number of bacteria recovered from the host, and in most instances, colonization was below our limit of detection (Fig. 5C). These data argue for a role of BinK in regulating the production of Syp polysaccharide and support assignment of BinK as a negative regulator of biofilm formation and squid colonization.
FIG 5.
Overexpression of BinK inhibits biofilm formation and squid colonization. (A) Wrinkled colony formation in the rscS* strain MJM1198 is inhibited upon overexpression of BinK. Bar, 0.5 cm. (B) Immunoblot analysis of Syp exopolysaccharide (EPS) in strain ES114 and in MJM1198-derived rscS* strains. Vector is pVSV104, the parent for the pBinK plasmid. The sypB and sypQ mutants function as controls. SypQ is required for EPS production, whereas SypB is specifically required for EPS secretion. (C) The effect of BinK overexpression on squid colonization. V. fischeri strain ES114 cells (2 × 103 CFU/ml) carrying the pVSV104 vector control or the pBinK derivative were inoculated into filter-sterilized Instant Ocean containing juvenile squid and assessed for CFU at 24 h postinoculation. The limit of detection is shown by the dotted line, and the median of each data set is shown by the red bar; the data were collected alongside those for control uncolonized animals (APO, aposymbiotic).
The standard wrinkled colony assay relies on a biofilm-induced genetic background that permits visual inspection of the phenotype on the agar surface. Characterization of positive regulators of wrinkled colony formation is straightforward in that mutants in such factors have wrinkling that is delayed or completely absent (25, 37). The colony biofilm phenotype is especially strong, and, therefore, nuanced approaches are required to examine negative regulation. For example, a kinetic approach has been used to reveal positive and negative activities within the response regulator SypE (43). We undertook a similar analysis. The rscS* strain MJM1198 begins to show evidence of wrinkling 19 to 20 h after preparation of 8-μl spots, whereas deletion of binK leads to an acceleration in the wrinkling phenotype (Fig. 6). Robust wrinkling in the mutant is already present by 18 h, consistent with BinK playing a role in inhibiting biofilm formation (Fig. 6).
FIG 6.

The ΔbinK strain exhibits accelerated wrinkled colony formation. Time course assay of wrinkled colony formation in the rscS* strain (MJM1198). Overnight cultures of the indicated strains were spotted onto LBS medium at room temperature, and wrinkled colony formation was imaged 18 to 24 h postspotting. Bar, 0.5 cm.
There were hints in the literature that overexpression of rscS induces a robust colony biofilm at 25°C but not at 28°C (i.e., from an rscS* allele carried on the plasmid-borne pKG11). To assess the temperature phenotype directly and to determine whether BinK contributed to a temperature-dependent effect, we assessed wrinkled colony formation and observed a prominent biofilm at 28°C only in the absence of BinK (Fig. 7). As expected, wrinkled colony formation was dependent on SypG (Fig. 7). We did not observe any significant effect of BinK on bacterial growth. These results argue that in the absence of BinK, biofilm signaling from RscS is derepressed at 28°C.
FIG 7.
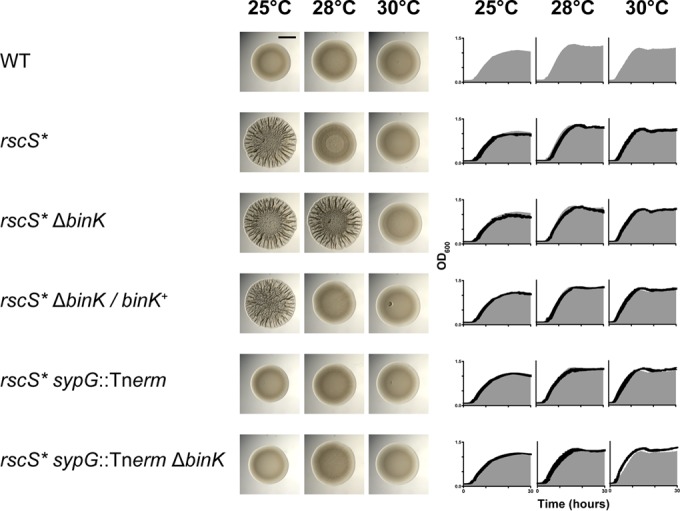
BinK is a negative regulator of Syp biofilm phenotypes. Wrinkled colony formation was assessed at 25°C, 28°C, and 30°C in the strains indicated. Only the ΔbinK mutant produced a wrinkled colony at both 25°C and 28°C. Wrinkled colony formation in the ΔbinK mutant was dependent on SypG. Growth curves were obtained for the strains indicated at each of the three temperatures assessed, with the wild-type (WT) curve for each temperature represented by the gray area. Images were taken 48 h postspotting.
BinK influences syp gene transcription.
SypG is a σ54-dependent activator of syp locus transcription (26, 44). There are characterized regulators that act upstream of SypG to influence syp transcription (e.g., RscS and SypF) and factors that act downstream of SypG to influence exopolysaccharide production by other means (e.g., SypE/SypA and DnaJ) (6, 24, 45). We therefore asked whether syp transcription was affected by the absence of BinK. We constructed a sypA′-gfp+ transcriptional fusion and assessed GFP activity in a strain lacking BinK. The reporter was constructed in the pTM267 backbone, in which constitutive mCherry (red) fluorescence normalizes for plasmid copy number, and the resulting transcriptional activity from the test promoter (green fluorescence) can be determined with high precision (32). Using this reporter, we noted that in the parent strain higher GFP levels from the sypA′-gfp+ fusion were observed at 25°C than at 28°C, consistent with the wrinkled colony phenotypes at the two temperatures. Strains lacking BinK exhibited elevated sypA′-gfp+ activity, at both 25°C and 28°C (Fig. 8). During the course of these assays, we additionally examined a higher temperature to determine what effect additional perturbations would have on syp expression. At 30°C, we observed that all samples had baseline levels of sypA′-gfp+ activity, including those lacking BinK. We examined the morphology of these strains and found that the spotted colonies were completely smooth (Fig. 7). Therefore, sypA′-gfp+ expression correlates with wrinkled colony biofilm formation across a broad set of temperature and mutant conditions, supporting a role for BinK in regulating Syp biofilm through its regulation of syp transcription.
FIG 8.
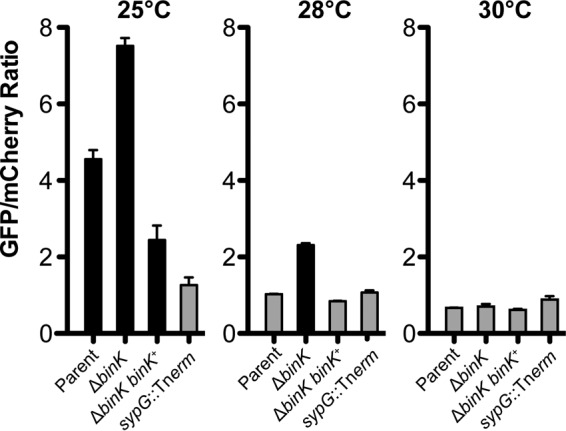
BinK is a negative regulator of sypA′-gfp+ at 25°C and 28°C. A sypA′-gfp+ transcriptional reporter was assessed in cultures grown at 25°C, 28°C, and 30°C. Strains that produce a wrinkled colony on LBS are shaded in black. The parent strain is MJM1198. The experiment was performed in triplicate, and error bars represent the standard errors of the mean.
BinK signals even in the presence of a locked SypE-SypA pathway.
The above results suggested that BinK signals at or upstream of SypG and that BinK has an effect on SypG-dependent transcription of the syp locus. We note that RscS-SypF signaling leads to activation of SypG and inhibition of SypE (and subsequent activation of SypA). Both activation of SypG and activation of SypA are required to observe wrinkled colony formation (45). We therefore asked whether BinK acts to inhibit SypG and/or to signal through the SypE-SypA pathway. First, we found that in strains lacking SypE, overexpression of BinK (i.e., pBinK) still interferes with wrinkled colony formation (Fig. 9A). Similarly, locking the pathway in an activated state with a constitutive SypA allele did not interfere with the ability of pBinK to inhibit wrinkled colony formation (Fig. 9A). We note that the rscS* allele in this panel is different, a plasmid-borne pKG11 (25), but this has no effect on the BinK phenotypes and provides additional assurance that the effects are general and not due to one specific induction system.
FIG 9.
pSypG is epistastic to BinK, and BinK signaling is independent of SypE-SypA. (A) Wrinkled colony formation in the ΔsypE, ΔsypA attTn7::sypA, and ΔsypA attTn7::sypAS56A strains carrying the plasmid pKG11 (rscS*) is inhibited upon overexpression of BinK. Wrinkled colony formation was assessed on LBS-tetracycline (Tet)-kanamycin (Kan). (B) Wrinkled colony formation in the ΔsypE, ΔsypA attTn7::sypA, and ΔsypA attTn7::sypAS56A strains carrying the plasmid pSypG (pEAH73) is not inhibited upon overexpression of BinK. Wrinkled colony formation was assessed on LBS-Tet-Kan. (C) Wrinkled colony formation is not observed in the wild-type or ΔbinK strains carrying the plasmid pSypG (pEAH73). Wrinkled colony formation was assessed on LBS-Tet. All images were taken 48 h postspotting. Bars, 0.5 cm.
We next changed to a system in which biofilm induction is accomplished directly by overexpression of SypG (i.e., pEAH73 [46]; here called pSypG for clarity). In the absence of SypE, pSypG stimulates wrinkled colony formation (Fig. 9B). In this background, BinK had no effect, suggesting that SypG overexpression is epistatic to BinK. This effect was independent of signaling through the SypE-SypA pathway (Fig. 9B). Furthermore, inactivation of wild-type SypA (by SypE; no wrinkling) and the constitutively active SypAS56A (wrinkled colony) yielded the predicted phenotypes regardless of the presence of pBinK (Fig. 9B).
The above data support BinK signaling through SypG (but not SypE-SypA), though they are based on the BinK overexpression construct. We next asked whether deletion of binK, which leads to wrinkled colony formation in the rscS* strain MJM1198, similarly leads to wrinkled colony formation in strains carrying pSypG and an intact SypE (i.e., pSypG sypE+). If a ΔbinK mutant exhibits wrinkled colony formation in a pSypG sypE+ background, then this result suggests that BinK can signal to dephosphorylate SypA. Instead, we observed that ΔbinK had no effect in the pSypG sypE+ background (Fig. 9C). Therefore, the results in Fig. 9 consistently argue that BinK is acting through SypG and not SypE.
DISCUSSION
In this report, we investigated the histidine kinase for which mutation revealed the greatest colonization advantage in an insertion sequencing study. This work further validated the global approach in the squid model, led to the identification of BinK as a novel colonization inhibitor, and revealed that BinK negatively regulates bacterial aggregation, Syp EPS, and syp gene transcription. We address these issues below.
INSeq as a discovery tool for mutants that exhibit a competitive advantage.
In this report, we present BinK as the first validated negative regulator of host colonization from the INSeq global approach. In our previous study, use of INSeq as a discovery tool was limited to a detailed analysis of mutants that were deficient in colonization. To identify novel colonization-deficient mutants, we used 37 previously identified mutants that exhibited competitive defects to train the data set. In contrast, previously identified mutants that exhibited a colonization advantage were poorly represented in the global data set (litR) or previously exhibited various competition phenotypes during the first 48 h (hnoX) (38, 39). Examination of the binK data in the INSeq data set is likely to serve as a useful marker for identification of other relevant negative regulators during host colonization. The magnitude of the effect (approximately 4-fold colonization advantage for the mutant at 48 h) is among the largest advantage that has been observed in the system and is comparable to the median reported previously for a mutant of the quorum-sensing regulator LitR (38). The tight agreement between the median INSeq result and the median competition result observed in 1:1 competitions with the wild type (Fig. 3) suggest that for mutants overrepresented in the output pool median may be a strong predictor of their colonization in defined colonization assays. We have therefore reordered the histidine kinase mutants in the INSeq data set by median value in Fig. 1B. As additional such candidates are characterized, we will assess the predictive value of the INSeq results.
INSeq mutants defective for colonization were enriched for the “initiation” stage of colonization. Colonization initiation includes biofilm formation in the host, and more than 30% of the validated mutants affected biofilm phenotypes in vitro (6). In this study, we started with the INSeq data set and then examined a previously uncharacterized gene for which the mutant exhibited a colonization advantage. We think it significant that biofilm formation and colonization initiation are also represented among the first colonization negative regulators identified from the screen, and this further bolsters the importance of biofilm regulation for V. fischeri.
BinK is highly conserved across V. fischeri strains.
V. fischeri forms tight aggregates in the squid mucus, and it seems likely that such behavior might be selected against during dispersal from the squid and in the ocean. Therefore, BinK inhibition of aggregation may provide a means for cells to remain planktonic when they are not undergoing acquisition by the squid host, both to facilitate access to nutrients and to enable uptake by another squid host. We note that BinK is conserved in the Mediterranean squid symbiont V. fischeri SR5, which colonizes squid yet lacks RscS (47, 48). Discovery of BinK presents the intriguing possibility that squid-specific symbionts lacking rscS promote syp expression by interfering with BinK activity. BinK is largely conserved in the fish symbiont MJ11, having 97% amino acid identity to ES114 (98% similarity) across the protein, with residues that differ in annotated signaling domains noted in Fig. 2D. It remains to be examined whether these sites have functional consequences for host colonization specificity.
BinK is an inhibitor of syp-dependent biofilm formation.
We have shown that BinK negatively regulates Syp EPS production and that it represses syp transcription. It seems likely that kinase and/or phosphatase activity is required for BinK function, and future work will test the role of BinK during biofilm formation by both genetic and biochemical methods. The data presented here support the hypothesis that BinK antagonizes the signal from RscS and that BinK exerts this effect through the SypG arm of the pathway, independent of SypE and SypA. It might perform this function by directly regulating the phosphorylation status of SypG. As a predicted membrane-bound hybrid histidine kinase, BinK requires an HPt domain-containing protein and a response regulator receiver domain (REC) for signaling. SypF (HPt) and SypG (REC) are possible candidates, respectively, especially given that previous analyses of response regulators did not identify candidates involved in wrinkled colony formation outside those encoded in the syp locus (17, 46). We noted that deletion of binK did not phenocopy the ΔsypE allele. Therefore, either BinK signals once the SypG pathway has diverged from the shared (i.e., SypF) portion of the pathway or BinK signaling in the shared portion of the pathway (e.g., RscS or SypF) disproportionately influences SypG activity. RscS serves as a phosphate donor for SypF-SypG, so future work will test whether BinK acts to dephosphorylate SypF under planktonic conditions. In this model, inhibition of BinK phosphatase activity might lead to a redirection of phosphates to SypG, leading to syp gene activation. We have not ruled out the possibility that BinK antagonizes RscS by other means (alternate pathways or not via phosphotransfer), and testing of the ideas described here will provide a clearer picture of how BinK activity is modulated during colonization, during dispersal, and in the environment.
Temperature influences biofilm development in V. fischeri.
We note that biofilm phenotypes in RscS-overexpressing strain MJM1198 are robust at 25°C, diminished at 28°C, and absent at 30°C. In many organisms, including Listeria monocytogenes, Yersinia pestis, Vibrio cholerae, and Pseudomonas aeruginosa, temperature has been noted to influence biofilm regulation (49–51). In many instances, there is a correlation between traits expressed at higher temperatures (e.g., 37°C) and pathogenicity in mammalian hosts; however, we point out that the squid host is ectothermic and does not internally regulate its body temperature. Therefore, a role for temperature regulation to discriminate the presence of this host is unlikely, recalling phenotypes observed in other invertebrate-associated microbes, e.g., bacterial colonizers and pathogens that influence coral bleaching pathogenesis (52, 53). In V. fischeri, at least one other temperature-responsive system, the heat shock chaperone DnaK-DnaJ has been shown to be required for bacterial aggregation in vivo and syp-dependent biofilm phenotypes in vitro (6). Taken together, these data suggest that the Syp biofilm can be experimentally manipulated through temperature regulation to reveal relevant regulation in the host. It will be interesting to examine whether the temperature phenotypes observed are representative of marine bacteria that live in an environment in which the temperature is variable or whether selection to resist host insults (i.e., reactive oxygen and nitrogen species) has manifested as a temperature-dependent biofilm phenotype.
Our previous work and studies from other groups have established biofilm formation as a critical developmental event during squid colonization. V. fischeri rapidly transitions from a single-celled planktonic lifestyle in seawater to an aggregated state in the host mucus. The molecular communication between the host and symbiont to control this lifestyle transition is poorly understood. This work has revealed a novel two-component histidine kinase that is important for biofilm signaling in vivo, bringing us closer to an understanding of the chemical communication at the microbe-host interface.
ACKNOWLEDGMENTS
We thank Jeremy Ritzert and Sarah Quillin for preliminary data; Timothy Miyashiro and Karen Visick for strains and reagents; Margaret McFall-Ngai, Ned Ruby, Michael Hadfield, and the Kewalo Marine Laboratory for Euprymna field resources; and Dhruv Arora and Ella Rotman for comments on the manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Visick KL, Ruby EG. 2006. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol 9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 2.McFall-Ngai MJ. 2014. The importance of microbes in animal development: lessons from the squid-Vibrio symbiosis. Annu Rev Microbiol 68:177–194. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nyholm SV, McFall-Ngai MJ. 2004. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol 2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 4.Mandel MJ. 2010. Models and approaches to dissect host-symbiont specificity. Trends Microbiol 18:504–511. doi: 10.1016/j.tim.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EAC, DeLoney-Marino CR, McFall-Ngai MJ, Ruby EG. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol 78:4620–4626. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks JF, Gyllborg MC, Cronin DC, Quillin SJ, Mallama CA, Foxall R, Whistler C, Goodman AL, Mandel MJ. 2014. Global discovery of colonization determinants in the squid symbiont Vibrio fischeri. Proc Natl Acad Sci U S A 111:17284–17289. doi: 10.1073/pnas.1415957111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol 68:5113–5122. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altura MA, Heath-Heckman EAC, Gillette A, Kremer N, Krachler AM, Brennan C, Ruby EG, Orth K, McFall-Ngai MJ. 2013. The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol 15:2937–2950. doi: 10.1111/1462-2920.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visick KL. 2009. An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol 74:782–789. doi: 10.1111/j.1365-2958.2009.06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–218. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brennan CA, DeLoney-Marino CR, Mandel MJ. 2013. Chemoreceptor VfcA mediates amino acid chemotaxis in Vibrio fischeri. Appl Environ Microbiol 79:1889–1896. doi: 10.1128/AEM.03794-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. 2013. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. Microbiologyopen 2:576–594. doi: 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyholm SV, McFall-Ngai MJ. 2003. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol 69:3932–3937. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Millikan DS, Ruby EG. 2003. FlrA, a σ54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J Bacteriol 185:3547–3557. doi: 10.1128/JB.185.12.3547-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koropatnick T, Goodson MS, Heath-Heckman EAC, McFall-Ngai M. 2014. Identifying the cellular mechanisms of symbiont-induced epithelial morphogenesis in the squid-Vibrio association. Biol Bull 226:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 17.Hussa EA, O'Shea TM, Darnell CL, Ruby EG, Visick KL. 2007. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J Bacteriol 189:5825–5838. doi: 10.1128/JB.00242-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu Rev Genet 41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 19.Jung K, Fried L, Behr S, Heermann R. 2012. Histidine kinases and response regulators in networks. Curr Opin Microbiol 15:118–124. doi: 10.1016/j.mib.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Igo MM, Ninfa AJ, Silhavy TJ. 1989. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev 3:598–605. doi: 10.1101/gad.3.5.598. [DOI] [PubMed] [Google Scholar]
- 21.West AH, Stock AM. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci 26:369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 22.Stephenson K, Hoch JA. 2002. Two-component and phosphorelay signal-transduction systems as therapeutic targets. Curr Opin Pharmacol 2:507–512. doi: 10.1016/S1471-4892(02)00194-7. [DOI] [PubMed] [Google Scholar]
- 23.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 24.Norsworthy AN, Visick KL. 2015. Signaling between two interacting sensor kinases promotes biofilms and colonization by a bacterial symbiont. Mol Microbiol 96:233–248. doi: 10.1111/mmi.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yip ES, Grublesky BT, Hussa EA, Visick KL. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 27.Bassis CM, Visick KL. 2010. The cyclic-di-GMP phosphodiesterase BinA negatively regulates cellulose-containing biofilms in Vibrio fischeri. J Bacteriol 192:1269–1278. doi: 10.1128/JB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. doi: 10.1016/S0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 29.McCann J, Stabb EV, Millikan DS, Ruby EG. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol 69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol 190:169–183. doi: 10.1007/s00203-008-0387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Septer AN, Wang Y, Ruby EG, Stabb EV, Dunn AK. 2011. The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ Microbiol 13:2855–2864. doi: 10.1111/j.1462-2920.2011.02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol Microbiol 77:1556–1567. doi: 10.1111/j.1365-2958.2010.07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollack-Berti A, Wollenberg MS, Ruby EG. 2010. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ Microbiol 12:2302–2311. doi: 10.1111/j.1462-2920.2010.02250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naughton LM, Mandel MJ. 2012. Colonization of Euprymna scolopes squid by Vibrio fischeri. J Vis Exp 61:e3758. doi: 10.3791/3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata S, Yip ES, Quirke KP, Ondrey JM, Visick KL. 2012. Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. J Bacteriol 194:6736–6747. doi: 10.1128/JB.00707-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh P, Brooks JF, Ray VA, Mandel MJ, Visick KL. 2015. CysK plays a role in biofilm formation and colonization by Vibrio fischeri. Appl Environ Microbiol 81:5223−5234. doi: 10.1128/AEM.00157-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol Microbiol 45:131–143. doi: 10.1046/j.1365-2958.2002.02996.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. 2010. H-NOX-mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci U S A 107:8375–8380. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Visick KL, Skoufos LM. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol 183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ray VA, Morris AR, Visick KL. 2012. A semi-quantitative approach to assess biofilm formation using wrinkled colony development. J Vis Exp 64:e4035. doi: 10.3791/4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morris AR, Darnell CL, Visick KL. 2011. Inactivation of a novel response regulator is necessary for biofilm formation and host colonization by Vibrio fischeri. Mol Microbiol 82:114−130. doi: 10.1111/j.1365-2958.2011.07800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ray VA, Eddy JL, Hussa EA, Misale M, Visick KL. 2013. The syp enhancer sequence plays a key role in transcriptional activation by the σ54-dependent response regulator SypG and in biofilm formation and host colonization by Vibrio fischeri. J Bacteriol 195:5402–5412. doi: 10.1128/JB.00689-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris AR, Visick KL. 2013. The response regulator SypE controls biofilm formation and colonization through phosphorylation of the syp-encoded regulator SypA in Vibrio fischeri. Mol Microbiol 87:509–525. doi: 10.1111/mmi.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hussa EA, Darnell CL, Visick KL. 2008. RscS functions upstream of SypG to control the syp locus and biofilm formation in Vibrio fischeri. J Bacteriol 190:4576–4583. doi: 10.1128/JB.00130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fidopiastis PM, von Boletzky S, Ruby EG. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J Bacteriol 180:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gyllborg MC, Sahl JW, Cronin DC, Rasko DA, Mandel MJ. 2012. Draft genome sequence of Vibrio fischeri SR5, a strain isolated from the light organ of the Mediterranean squid Sepiola robusta. J Bacteriol 194:1639. doi: 10.1128/JB.06825-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemon KP, Freitag NE, Kolter R. 2010. The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J Bacteriol 192:3969–3976. doi: 10.1128/JB.00179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Townsley L, Yildiz FH. 2015. Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ Microbiol 17:4290−4305. doi: 10.1111/1462-2920.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog 6:e1000783. doi: 10.1371/journal.ppat.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Case RJ, Longford SR, Campbell AH, Low A, Tujula N, Steinberg PD, Kjelleberg S. 2011. Temperature induced bacterial virulence and bleaching disease in a chemically defended marine macroalga. Environ Microbiol 13:529–537. doi: 10.1111/j.1462-2920.2010.02356.x. [DOI] [PubMed] [Google Scholar]
- 53.Rosenberg E, Ben-Haim Y. 2002. Microbial diseases of corals and global warming. Environ Microbiol 4:318–326. doi: 10.1046/j.1462-2920.2002.00302.x. [DOI] [PubMed] [Google Scholar]
- 54.Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol 172:3701–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandel MJ, Stabb EV, Ruby EG. 2008. Comparative genomics-based investigation of resequencing targets in Vibrio fischeri: focus on point miscalls and artefactual expansions. BMC Genomics 9:138. doi: 10.1186/1471-2164-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dunn AK, Martin MO, Stabb EV. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114–134. doi: 10.1016/j.plasmid.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene 109:167–168. doi: 10.1016/0378-1119(91)90604-A. [DOI] [PubMed] [Google Scholar]