ABSTRACT
In bacteria, the functions of polyamines, small linear polycations, are poorly defined, but these metabolites can influence biofilm formation in several systems. Transposon insertions in an ornithine decarboxylase (odc) gene in Agrobacterium tumefaciens, predicted to direct synthesis of the polyamine putrescine from ornithine, resulted in elevated cellulose. Null mutants for odc grew somewhat slowly in a polyamine-free medium but exhibited increased biofilm formation that was dependent on cellulose production. Spermidine is an essential metabolite in A. tumefaciens and is synthesized from putrescine in A. tumefaciens via the stepwise actions of carboxyspermidine dehydrogenase (CASDH) and carboxyspermidine decarboxylase (CASDC). Exogenous addition of either putrescine or spermidine to the odc mutant returned biofilm formation to wild-type levels. Low levels of exogenous spermidine restored growth to CASDH and CASDC mutants, facilitating weak biofilm formation, but this was dampened with increasing concentrations. Norspermidine rescued growth for the odc, CASDH, and CASDC mutants but did not significantly affect their biofilm phenotypes, whereas in the wild type, it stimulated biofilm formation and depressed spermidine levels. The odc mutant produced elevated levels of cyclic diguanylate monophosphate (c-di-GMP), exogenous polyamines modulated these levels, and expression of a c-di-GMP phosphodiesterase reversed the enhanced biofilm formation. Prior work revealed accumulation of the precursors putrescine and carboxyspermidine in the CASDH and CASDC mutants, respectively, but unexpectedly, both mutants accumulated homospermidine; here, we show that this requires a homospermidine synthase (hss) homologue.
IMPORTANCE Polyamines are small, positively charged metabolites that are nearly ubiquitous in cellular life. They are often essential in eukaryotes and more variably in bacteria. Polyamines have been reported to influence the surface-attached biofilm formation of several bacteria. In Agrobacterium tumefaciens, mutants with diminished levels of the polyamine spermidine are stimulated for biofilm formation, and exogenous provision of spermidine decreases biofilm formation. Spermidine is also essential for A. tumefaciens growth, but the related polyamine norspermidine exogenously rescues growth and does not diminish biofilm formation, revealing that the growth requirement and biofilm control are separable. Polyamine control of biofilm formation appears to function via effects on the cellular second messenger cyclic diguanylate monophosphate, regulating the transition from a free-living to a surface-attached lifestyle.
INTRODUCTION
Polyamines are organic compounds with two or more primary amino groups and are positively charged at neutral pH (1). This class of compounds includes many that play important roles in both eukaryotic and prokaryotic cells, such as putrescine [H2N(CH2)4NH2], spermidine [H2N(CH2)4NH(CH2)3NH2], and norspermidine [H2N(CH2)3NH(CH2)3NH2]. Polyamines are ubiquitous in life and are essential in eukaryotes and many bacteria.
In bacteria, putrescine is synthesized either indirectly from arginine via arginine decarboxylase or directly from decarboxylation of ornithine (1). Spermidine synthesis from putrescine can also occur via two discrete mechanisms. One of these utilizes an aminopropyl group from decarboxylated S-adenosyl-l-methionine (SAM) to aminopropylate putrescine, a reaction catalyzed by spermidine synthase (SpeE) (2). In an alternative pathway, putrescine is first converted into carboxyspermidine with the precursor l-aspartate β-semialdehyde by the enzyme carboxyspermidine dehydrogenase (CASDH), and then carboxyspermidine is converted to spermidine by carboxyspermidine decarboxylase (CASDC) (3). Norspermidine can be synthesized by essentially the same enzymes from l-aspartate β-semialdehyde and 1,3-diaminopropane (4).
Cellular polyamines are tightly regulated via synthesis, import, export, and interconversion pathways (1). If polyamine synthesis decreases, cell growth in eukaryotes can be severely inhibited, but it is restored by supplementation with exogenous polyamines. The essentiality of polyamines in eukaryotes is due to the requirement of spermidine for the posttranslational modification of a specific lysine residue on eukaryotic translation initiation factor 5A (eIF5A) to create a hypusine residue (5). In bacteria, polyamines have diverse roles and may be essential or nonessential depending on the bacterial species or strain and on growth conditions. More generalized roles in DNA and RNA stability have been proposed for bacteria as well as more specific functions. For example, in Escherichia coli polyamines initiate the SOS responsive expression of the ColE7 colicin operon and, in parallel, inhibit the uptake of colicin (6). In several rhizobia, polyamines are the precursors for the catechol-type siderophore known as agrobactin, which is involved in iron acquisition (7). However, none of these functions would seem likely to be the basis for essentiality.
Recently, polyamines have been reported to modulate bacterial biofilm formation as extracellular and metabolic signals. Endogenous putrescine and, most likely, spermidine are essential for biofilm development in Yersinia pestis (8). Y. pestis mutants that are defective in putrescine synthesis are unable to form biofilms, but this defect can be restored by exogenous provision of putrescine in a dose-dependent manner. In Bacillus subtilis, the polyamine spermidine is not required for normal growth but is essential for robust colony biofilm and pellicle formation (9). The spermidine structural analogue norspermidine functions in regulating the biofilm formation of Vibrio cholerae, stimulating this process through the periplasmic sensor protein NspS and the transmembrane diguanylate cyclase/phosphodiesterase (PDE) MbaA, which affects levels of the intracellular second messenger cyclic diguanylate monophosphate (c-di-GMP) (10). It is hypothesized that norspermidine acts in the periplasm to exert its effect on NspS, countering the inhibitory activity of spermidine.
Our recent studies have revealed that the polyamine spermidine is essential for growth in Agrobacterium tumefaciens and that this is due to a requirement for its 1,3-diaminopropane moiety (11). In the current study, we isolated Agrobacterium tumefaciens transposon mutants that led to the elevated production of cellulose and increased biofilm formation and found that they were disrupted for the polyamine biosynthetic pathway. Examination of these mutants led to the discovery that spermidine limitation stimulates increased adherence and biofilm formation and that this activity is functionally separable from the essential role of spermidine in growth per se.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
All of the strains and plasmids used in this study are listed in Table S1 in the supplemental material. DNA was manipulated using standard protocols (12). All restriction enzymes and molecular biology reagents were purchased from NEB (Ipswich, MA). DNA was purified using QIAquick Spin kits (Qiagen, Germantown, MD) by following the manufacturer protocols, and DNA sequencing was performed on the ABI 3730 system at the Indiana Molecular Biology Institute (Bloomington, IN). Plasmid introduction into A. tumefaciens was performed via electroporation (13). E. coli strains were usually grown on LB medium. AT minimal medium (14) supplemented with 0.5% (wt/vol) glucose and 15 mM ammonium sulfate (ATGN medium) was used for A. tumefaciens cultivation. Sucrose (5%) was used to replace glucose as the sole carbon source (yielding ATSN medium) during sacB counterselection. The following antibiotic concentrations were used for E. coli strains: 100 μg/ml ampicillin (Ap), 100 μg/ml spectinomycin (Sp), and 25 μg/ml kanamycin (Km). For A. tumefaciens strains, 300 μg/ml Km and 300 μg/ml Sp were used. For induction of the lacZYA promoter (Plac), media were supplemented with 100 or 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) and are listed in Table S2 in the supplemental material. Unless otherwise noted, reagents, antibiotics, and microbiological media were obtained from Fisher Scientific (Pittsburgh, PA) and Sigma-Aldrich (St. Louis, MO).
Statistical analysis.
For measurements, assays were performed on at least three biological replicates. Standard deviations were determined and are presented as error bars. Where appropriate, significance was conservatively evaluated using the t test function in Microsoft Excel, set for a paired test with a two-tailed distribution.
Transposon mutagenesis.
Transposon mutagenesis of an A. tumefaciens strain with all known polysaccharides mutated except for cellulose (exopolysaccharide-negative Cel-positive [EPS− Cel+] strain C58-YW2) was performed with the mariner minitransposon Himar1 (15). Overnight, LB cultures of the donor E. coli SM10 λpir/pFD1 (Himar1) and the A. tumefaciens EPS− Cel+ strain were inoculated at a 1:10 dilution in 2 ml of LB broth and incubated for 4 h at 37°C and 27°C, respectively. Aliquots of 1 ml from each culture were collected by centrifugation (6,000 × g) and resuspended in 50 μl of LB broth. E. coli and EPS− Cel+ cell suspensions were mixed together, and this suspension was spotted on 0.2-μm cellulose acetate filters on an LB broth plate without antibiotics. After the inoculant soaked in, these plates were incubated overnight at 28°C to allow mating. Following incubation, cells were suspended off the filter into 1 ml sterile 30% glycerol, and then aliquots were serially diluted (10−1 to 10−5), with 100 μl plated on Congo red ATGN (ATGN-CR) medium with appropriate levels of Km, followed by incubation at 28°C. The unused mating mixture was frozen at −80°C for subsequent plating.
Touchdown PCR and sequencing.
Genomic DNA was isolated from mariner-mutagenized A. tumefaciens derivatives using a Wizard genomic DNA purification kit (Promega Corp.). DNA concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Touchdown PCR was performed using 80 to 120 ng genomic DNA with primer pair MarRSeq and MarTDL2 or MarLSeq and MarTDR1 and NEB high-fidelity Phusion polymerase, followed by a standard touchdown PCR program: (i) 54 min at 95οC; (ii) 24 cycles of 95οC for 45 s, 60οC for 45 s decreasing by 5 degrees with each cycle, and 2 min at 72οC; and (iii) 24 cycles of 95οC for 45 s, 60οC for 45 s, and 2 min at 72οC. PCRs that yielded one or several strong bands were purified using the Qiagen QIAquick purification kit and were used as the templates for cycle sequencing using the BigDye reagent, primers MarRSeq or MarLseq, and the recommended protocol of the Indiana Molecular Biology Institute.
Construction of in-frame deletions and expression constructs.
We generated in-frame deletions of odc (Atu3196), the CASDC gene (Atu4169), the CASDH gene (Atu4170), and hss (Atu3768) by allelic replacement as described previously (16). Briefly, approximately 500 to 1,000 bp of flanking sequence upstream (primers P1 and P2) and downstream (primers P3 and P4) of the reading frame targeted for deletion were amplified by PCR. Primers were designed to remove as much of the coding sequence as possible without disrupting any possible translational coupling. Primers P2 and P3 were designed with 18-bp complementary sequences at their 5′ ends (see the lowercase, italicized nucleotides in Table S1 in the supplemental material) to facilitate splicing by overlapping extension (SOE) (17). The final PCR products, with the fragments flanking the deletion segment fused, were introduced into the suicide vector pNPTS138. The pNPTS138 suicide plasmid derivatives, conferring Km resistance (Kmr) and sucrose sensitivity (Sucs), were introduced into A. tumefaciens C58 by conjugal transfer, and transconjugants with the integrated plasmid were selected on ATGN plus Km. Allelic replacement recombinants were subsequently selected using the pNPTS138 sacB marker and Sucs by plating them on ATSN. Plasmid excision was verified by patching Sucr clones onto ATGN plus Km to identify derivatives that had lost the plasmid Kmr marker. Appropriate deletion of the target genes was confirmed by diagnostic PCR and DNA sequencing of the products (with primers P5 and P6, which flank the target region, and primer P7, which is within the target region).
Expression plasmids for the corresponding genes that were to be deleted were constructed in pSRK-Km (18), fusing the intact genes generated by PCR amplification to the Plac promoter and the lacZ start codon under the control of the Lac repressor (lacI) and the gratuitous inducer IPTG.
Static biofilm assays.
Biofilm assays were performed as previously described (16). Polyvinyl chloride (PVC) coverslips were placed upright in 12-well polystyrene dishes. Overnight cultures were diluted to an initial optical density at 600 nm (OD600) of 0.05 in ATGN. Each well was inoculated with 3 ml of culture, and the dishes were incubated at room temperature for 12 to 96 h. Coverslips were stained with 0.1% crystal violet (CV) dye. Adherent dye was solubilized with 33% acetic acid. The OD600 of the 48-h planktonic cultures and the A600 of the solubilized adherent CV were measured with a BioTek Synergy HT plate reader using BioTek Gen5 (version 1.07) software.
Determination of polyamine profiles.
A. tumefaciens C58 and derivative strains were grown in 5 ml LB medium in 50-ml plastic tubes at 28°C overnight and were then subcultured to 5 ml ATGN minimal medium for growth. The 5 ml of cell culture was transferred to a 15-ml Falcon tube and centrifuged (10,000 × g for 5 min at 4°C) to pellet cells. The pellet was resuspended with 0.25 ml H2O, and then 40% trichloroacetic acid was added to a final concentration of 10% and incubated on ice for 30 min. This pellet suspension was subjected to three cycles of freezing/thawing. After centrifugation (18,000 × g for 5 min at 4°C), the resulting supernatant was used for immediate analysis or stored at −20°C until needed.
Analysis of trichloroacetic acid-extracted polyamines was performed by precolumn derivatization with an AccQ-Fluor reagent labeling kit (catalog no. WAT052880; Waters). For each sample (5 μl), derivatization was carried out in a total volume of 100 μl AccQ-Fluor borate buffer and heated to 55°C for 10 min. Pure chemical standards were used to identify polyamines and were purchased from Sigma-Aldrich. Syntheses of carboxyspermidine and homospermidine were described previously (4, 19). Polyamines labeled with the AccQ-Fluor reagent were separated by high-pressure liquid chromatography (HPLC) with a hydrolysate amino acid analysis column (AccQ-Tag column; 60 Å, 4 μm, 3.9 by 150 mm; Waters) on a Beckman Coulter System Gold with fluorescence detection (excitation, 248 nm; emission, 398 nm [ProStar]). The solvent system consisted of solvent A (520 mM sodium acetic acid, 17 mM triethylamine, and 0.01% sodium azide [pH 4.65]) and solvent B (60% acetonitrile, 0.01% acetone at a flow rate of 1.0 ml/min). Elution was performed using a linear gradient of buffer B: 2 to 6 min, 0% to 20%; 6 to 11 min, 20%; 11 to 27 min, 20% to 50%; 27 to 34 min, 50%; 34 to 37 min, 100%; and 40 to 50 min, 0%.
Measurement of cellular c-di-GMP.
A. tumefaciens derivatives were cultivated in ATGN medium at 28°C to stationary phase (OD, >1.0). Culture densities were normalized by collecting cells by centrifugation and adjusting the volume so that all samples had the same cell density once resuspended in extraction buffer. Culture aliquots were centrifuged (10,000 × g for 5 min at 4°C), and the pellet was immediately resuspended in 250 μl extraction buffer (methanol-acetonitrile-distilled water, 40:40:20 plus 0.1 N formic acid cooled at −20°C) by vigorous vortexing and pipetting. The extractions were incubated at −20°C for 30 min, followed by transfer to a new microcentrifuge tube on ice. Cell debris was removed by centrifugation (10,000 × g for 3 min at 4°C), and 200 μl of supernatant was transferred into a new tube on ice. Samples were neutralized within 1 h of preparation by adding 4 μl of 15% NH4HCO3 per 100 μl of sample, aimed to set a pH of ∼7 to ∼7.5.
Prior to analysis, the sample was subjected to vacuum centrifugation to remove the extraction buffer and resuspended in an equal volume of water. Ten microliters of each sample was then analyzed using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) on a Quattro Premier XE mass spectrometer (Waters Corporation) coupled with an Acquity UltraPerformance LC system (Waters Corporation). Detection and quantification of c-di-GMP was performed as previously described (20). To calculate the c-di-GMP concentration, chemically synthesized c-di-GMP (Axxora; Enzo Life Sciences) was dissolved in water at concentrations of 250, 125, 62.5, 31.2, 15.6, 7.8, 3.9, and 1.9 nM and analyzed using LC-MS/MS to generate a standard curve.
RESULTS
An unbiased screen for negative regulators of cellulose production implicates polyamines as regulators of biofilm formation.
In A. tumefaciens, elevated intracellular c-di-GMP promotes surface attachment and stimulates production of adhesive polysaccharides, including cellulose and the polar adhesin described as the unipolar polysaccharide (UPP) (21). Overexpression of either cellulose or the UPP or both in A. tumefaciens colonies resulted in dark-red colony pigmentation on ATGN medium with 80 μg/ml Congo red (ATGN-CR). This is described as the elevated-Congo red (ECR) phenotype. In order to identify genes that were specifically involved in the negative regulation of cellulose production, we screened for the ECR phenotype in roughly 25,000 transposon mutants of an A. tumefaciens mutant strain in which the UPP and other known exopolysaccharides were genetically disabled (EPS− Cel+ strain C58-YW2). Several transposon mutants that formed dark-red colonies on ATGN-CR medium due to cellulose overproduction were identified, and the mutant genes included genes involved in transcriptional regulation and synthesis of c-di-GMP, already known to regulate cellulose synthesis in A. tumefaciens (21, 22). Among these mutants, two independent isolates had unique transposon insertions in the Atu3196 gene, annotated as an ornithine decarboxylase gene (odc) presumptively required for the biosynthesis of polyamines, such as putrescine and spermidine. We recently described a polyamine biosynthesis pathway for A. tumefaciens (Fig. 1) (11). The Odc ornithine decarboxylase catalyzes decarboxylation of ornithine to form putrescine, which is a precursor for spermidine and homospermidine.
FIG 1.
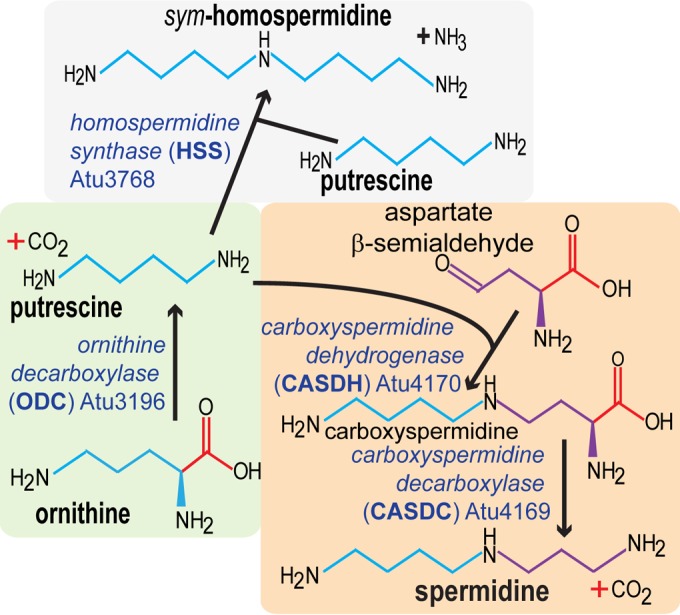
Polyamine biosynthesis pathway in A. tumefaciens C58.
An odc null mutant displays polyamine auxotrophy, elevated Congo red binding, and increased biofilm formation.
An in-frame deletion of the odc gene in the otherwise wild-type A. tumefaciens C58 strain resulted in a mutant with significantly increased Congo red binding (Fig. 2A), which is consistent with the original transposon mutant phenotype. Our prior studies have established a strong correlation between the ECR colony phenotype and increased biofilm formation in A. tumefaciens (21). Based on a time course in a static biofilm assay in ATGN minimal medium with no added polyamines, the Δodc mutant displayed a growth deficiency in the planktonic phase, with a lag and a lower growth yield (∼60%) than that of the wild type (Fig. 2B). However, despite this growth deficiency, the Δodc mutant was significantly elevated for biofilm formation when normalized for planktonic culture turbidity. Biofilm formation in the Δodc mutant was 4- to 5-fold greater than in the wild type after 48 h of incubation (Fig. 2C). An expression plasmid with the odc gene under the control of the Plac promoter (Plac-odc) was introduced into the mutant, and under full induction (500 μM IPTG), the Δodc mutant phenotypes were effectively complemented. The Δodc strain did not exhibit a growth defect or elevated biofilm formation in LB medium, which naturally contains putrescine and spermidine (see Fig. S1 in the supplemental material).
FIG 2.
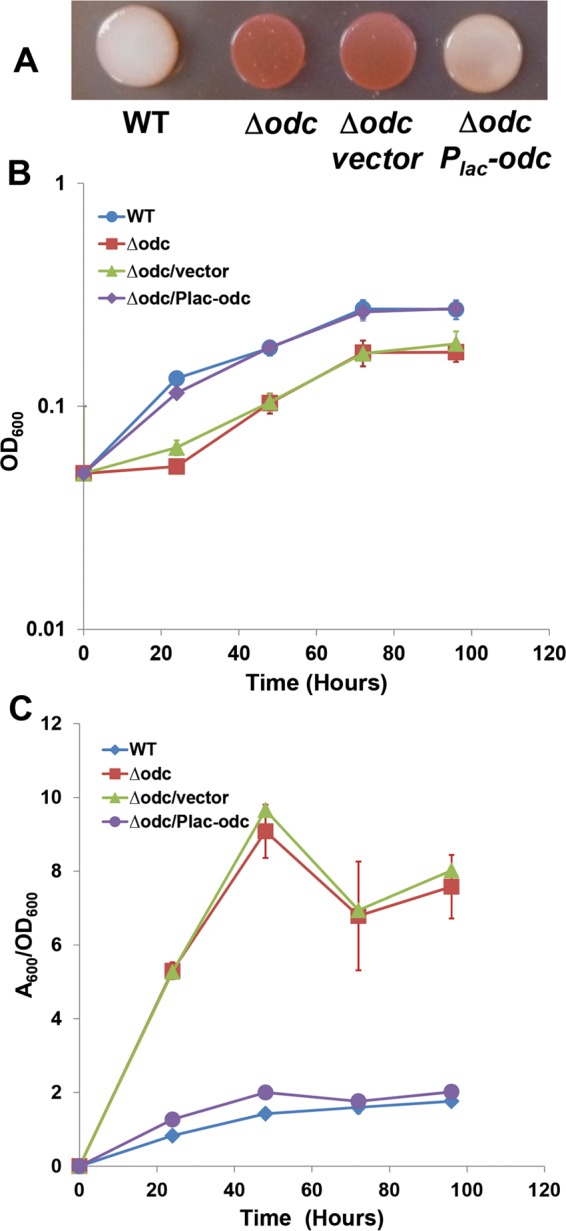
The odc mutant has elevated Congo red staining, growth deficiency, and elevated biofilm formation. (A) Congo red staining on an ATGN-CR agar plate; (B) growth curve from cultures used in the biofilm assay; (C) biofilm assays on coverslips. A. tumefaciens C58 and derivatives were incubated in 12-well plates with PVC coverslips for 0 to 96 h at 28°C. After being rinsed, coverslips were stained with 0.1% crystal violet and adherent biomass was measured as the absorbance of acetic acid-solubilized crystal violet, normalized for planktonic culture growth (A600/OD600) from the same well. Derivatives harboring the Plac-odc plasmid and the vector control were induced with 500 μM IPTG. Error bars show the standard deviation from a minimum of three biological replicates. WT, wild type.
Cellulose biosynthetic functions are required for increased biofilm formation of the odc mutant.
We previously screened a transposon mutant library in a UPP-proficient, cellulose-negative background for the ECR phenotype and did not isolate odc gene mutants (21). The ECR mutants isolated in the current study were isolated in a genetic background (C58-YW2) that produced cellulose (Cel+) but not UPP (UPP−). We engineered the odc deletion into a previously generated Δcel C58 mutant (Cel− UPP+) and an isogenic Δupp C58 mutant (Cel+ UPP−) and then examined these mutants (C58-RN12 and C58-RN13, respectively) for Congo red binding and for biofilm formation. Mutation of the cel locus in the Δodc mutant resulted in reversal of the ECR phenotype, whereas the UPP− Δodc mutant maintained dark-red-pigmented growth (Fig. 3A). Consistent with its Congo red colony phenotype, the Cel− Δodc mutant was reduced to nearly wild-type levels of biofilm formation (Fig. 3B). As we have observed in multiple prior studies (21, 23), a functional UPP adhesin is absolutely required for biofilm formation in vitro under standard growth conditions, and the UPP− Δodc mutant is completely debilitated in this respect. Thus, the Δodc mutation does not circumvent the UPP requirement.
FIG 3.
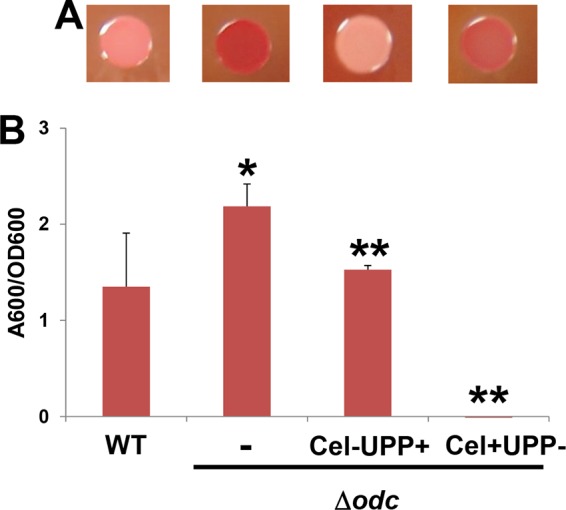
The odc-elevated Congo red and biofilm phenotypes require the cel genes. (A) Congo red staining on ATGN-CR agar plates; (B) biofilm assays on coverslips (prepared as described in the legend to Fig. 2). Additional deletions in the cellulose biosynthesis genes (Cel−) or the UPP biosynthesis genes (UPP−) were introduced into the Δodc mutant to create Cel− UPP+ and Cel+ UPP− mutant derivatives. Error bars show the standard deviations from a minimum of three biological replicates. *, P < 0.08 compared to the wild type; **, P < 0.03 compared to the Δodc mutant.
Exogenous putrescine and spermidine but not norspermidine decrease biofilm formation.
Our recent findings indicate that the A. tumefaciens odc mutant has depressed spermidine levels and accumulates ornithine (11). The mutant does produce low levels of spermidine (see Fig. S2 in the supplemental material), apparently sufficient for diminished, but significant, growth. A. tumefaciens can transport various polyamines, and the weak growth of this mutant can be rescued with the addition of exogenous polyamines that, like spermidine, possess a terminal 1,3-diaminopropane moiety (Fig. 1), including norspermidine, a symmetrical polyamine one methylene group shorter than spermidine (11). Most organisms have dedicated systems for the uptake of polyamines from the environment, and there are several presumptive polyamine transporters in the A. tumefaciens C58 genome (24, 25). To identify whether exogenous polyamines rescue the Δodc mutant biofilm phenotypes, putrescine, spermidine, and norspermidine (not synthesized naturally by A. tumefaciens) were added exogenously to ATGN cultures diluted at inoculation to deplete polyamines, and growth and biofilm formation were evaluated in parallel over a range of concentrations. Exogenous putrescine was able to rescue the growth defect in the Δodc mutant but required more than 1 mM to completely return the growth of the mutant back to wild-type levels (Fig. 4A; see also Fig. S3A in the supplemental material). Spermidine and norspermidine were also able to rescue the growth deficiency and at much lower concentrations, 200 μM and 20 μM, respectively (Fig. 4B and C; see also Fig. S3B and C). Putrescine and spermidine were able to effectively reverse the increased biofilm formation of the Δodc mutant to normal levels, and this occurred at notably lower concentrations than those required to fully rescue growth (Fig. 4A and B; see also Fig. S3D and E). Interestingly, although adding norspermidine to ATGN medium completely restored the growth defect of the Δodc mutant, it could not reverse its elevated biofilm formation even at the highest concentration tested (Fig. 4C; see also Fig. S3F).
FIG 4.
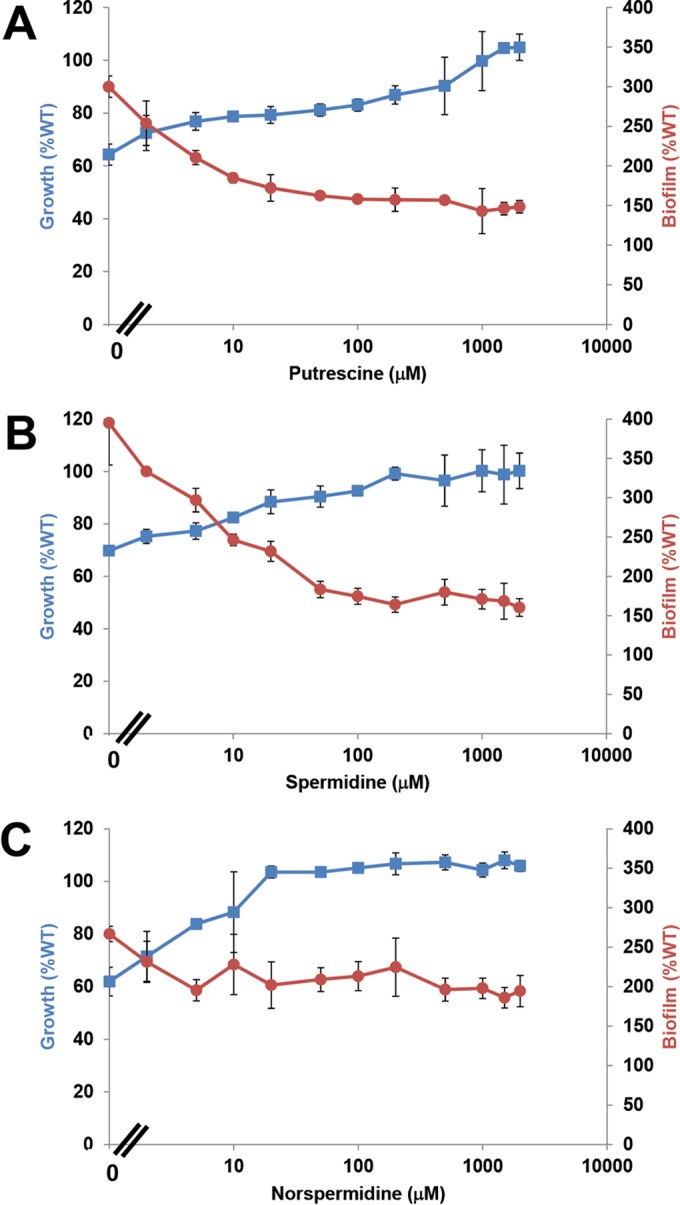
Rescue of growth and reversal of increased biofilm formation in the Δodc mutant by exogenous polyamines. Exogenous putrescine (A), spermidine (B), and norspermidine (C) were tested across the indicated concentration range for their impact on growth (blue) and biofilm formation (red) of the Δodc mutant (prepared as described in the legend to Fig. 2). Plotted are percentages of the wild type's growth and biofilm formation in the absence of exogenous polyamines. Error bars show the standard deviations from a minimum of three biological replicates.
Exogenous norspermidine promotes biofilm formation in wild-type A. tumefaciens.
We also tested the impact of exogenous norspermidine on biofilm formation in wild-type A. tumefaciens. Consistent with our prior findings, exogenous norspermidine did not influence the growth of the wild type (11) but promoted its biofilm formation (Fig. 5A). Biofilm formation increased from 0 to 10 μM norspermidine up to 2 times the amount in the untreated culture but did not increase further above that concentration.
FIG 5.
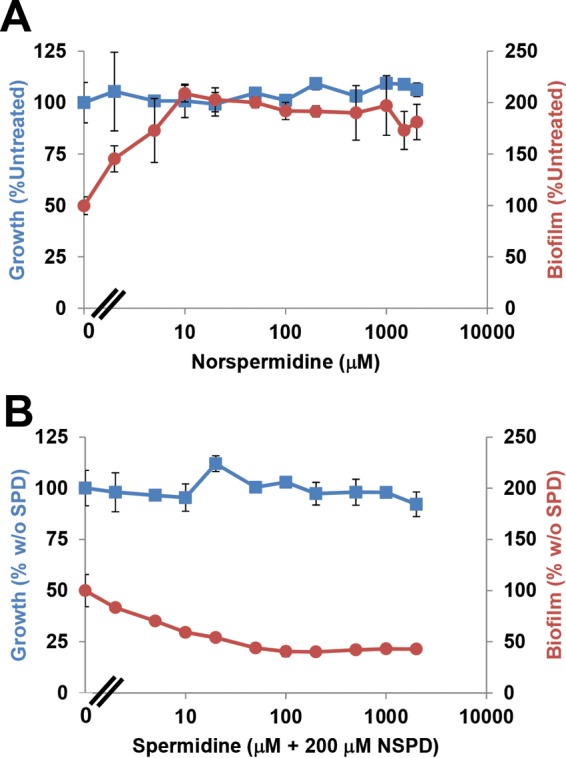
Exogenous norspermidine enhances biofilm formation in the wild type, and spermidine reverses this effect. Shown are growth (blue) and biofilm formation (red) with exogenous norspermidine (NSPD) across the indicated concentration range (A) and 200 μM norspermidine plus spermidine (SPD) across the indicated concentration range (B). Values are presented as percentages of the wild type's growth and biofilm formation in the absence of added polyamines (A) and in the presence of 200 μM norspermidine without added spermidine (w/o SPD) (B). Error bars show the standard deviations from a minimum of three biological replicates.
Both norspermidine and spermidine possess terminal 1,3-diaminopropane moieties. Norspermidine effectively replaced spermidine for growth and also depressed the intracellular levels of spermidine (11). If the depressed levels of spermidine were responsible for the stimulation of biofilm formation, the effect of norspermidine should be titratable by exogenous spermidine. To test this hypothesis, 200 μM norspermidine was added to A. tumefaciens C58 to stimulate biofilm formation, and a range of spermidine concentrations was added to independent assays (0 to 2 mM). The mixture of norspermidine and spermidine, at any concentration tested, did not affect growth (Fig. 5B). Interestingly, as the concentration of spermidine in the culture increased, the elevated biofilm caused by 200 μM norspermidine was proportionally reversed to the level of untreated culture (Fig. 5B).
Biofilm phenotypes of mutants blocked for conversion of putrescine to spermidine.
The A. tumefaciens genome encodes homologues of carboxyspermidine dehydrogenase (CASDH, Atu4170) and carboxyspermidine decarboxylase (CASDC, Atu4169), enzymes required to synthesize spermidine in Campylobacter jejuni (19) and norspermidine in V. cholerae and other bacteria (26). In-frame CASDH and CASDC gene deletion mutants have severe growth defects in minimal medium but are rescued in this respect by exogenous addition of exogenous polyamines that supply the 1,3-diaminopropane group, including spermidine (11). Not surprisingly, the severe growth defect of the CASDH and CASDC mutants in ATGN also manifested itself as a severe biofilm deficiency (Fig. 6). Ectopic expression of plasmids carrying Plac-CASDH or Plac-CASDC was able to correct the growth of the corresponding mutants, and this also returned biofilm formation to wild-type levels. The Plac-CASDH or Plac-CASDC plasmid could not cross-complement mutations in the other gene (see Fig. S4 in the supplemental material), which is consistent with their discrete enzymatic functions.
FIG 6.
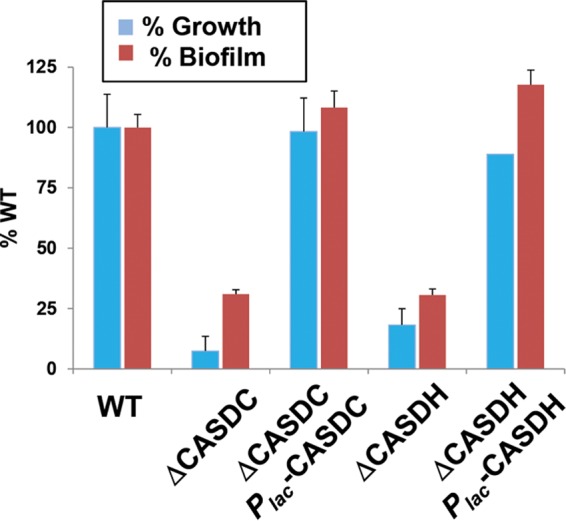
Mutations in the CASDH and CASDC homologues block growth and biofilm formation. Growth was measured by determining OD600 (blue bars), and biofilm formation was measured as A600/OD600 (red bars), assayed at 49 h postinoculation as described in the legend to Fig. 2 for A. tumefaciens C58 and derivatives. Plasmid-harboring strains were induced with 500 μM IPTG. Values are expressed as percentages of the wild type's growth and biofilm formation. Error bars show the standard deviations from a minimum of three biological replicates.
Exogenous spermidine and norspermidine restore prototrophic growth for CASDH and CASDC mutants, but only spermidine inhibits biofilm formation.
Biofilm formation of the wild type, ΔCASDH mutant, and ΔCASDC mutant was examined in the presence of exogenous polyamines. As expected, exogenous putrescine was unable to suppress the growth defect in either mutant (data not shown). In contrast, exogenous spermidine (50 to 100 μM) and norspermidine (5 to 10 μM) fully rescued the growth of the ΔCASDH and ΔCASDC mutants (Fig. 6A and B; see also Fig. S5A and B in the supplemental material). The decreased biofilm formation observed in the two mutants in the absence of exogenous polyamines was clearly due to their severe growth deficiency (cultures that do not grow cannot form dense biofilms). Exogenous putrescine had no effect on these mutants (data not shown). The addition of spermidine to these mutants corrected the growth deficiency and accordingly increased biofilm formation at intermediate (2 to 100 μM) levels (Fig. 7A; see also Fig. S5A). However, at spermidine concentrations above 100 μM, the biofilm formation of the ΔCASDH and ΔCASDC mutants decreased to the wild-type level. In contrast, increasing norspermidine concentrations, while fully restoring growth, did not return biofilm formation to wild-type levels, which is consistent with the observation that this polyamine effectively replaces the spermidine growth requirement but does not regulate surface interactions (Fig. 7B; see also Fig. S5B).
FIG 7.
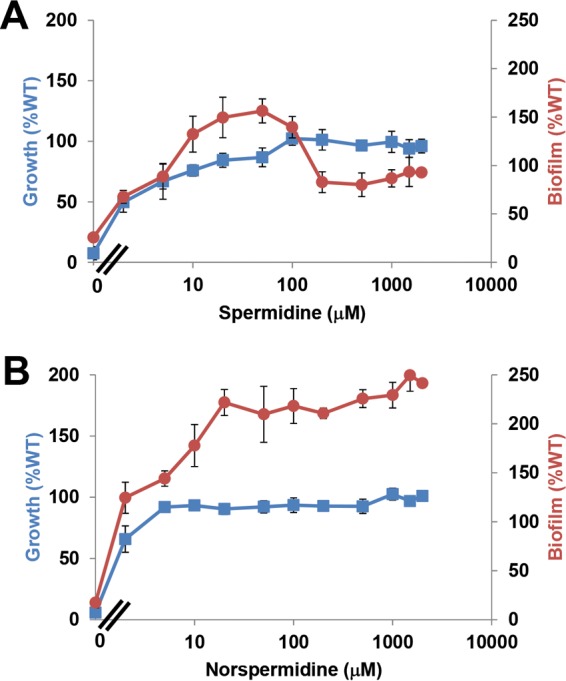
Growth rescue and biofilm control of the CASDC mutant by spermidine and norspermidine. Growth of the ΔCASDC mutant with spermidine (A) and norspermidine (B) measured by OD600 and biofilm formation (A600/OD600) were assayed as described in the legend to Fig. 2. All values are expressed as percentages of the wild type's growth and biofilm formation, and error bars show the standard deviations from a minimum of three biological replicates. For the samples with no supplemental polyamines, A600 readings were used to calculate the ratios instead of A600/OD600 readings to avoid the very low growth due to polyamine auxotrophy from artificially inflating the calculated value.
CASDH and CASDC mutants accumulate homospermidine via a homospermidine synthase homologue.
Analysis of polyamine profiles in odc, CASDH, and CASDC mutants of A. tumefaciens revealed the predicted precursor accumulations of ornithine, putrescine, and carboxyspermidine, respectively (see Fig. S2 in the supplemental material). Unexpectedly, however, the CASDH mutant, and to a lesser extent the CASDC mutant, produced homospermidine (Hspd) (Fig. 8), a symmetrical polyamine with two aminobutyl moieties (Fig. 1), whereas this was undetectable in the wild type and the odc mutant. A. tumefaciens C58 encodes a homospermidine synthase (hss) homologue (Atu3768). Deletion of hss had no effect on polyamine levels in an otherwise-wild-type strain, but in the CASDH mutant, it completely abolished the increased Hspd levels (Fig. 8). The Δhss mutant is also unaffected for prototrophic growth in the wild type, and this mutation does not alter the growth phenotypes of the ΔCASDH mutant (data not shown). Our published work revealed that exogenous Hspd does not rescue the spermidine growth requirement of either the ΔCASDH or the ΔCASDC mutant (11), and correspondingly, the Hspd produced in these mutants is also incapable of rescuing their spermidine auxotrophy. Finally, the Hspd produced in the ΔCASDH and ΔCASDC mutants is not likely to be involved in enhanced biofilm formation under conditions of limiting spermidine, as the Δodc mutant that is intrinsically elevated for biofilm formation does not produce Hspd (see Fig. S2).
FIG 8.
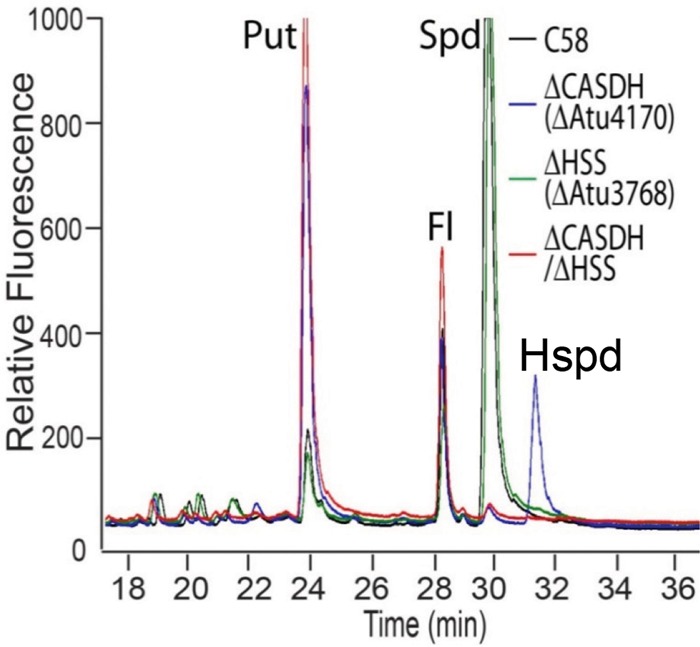
Polyamine profiles from A. tumefaciens mutants for CASDH and homospermidine synthase (hss). Polyamines were prepared from whole-cell extracts, labeled with AccQ-Fluor, and fractionated over a hydrolysate amino acid analysis HPLC column with an acetonitrile gradient. Polyamine and ornithine peaks were detected using an in-line fluorescence detector (excitation, 248 nm; emission, 398 nm). Put, putrescine; Fl, unconjugated AccQ-Fluor; Spd, spermidine; Hspd, homospermidine.
The CASDH mutant accumulates high levels of the 1,4-diaminobutane polyamine putrescine (11), but the CASDC mutant accumulates far less putrescine and predominantly accumulates carboxyspermidine, the pathway intermediate produced by CASDH (Fig. 1). The highly elevated putrescine in the CASDH mutant may conceivably activate the expression of the hss gene. A plasmid-borne translational fusion of the hss region upstream of lacZ (fused at the hss start codon) was tested for expression in the different mutant backgrounds in media depleted of spermidine, and its expression clearly did not significantly increase in the mutant and in fact decreased significantly in the ΔCASDH Δhss double mutant (see Fig. S6 in the supplemental material) (P value < 0.06). This suggests that activation of Hspd biosynthesis in the CASDH mutant does not occur via stimulation of hss transcription or translational initiation.
The Δodc mutant exhibits elevated levels of cyclic di-GMP and can be suppressed for increased attachment by expression of a c-di-GMP PDE.
Several of the mutants that we have isolated that stimulate cellulose and UPP biosynthesis do so by affecting pools of the cellular c-di-GMP second messenger (21). The signal is synthesized by diguanylate cyclases (DGCs) with a GGDEF catalytic motif and is usually broken down by phosphodiesterases (PDEs) with an EAL catalytic motif (27). Comparison of cytoplasmic c-di-GMP levels in the A. tumefaciens Δodc mutant with those in the wild type indicates that the second messenger is elevated 2- to 3-fold in the mutant (P value by t test, <0.1), and this is completely reversed by the plasmid-borne odc gene (P value < 0.1 compared to the mutant), which in fact decreases these levels modestly lower than those in the wild type (Fig. 9A). Exogenous addition of norspermidine (100 μM), which decreases cellular spermidine levels (11), tends toward a slight, observable, but statistically insignificant, c-di-GMP increase in the wild type (P value = 0.19). Upon the addition of exogenous spermidine (100 μM), this increase is reversed significantly less than that observed after norspermidine addition (P value = 0.1)
FIG 9.
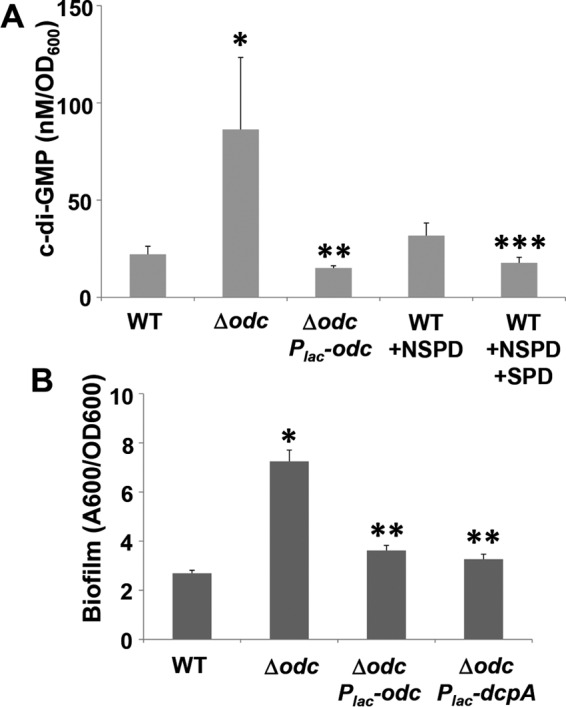
Cyclic di-GMP levels and biofilm formation in the A. tumefaciens odc mutant and in the presence of exogenous spermidine and norspermidine. (A) Cyclic di-GMP levels in whole-culture extracts as measured by HPLC fraction and LC-MS/MS as described in Materials and Methods. Norspermidine and spermidine were added to 100 μM (final concentration) cultures where indicated. Values are results of three independent biological replicates consisting of three technical replicates each. Error bars show standard deviations. *, P < 0.1 compared to the wild type; **, P < 0.1 compared to the Δodc mutant; ***, P value compared to Nspd treatment. (B) Biofilm measurements of specific derivative strains grown for 48 h in the presence of 200 μM IPTG were assayed as described in the legend to Fig. 2. Error bars show the standard deviations from a minimum of three biological replicates. *, P < 0.01 compared to the wild type; **, P < 0.04 compared to the Δodc mutant.
In A. tumefaciens, there are upwards of 30 proteins that contain DGC domains, in some cases also with PDE domains, and several impact cellulose synthesis (21). DcpA is an A. tumefaciens protein with both a DGC domain and a PDE domain that exhibits a predominant PDE activity in the wild-type background under laboratory conditions (28). Ectopic expression of dcpA can often suppress the production of cellulose and other adhesins, such as the UPP, coincidently with lowering c-di-GMP levels. A plasmid-borne Plac-dcpA fusion was introduced into the Δodc mutant, and when fully induced, dcpA had no effect on the diminished growth phenotype but returned biofilm formation to normal levels (Fig. 9B). In parallel, the Plac-odc plasmid complemented the elevated biofilm phenotype and the growth defect in the Δodc mutant.
DISCUSSION
In this study, we have reported the impact of specific polyamines on the attachment of A. tumefaciens to surfaces. Our genetic analyses are complicated by our recent discovery that A. tumefaciens requires spermidine for growth (11). Mutations and the addition of exogenous polyamines that impact spermidine levels can influence both growth and attachment processes. We benefitted from the identification of the odc gene in an unbiased genetic screen for elevated cellulose production in A. tumefaciens, and were particularly fortunate that odc mutants remain viable, even though they grow more slowly. Our analyses indicate that although spermidine is required for growth, decreases in endogenous spermidine levels stimulate production of cellulose and increased adherence via regulatory effects mediated through the second messenger c-di-GMP.
Polyamine synthesis in A. tumefaciens.
The synthesis of spermidine in A. tumefaciens C58 proceeds from ornithine primarily via the activity of the odc gene product to form putrescine. It should be noted that the ODC encoded by A. tumefaciens is homologous to eukaryotic ODCs and not the E. coli speC-encoded ODC, which adopts a different protein fold. The A. tumefaciens Δodc mutant accumulated a large amount of ornithine, the first documentation of such an accumulation (11). The enzymes CASDH and CASDC generate spermidine by a two-step reaction from putrescine and the aminopropyl group donor l-aspartate β-semialdehyde via a predicted carboxyspermidine intermediate (Fig. 1) (11).
The ODC-driven reaction cannot however be the only source of putrescine in A. tumefaciens. Although the growth of the Δodc mutant in the absence of exogenous polyamines is diminished, it is not abolished, as with the auxotrophic CASDC and CASDH mutants (catalyzing conversion of putrescine to spermidine) (Fig. 1), and mutants lacking odc produce a small amount of spermidine (see Fig. S2 in the supplemental material). A potential secondary source was the Atu1673 gene, encoding an alanine racemase fold meso-diaminopimelate decarboxylase-like protein with homology to ODC, although our mutational analysis suggests that it is not responsible for the low level of putrescine observed in the odc mutant (data not shown). After repeated passaging of the odc mutant, suppressors that restore near-normal spermidine synthesis and growth develop. Analysis of these mutants in the future may reveal the alternate source of putrescine.
Our earlier study revealed the presence of homospermidine (Hspd) in cell extracts in the A. tumefaciens CASDH mutant and to a lesser extent in the CASDC mutant, suggesting feedback regulation of the HSS enzyme. Under laboratory conditions, this polyamine is undetectable in the wild type. Such an induction of homospermidine biosynthesis has not been observed previously in bacteria. Some bacteria synthesize Hspd via the activity of a homospermidine synthase (HSS) (or, in other cases, via a deoxyhypusine synthase-like enzyme) that generates the polyamine from two putrescine molecules (29). We found that the A. tumefaciens hss homologue is required for Hspd synthesis in the CASDH mutant. This suggests that blocking spermidine synthesis at the CASDH step strongly directs the flow of putrescine toward Hspd, although analysis of hss expression fails to reveal any increase at the level of hss transcription and translation. Unlike with norspermidine, the Hspd generated in the CASDH mutant cannot replace the spermidine in its growth requirement, as this mutant still manifests a severe growth deficiency. It is noteworthy that several rhizobia do not synthesize spermidine at all but rather synthesize Hspd generated via HSS enzymes (29).
Influence of polyamines on biofilm formation.
Our findings suggest that the limitation for spermidine stimulates production of cellulose and biofilm formation in A. tumefaciens. We were fortunate that our screen identified the odc mutant, which has severely diminished spermidine levels but retains a significant, albeit diminished, level of growth. These findings coincide with several recent studies that indicate a role for polyamines in biofilm formation. In most of these studies, however, spermidine or related polyamines are required for biofilm formation. Spermidine is required for biofilm formation in B. subtilis and Yersinia pestis (8, 9). In B. subtilis, norspermidine can replace spermidine in promoting biofilm formation. For V. cholerae, biofilm formation is stimulated by norspermidine biosynthesis, and this cannot be replaced by spermidine in promoting biofilm formation (4). In contrast, putrescine biosynthesis inhibits biofilm formation in Shewanella oneidensis, a gammaproteobacterium that does not synthesize spermidine (30, 31). In the current study, we found that in A. tumefaciens, spermidine limitation resulted in enhanced attachment, although a complete block of spermidine production prevented growth and thereby diminished biofilm formation overall. Intriguingly, although is it not synthesized de novo in A. tumefaciens, norspermidine could substitute for spermidine, which has an essential role in growth, but could not reverse the spermidine limitation effect on biofilm formation. In fact, the addition of norspermidine on its own to wild-type A. tumefaciens stimulated attachment. Subsequent analysis indicates that the addition of norspermidine depresses spermidine levels (11), and thus, with respect to attachment and biofilm formation, the addition of norspermidine equates to spermidine limitation, such as that in the odc mutant.
For V. cholerae, norspermidine stimulates biofilm formation acting through a periplasmic polyamine-binding protein called NspS (10). Exogenous spermidine has the opposite effect, inhibiting biofilm formation, and this also requires NspS (32). These responses are mediated through a transmembrane protein called MbaA, originally identified as a repressor of biofilm formation (33). MbaA is a GGDEF-EAL protein that directs the turnover of c-di-GMP in V. cholerae and thereby influences production of the Vibrio exopolysaccharide (VPS). MbaA is thought to provide a response to polyamines through association with the NspS periplasmic binding protein (30). Our findings show a similar pattern in A. tumefaciens, in which norspermidine can stimulate biofilm formation, at least partially due to the elevation of cellulose production. The presence of norspermidine causes a significant decrease in spermidine levels, probably due to feedback inhibition (11). Addition of exogenous spermidine reverses the stimulatory effect of norspermidine on biofilm formation down to levels seen in untreated cells. Furthermore, limitation of endogenously produced spermidine, such as in the odc mutant, also stimulates biofilm formation. This mutant exhibits significantly higher levels of c-di-GMP than the wild type, and exogenous polyamines also modulate these levels in the wild type, albeit less dramatically than in the odc mutant. Concordantly, we find that the odc mutant with reduced spermidine levels can be suppressed by ectopic expression of DcpA, a GGDEF-EAL protein with a dominant PDE activity in wild-type A. tumefaciens (28). This suggests that the influence of polyamines on biofilm formation in A. tumefaciens may also function through the modulation of c-di-GMP pools. As with MbaA in V. cholerae, DcpA is predicted to have a large periplasmic domain and may be regulated through interactions with periplasmic binding proteins. NspS is homologous to the periplasmic binding proteins that facilitate the uptake of polyamines, called PotD-type proteins (10). A. tumefaciens C58 encodes several PotD-type proteins in the genome, and it is conceivable that these proteins facilitate interactions with exogenous polyamines or intracellularly produced polyamines that are effluxed from the cytoplasm into the periplasm. Exogenous putrescine, either self-produced or released by other bacteria, has also been shown to function in bacterial cell-cell communication to regulate swarming motility in Proteus mirabilis (34). How and why spermidine and norspermidine function to influence surface interactions in A. tumefaciens and the mechanism by which this is integrated with the c-di-GMP pool is not yet understood, and more work will be required to decipher this process. However, it is well established that plants produce polyamines and can release them into the environment (35). It may be that in A. tumefaciens, the modulation of attachment processes by specific polyamines, either directly or indirectly through a stress response, is integrated into acclimation to the host environment.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Institutes of Health (NIH) under grants GM080546 (C.F.) and GM109259 (C.M.W.) and by a High Impact/High Risk award from UT Southwestern Medical Center (A.J.M). J.E.H. was supported by Ruth L. Kirchstein National Research Service award F32 GM100601.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00265-16.
REFERENCES
- 1.Tabor CW, Tabor H. 1984. Polyamines. Annu Rev Biochem 53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 2.Anton DL, Kutny R. 1987. Escherichia coli S-adenosylmethionine decarboxylase. Subunit structure, reductive amination, and NH2-terminal sequences. J Biol Chem 262:2817–2822. [PubMed] [Google Scholar]
- 3.Tait GH. 1976. A new pathway for the biosynthesis of spermidine. Biochem Soc Trans 4:610–612. doi: 10.1042/bst0040610. [DOI] [PubMed] [Google Scholar]
- 4.Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA, Michael AJ. 2009. An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J Biol Chem 284:9899–9907. doi: 10.1074/jbc.M900110200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki K, Abid MR, Miyazaki M. 1996. Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett 384:151–154. doi: 10.1016/0014-5793(96)00310-9. [DOI] [PubMed] [Google Scholar]
- 6.Pan YH, Liao CC, Kuo CC, Duan KJ, Liang PH, Yuan HS, Hu ST, Chak KF. 2006. The critical roles of polyamines in regulating ColE7 production and restricting ColE7 uptake of the colicin-producing Escherichia coli. J Biol Chem 281:13083–13091. doi: 10.1074/jbc.M511365200. [DOI] [PubMed] [Google Scholar]
- 7.Ong SA, Peterson T, Neilands JB. 1979. Agrobactin, a siderophore from Agrobacterium tumefaciens. J Biol Chem 254:1860–1865. [PubMed] [Google Scholar]
- 8.Patel CN, Wortham BW, Lines JL, Fetherston JD, Perry RD, Oliveira MA. 2006. Polyamines are essential for the formation of plague biofilm. J Bacteriol 188:2355–2363. doi: 10.1128/JB.188.7.2355-2363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrell M, Hanfrey CC, Murray EJ, Stanley-Wall NR, Michael AJ. 2010. Evolution and multiplicity of arginine decarboxylases in polyamine biosynthesis and essential role in Bacillus subtilis biofilm formation. J Biol Chem 285:39224–39238. doi: 10.1074/jbc.M110.163154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karatan E, Duncan TR, Watnick PI. 2005. NspS, a predicted polyamine sensor, mediates activation of Vibrio cholerae biofilm formation by norspermidine. J Bacteriol 187:7434–7443. doi: 10.1128/JB.187.21.7434-7443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim SH, Wang Y, Khomutov M, Khomutov A, Fuqua C, Michael AJ. 2016. The essential role of spermidine in growth of Agrobacterium tumefaciens is determined by the 1,3-diaminopropane moiety. ACS Chem Biol 11:491–499. doi: 10.1021/acschembio.5b00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 13.Mersereau M, Pazour GJ, Das A. 1990. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 90:149–151. doi: 10.1016/0378-1119(90)90452-W. [DOI] [PubMed] [Google Scholar]
- 14.Tempé J, Petit A, Holsters M, Van Montagu M, Schell J. 1977. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc Natl Acad Sci U S A 74:2848–2849. doi: 10.1073/pnas.74.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci U S A 96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Haitjema CH, Fuqua C. 2014. The Ctp type IVb pilus locus of Agrobacterium tumefaciens directs formation of the common pili and contributes to reversible surface attachment. J Bacteriol 196:2979–2988. doi: 10.1128/JB.01670-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Warrens AN, Jones MD, Lechler RI. 1997. Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186:29–35. doi: 10.1016/S0378-1119(96)00674-9. [DOI] [PubMed] [Google Scholar]
- 18.Khan SR, Gaines J, Roop RM II, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl Environ Microbiol 74:5053–5062. doi: 10.1128/AEM.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanfrey CC, Pearson BM, Hazeldine S, Lee J, Gaskin DJ, Woster PM, Phillips MA, Michael AJ. 2011. Alternative spermidine biosynthetic route is critical for growth of Campylobacter jejuni and is the dominant polyamine pathway in human gut microbiota. J Biol Chem 286:43301–43312. doi: 10.1074/jbc.M111.307835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc Natl Acad Sci U S A 109:12746–12751. doi: 10.1073/pnas.1115663109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, Kim J, Koestler BJ, Choi JH, Waters CM, Fuqua C. 2013. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol 89:929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amikam D, Benziman M. 1989. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol 171:6649–6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Kim J, Danhorn T, Merritt PM, Fuqua C. 2012. Phosphorus limitation increases attachment in Agrobacterium tumefaciens and reveals a conditional functional redundancy in adhesin biosynthesis. Res Microbiol 163:674–684. doi: 10.1016/j.resmic.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. 2001. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science 294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 25.Wood DW, Setulab JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NFJ, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Dovee DS, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li M-J, McClellund E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao Z-Y, Dolan M, Chumley F, Tingey SV, Tomb J-F, Gordon M, Olson MV, et al. . 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 26.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol 75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: c-di-GMP signaling. Curr Opin Microbiol 15:140–146. doi: 10.1016/j.mib.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feirer N, Xu J, Allen KD, Koestler BJ, Bruger EL, Waters CM, White RH, Fuqua C. 2015. A pterin-dependent signaling pathway regulates a dual-function diguanylate cyclase-phosphodiesterase controlling surface attachment in Agrobacterium tumefaciens. mBio 6:e00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw FL, Elliott KA, Kinch LN, Fuell C, Phillips MA, Michael AJ. 2010. Evolution and multifarious horizontal transfer of an alternative biosynthetic pathway for the alternative polyamine sym-homospermidine. J Biol Chem 285:14711–14723. doi: 10.1074/jbc.M110.107219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cockerell SR, Rutkovsky AC, Zayner JP, Cooper RE, Porter LR, Pendergraft SS, Parker ZM, McGinnis MW, Karatan E. 2014. Vibrio cholerae NspS, a homologue of ABC-type periplasmic solute binding proteins, facilitates transduction of polyamine signals independent of their transport. Microbiology 160:832–843. doi: 10.1099/mic.0.075903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding Y, Peng N, Du Y, Ji L, Cao B. 2014. Disruption of putrescine biosynthesis in Shewanella oneidensis enhances biofilm cohesiveness and performance in Cr(VI) immobilization. Appl Environ Microbiol 80:1498–1506. doi: 10.1128/AEM.03461-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGinnis MW, Parker ZM, Walter NE, Rutkovsky AC, Cartaya-Marin C, Karatan E. 2009. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol Lett 299:166–174. doi: 10.1111/j.1574-6968.2009.01744.x. [DOI] [PubMed] [Google Scholar]
- 33.Bomchil N, Watnick P, Kolter R. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J Bacteriol 185:1384–1390. doi: 10.1128/JB.185.4.1384-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sturgill G, Rather PN. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol Microbiol 51:437–446. doi: 10.1046/j.1365-2958.2003.03835.x. [DOI] [PubMed] [Google Scholar]
- 35.Kusano T, Berberich T, Tateda C, Takahashi Y. 2008. Polyamines: essential factors for growth and survival. Planta 228:367–381. doi: 10.1007/s00425-008-0772-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


