Summary
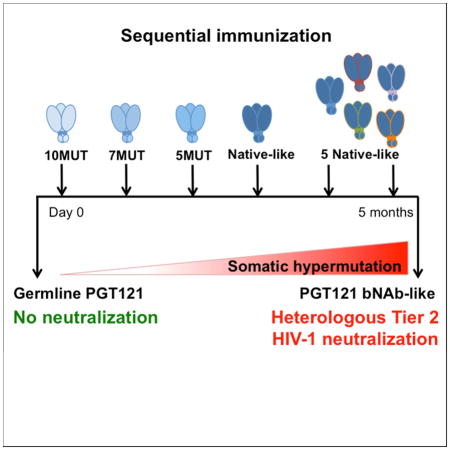
A vaccine that elicits broadly neutralizing antibodies (bNAbs) against HIV-1 is likely to be protective, but this has not been achieved. To explore immunization regimens that might elicit bNAbs, we produced and immunized mice expressing the predicted germline of PGT121, a bNAb specific for the V3-loop and surrounding glycans on the HIV-1 spike. Priming with an epitope modifiied immunogen designed to activate germline antibody-expressing B cells, followed by ELISA-guided boosting with a sequence of directional immunogens, native-like trimers with decreasing epitope modification, elicited heterologous tier-2 neutralizing responses. In contrast, repeated immunization with the priming immunogen did not. Antibody cloning confirmed elicitation of high levels of somatic mutation and tier-2 neutralizing antibodies resembling the authentic human bNAb. Our data establishes that sequential immunization with specifically designed immunogens can induce high levels of somatic mutation and shepherd antibody maturation to produce bNAbs from their inferred germline precursors.
eTOC
Generation of HIV broadly neutralizing antibodies - a major goal in vaccine research - can be achieved through sequential immunization with HIV envelope glycoproteins designed to engage PGT121 antibody precursors and their intermediates.
Introduction
HIV-1 infected humans occasionally develop antibodies that are capable of broad and potent viral neutralization. Single cell antibody cloning methods revealed that this serologic activity is due to one or a combination of monoclonal antibodies that target several different epitopes on the HIV-1 spike protein gp160 (Scheid et al., 2009; Walker et al., 2009; West et al., 2014; Wu et al., 2010). When passively transferred into humanized mice or macaques, bNAbs protect against infection for prolonged periods of time (Gautam et al., 2016; Klein et al., 2012; Mascola et al., 2000; Moldt et al., 2012; Shibata et al., 1999; Shingai et al., 2013) Therefore, it is generally accepted that a vaccine that elicits such antibodies would be protective (Pantaleo and Koup, 2004). However, after more than 25 years and innumerable pre-clinical studies and human vaccine trials, bNAbs have not been elicited by immunization (Burton et al., 2004; Cohen and Dolin, 2013; Schiffner et al., 2013).
In addition to uncovering new sites of vulnerability on the HIV-1 spike, antibody cloning revealed several unusual features of anti-HIV-1 antibodies. These include long complementarity determining region 3s (CDR3s), a higher than expected propensity to polyreactivity, and an unusually high level of somatic hypermutation (SHM) (Dimitrov, 2010; Kwong et al., 2013; Mascola and Haynes, 2013; Scheid et al., 2009; West et al., 2014). Irrespective of their targets on the HIV-1 spike, the high level of somatic hypermutation is the most conserved amongst these unusual features of bNAbs. Moreover, mutation is essential for bNAb activity (Bonsignori et al., 2011; Klein et al., 2013a; Mouquet et al., 2012; Mouquet et al., 2010; Scheid et al., 2011; Sok et al., 2013; Zhou et al., 2010) and within longitudinal antibody lineages, the level of somatic mutation is directly correlated with antibody activity (Bhiman et al., 2015; Doria-Rose et al., 2014; Klein et al., 2013a; Liao et al., 2013; Sok et al., 2013; Wu et al., 2015; Wu et al., 2011).
Hypermutation occurs in germinal centers where B-lymphocytes undergo clonal expansion and affinity based selection in response to antigen (Victora and Nussenzweig, 2012). Under normal circumstances, the germinal center reaction is limited by antigen availability and by an antibody affinity ceiling (Baumjohann et al., 2013; Foote and Eisen, 1995; Goodnow et al., 2010). However, longitudinal studies of HIV-1 antibody evolution have shown that HIV-1 can escape immune pressure by mutation and selection, resulting in a continually evolving antigen that produces a persistent immune response to a highly variable infection (Bhiman et al., 2015; Doria-Rose et al., 2014; Kwong et al., 2002; Liao et al., 2013; Stewart-Jones et al., 2016). Consistent with this notion, prospective studies of HIV-1 infected humans that develop bNAbs revealed interdependence between changes in viral and the antibody sequences (Bhiman et al., 2015; Doria-Rose et al., 2014; Liao et al., 2013; Richman et al., 2003; Wei et al., 2003). Thus, as initially proposed (Scheid et al., 2009), the high levels of somatic mutation found in bNAbs are most consistent with continuing parallel evolution of the virus and the antibody response (Klein et al., 2013a; Klein et al., 2013b; Mouquet et al., 2012; Mouquet et al., 2010; Scheid et al., 2009).
PGT121 and related antibodies target a region of the HIV-1 spike that is heavily glycosylated (Kong et al., 2013). Their target epitopes are comprised of the GlyAspIleArg (GDIR) sequence at the base of the V3 loop, and surrounding glycans at positions Asn332, Asn156, Asn301, and Asn137 (N332, N156, N301, N137) (Garces et al., 2015; Garces et al., 2014; Julien et al., 2013; Mouquet et al., 2012; Sok et al., 2014; Walker et al., 2011; Walker et al., 2009). These antibodies are commonly found in individuals that develop bNAbs during natural infection (Mouquet et al., 2012; Walker et al., 2011; Walker et al., 2010). In addition, they are also amongst the most potent bNAbs discovered to date (Kwong et al., 2013).
PGT121 antibodies show high levels of somatic mutations and arise from a singular precursor that diverges into two distinct families, exemplified by PGT121 (Walker et al., 2011) and 10-1074 or PGT124 (Mouquet et al., 2012; Sok et al., 2013). Somatic mutations differentiate the PGT121 lineage that recognizes glycans at N301, N137, and N156 from the 10-1074 or PGT124 lineage that is highly dependent on N332 but less so on the other glycans surrounding the base of the V3 loop (Garces et al., 2015; Garces et al., 2014; Mouquet et al., 2012; Sok et al., 2014; Sok et al., 2013). Finally, like other bNAbs, the inferred germline precursor of PGT121 antibodies shows no appreciable affinity for all HIV-1 Env antigens tested, and no neutralizing activity (Andrabi et al., 2015; Doria-Rose et al., 2014; Gorman et al., 2016; Klein et al., 2013a; Liao et al., 2013; Mouquet et al., 2012; Sok et al., 2013).
Based on the requirements for unusually high levels of somatic mutation in bNAbs, and the inability of germline reverted antibodies to bind to native-like antigens, it was proposed that a vaccine to elicit these antibodies would necessitate a new paradigm involving a germline prime followed by sequential immunization with different antigens (Dimitrov, 2010; Haynes et al., 2012; Jardine et al., 2013; Klein et al., 2013a; Klein et al., 2013b; McGuire et al., 2013; Mouquet et al., 2012; Pancera et al., 2010; Scheid et al., 2009; Scheid et al., 2011; Zhou et al., 2010). To test this idea, we have sequentially immunized mice that carry the germline reverted version of the PGT121 family precursor with a series of specifically engineered antigens (Steichen et al., 2016 submitted).
Results
PGT121 Mice
Mice carrying the Ig V(D)Js encoding the inferred germline or fully mature IgH, and the predicted germline IgL of PGT121 (GLH121, MutH121, and GLL121 respectively) were produced by gene targeting Albino B6 (B6 (Cg)-Tyrc-2J/J) embryonic stem cells (Figure S1A)(Pelanda et al., 1997; Shih et al., 2002). The GL121 sequence contains germline V, D and J genes, and the naturally occurring CDRH3 sequence corresponding to the least mutated antibodies in this lineage (Kepler, 2013; Mouquet et al., 2012). Mice carrying GLH121 and MutH121 were bred with GLL121 to produce GLHL121 and MutHGLL121 mice (Mouquet et al., 2012). MutHGLL121 represents a synthetic intermediate of the PGT121 antibody wherein the IgH is fully mature. Nevertheless, induction of IgL maturation by immunization is ambitious because many of the essential contacts between PGT121 and the HIV-1 spike are dependent on IgL somatic mutations.
GLHL121 and MutHGLL121 mice showed normal numbers of splenocytes and near normal frequencies of marginal zone and follicular B cells in the spleen as measured by flow cytometry (Figure S1B,C). To examine the antibody repertoire expressed by naïve B cells in GLHL121 and MutHGLL121 mice, we purified single B cells by cell sorting and cloned their expressed IgVH and IgVL from cDNA (Dosenovic et al., 2015); von Boehmer et al, in press). In both genotypes the antibodies were composed exclusively of products of the knocked-in antibody genes (Figure S1D). We conclude that the majority of the B cells in GLHL121 and MutHGLL121 mice express the respective knock-in gene and develop normally.
Activation of GLHL121 B cells in vivo
Germline reverted PGT121 does not bind to native-like Env antigens and shows no neutralizing activity for all HIV-1 isolates tested (Klein et al., 2013a; Mouquet et al., 2012; Sok et al., 2013). Consistent with this observation, the serum of naïve GLHL121 and MutHGLL121 mice did not bind to the native-like Env protein BG505 SOSIP in ELISA (Figure S2A,B). Moreover, repeated immunization with native-like Env proteins YU2 SOSIP, BG505 SOSIP, or other Env-based immunogens such as BG505-5MUT and BG505-3MUT (5- and 3-MUT respectively (Steichen et al., submitted)) failed to elicit antibody responses in GLHL121 mice (Figure S2A,B). These results are in agreement with the observation that native-like Env immunogens failed to induce immune responses in knock-in mice that carry the inferred germline heavy chains of 3BNC60 or VRC01, and with the idea that activating germline bNAb expressing B cells requires specifically engineered antigens (Dosenovic et al., 2015; Hoot et al., 2013; Jardine et al., 2013; Jardine et al., 2016; Jardine et al., 2015; McGuire et al., 2013; Mouquet et al., 2010; Scheid et al., 2009; Xiao et al., 2009).
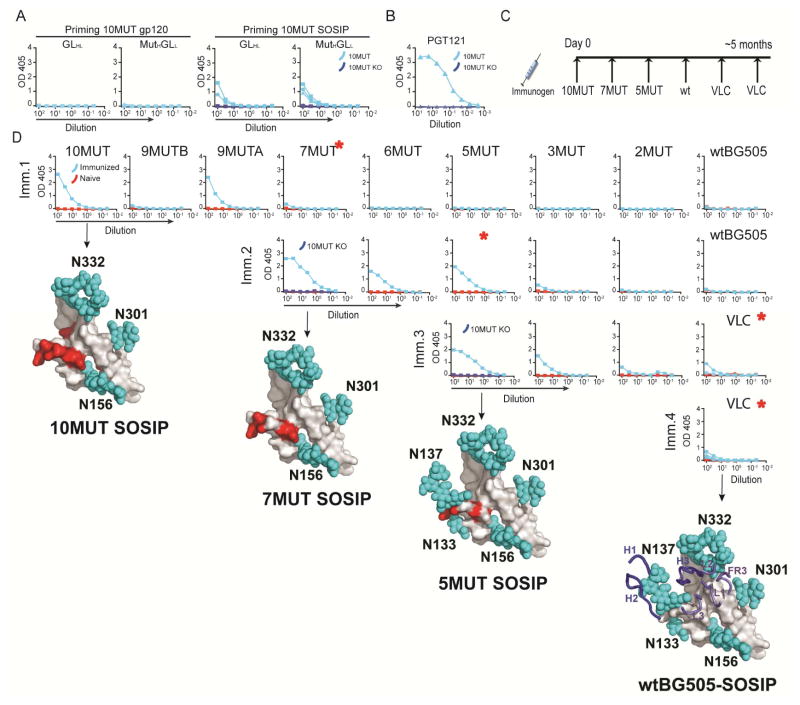
To attempt to activate B cells that carry PGT121 antibody precursors, we immunized GLHL121 and MutHGLL121 mice with BG505-10MUT (10MUT), a BG505 variant that was specifically engineered to bind to germline reverted PGT121 antibodies (Steichen et al., 2016). 10MUT binds to MutHGLL with a Kd =47000nM whereas it has no measurable affinity to GLHL121 (Figure S2C) by surface plasmon resonance. Despite the lack of measurable affinity to GLHL121, a single immunization with 10MUT trimer, but not the corresponding monomer, elicited antibody responses in both GLHL121 and MutHGLL121 mice as measured by ELISA (Figure 1A). Moreover, the antibodies induced by immunization of both types of mice specifically targeted the PGT121 epitope on Env, because binding to 10MUT was eliminated by mutation at positions N332, N301 and H330A (10MUT KO) (Figure 1A, and B). Despite the specificity of the response, immunization with 10MUT did not elicit neutralizing activity (Figure S2D.). We conclude that an engineered Env based BG505 SOSIP immunogen, 10MUT, can initiate antibody responses in GLHL121 and MutHGLL121 mice but it does not elicit bNAbs.
Figure 1.
ELISA-guided sequential immunization. (A) ELISAs against 10MUT gp120 and a mutated version of the same protein that is missing the PGT121 epitope (10MUT gp120 KO) using serum (1/100 starting dilution) from GLHL121 and MutHGLL121 mice primed with monomeric 10MUT (10MUT gp120, left) or trimeric 10MUT (10MUT SOSIP, right). (B) ELISA using human PGT121 (5μg/ml) against 10MUT gp120 and 10MUT gp120 KO. (C) Diagram shows the sequential immunization protocol delivered over a period of 5–6 months. (D) ELISAs of serum from a control and a sequentially immunized GLHL121 mouse after immunization steps 1–4. Test proteins are indicated above each column. ELISAs are ordered from left to right according to level of capture antigen modification compared to wtBG505. When indicated, the specificity of the serologic response for the PGT121 epitope was also evaluated by ELISA against the 10MUT gp120 KO protein. Each row of ELISA graphs represents the indicated step in the immunization protocol. A model of the corresponding immunogen is shown below each row (Steichen et al., submitted). Blue spheres represent glycans and red spheres indicate mutated residues. IgH (H1-3) and IgL (L1-3) loops are represented in the model of wtBG505. Red asterisks indicate the boosting immunogens. VLC= 5 native-like SOSIPs differing in the variable loops (Steichen et al, submitted). See also Figure S1 and S2.
Sequential immunization in GLHL121 and MutHGLL121 mice
To test whether a sequential immunization strategy can shepherd GLHL121 and MutHGLL121 antibodies to develop neutralizing activity, we immunized with a succession of Env-based immunogens that were developed as intermediates between 10MUT and native-like antigens and showed varying affinities (1–10 nM) for the mature PGT121 antibody (Figure 1C,D) (Steichen et al., submitted). ELISA testing of mouse serum on these proteins after each immunization, and consideration of the directionality engineered into the immunogens, determined the sequence of boosting immunogens (Figure 1C,D). For example, we selected 7MUT for boosting after immunization with 10MUT because 7MUT was the least mutated immunogen that showed detectable binding to the serum of one of the immunized mice by ELISA (Figure 1D). Subsequent immunogens were selected in the same manner (Figure 1D).
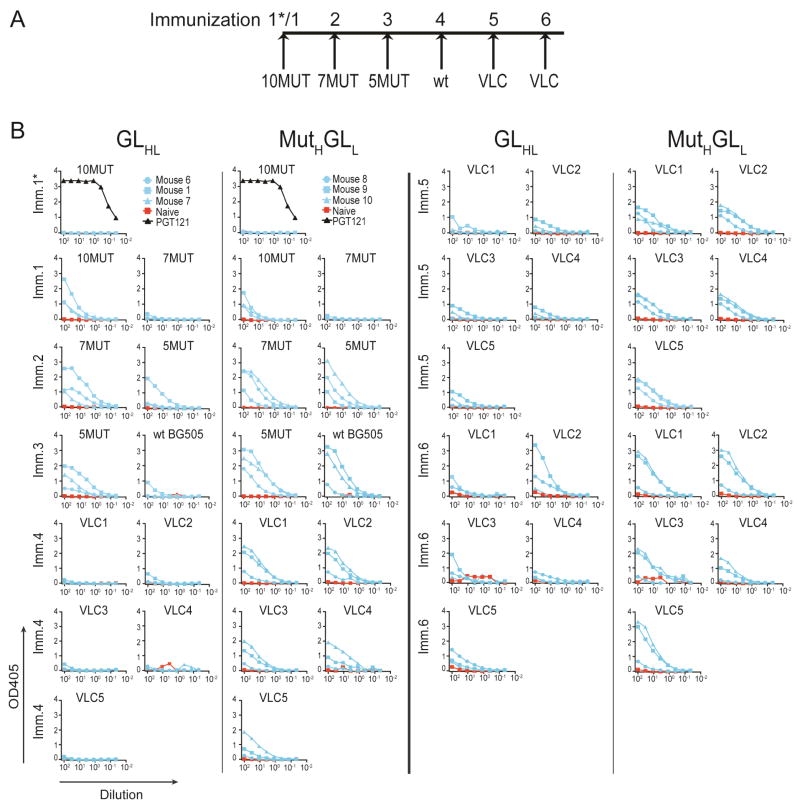
Longitudinal ELISA testing of the serum from sequentially immunized mice revealed that antibody responses were boosted in response to each of the immunogens while remaining specific for the PGT121 epitope (Figure 1D, Figure 2A,B, Figure S2E,F and data not shown). After immunization with 5MUT, the serum of 1 GLHL121 and all 7 MutHGLL121 mice showed binding to BG505 SOSIP in ELISAs (Figure 2B and data not shown), and though serum reactivity to 3MUT and 2MUT was also detected, immunization with either of those proteins at this stage would have violated directionality because both lack native PGT121-epitope glycans that are present in 5MUT and BG505 SOSIP (Steichen et al., 2016). Thus the BG505 SOSIP was used as a boost after 5MUT and then followed by a cocktail of 5 native-like SOSIPs differing in their variable loops (Variable Loop Cocktail or VLC)(Steichen et al., submitted). After the final immunization, all but one of the GLHL121 and all of the MutHGLL121 mice showed binding to the 5 native-like VLCs (Figure 2B and data not shown).
Figure 2.
Longitudinal analysis of the serum from sequentially immunized GLHL121 and MutHGLL121 mice by ELISA. (A) Diagram of the sequential immunization protocol. (B) Graphs show the results of ELISAs on serum (1/100 starting dilution) collected from naïve and sequentially immunized GLHL121 (3 mice) and MutHGLL121 (3 mice). ELISAs show reactivity against the immunogen and the boosting protein after each step. When indicated, ELISA binding of the human PGT121 bNAb is also shown (1/100 starting dilution of a 500μg/ml solution=5μg/ml). VLC= 5 different native-like SOSIPs. See also Figure S2.
Broadly Neutralizing Antibodies
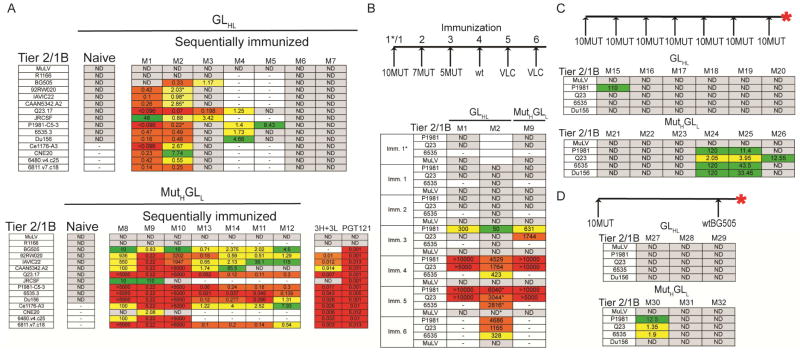
To determine whether sequential immunization elicited anti-HIV-1 bNAbs, purified serum antibodies from control, and immunized mice, were tested for neutralizing activity by TZM-bl assay using a panel of 12 tier 2, and 2 tier 1B viruses (Figure 3A). Igs from naïve control mice showed no neutralizing activity. In contrast, 5/7 GLHL121 and all 7 of the MutHGLL121 mice showed cross clade neutralization against heterologous tier 2 isolates (Figure 3A). As expected, none of the purified serum Igs samples neutralized HIV-1R1166, which lacks the N332 N-linked glycosylation site that is critical for the binding and in vivo activity of PGT121 antibodies (Figure 3A)(Klein et al., 2013a; Klein et al., 2012; Mouquet et al., 2012; Walker et al., 2011).
Figure 3.
Neutralization in the serum of immunized GLHL121 and MutHGLL121 knock-in mice. (A) TZM-bl neutralization activity (IC50 or 1/ID50 in mice M8 and M10) in the serum of naïve and sequentially immunized GLHL121 (M1–M7, top) and MutHGLL121 (M8–12, bottom) mice against a panel of 12 Tier 2 and 2 tier 1B HIV-1 isolates. Samples with asterisk correspond to step 5 in the immunization, all others were measured after complete protocol. The activity of the human PGT121 bNAb and the inferred intermediate 3H+3L against the same panel of viruses is shown. (B) Longitudinal analysis of the neutralization activity in the serum of sequentially immunized GLHL121 and MutHGLL121 mice. Table shows TZM-bl neutralization activity (1/ID50) against MuLV control and 3 Tier2/1B HIV-1 isolates for 1/50 dilutions of serum from 2 GLHL121 (M1–M2) and 1 MutHGLL121 (M9) mice collected after each immunization as indicated (left). Samples with an asterisk correspond to purified Igs and not serum. (C) Diagrammatic representation of the immunization protocol (top). Table shows TZM-bl neutralization activity (IC50) in the serum of GLHL121 and MutHGLL121 mice immunized 7 times with 10MUT. (D) Diagrammatic representation of the immunization protocol (top). Table shows TZM-bl neutralization activity (IC50) in the serum of GLHL121 and MutHGLL121 mice immunized with 10MUT followed by a boost with wtBG505. IC50 < 0.096 in red; 0.096- 0.5 in orange; 0.5- 4 in yellow; > 4 green; 1/ID50 > 5000 in red; 5000-1000 in orange; 1000-100 in yellow; <100 in green; not detectable (ND) in gray; not tested indicated by a dash. Red asterisk indicates the time point of analysis. See also Figure S3 and S4.
To further evaluate the breadth of the serologic responses, purified Ig from one GLHL121 and one MutHGLL121 mouse was assayed against a panel of 54 tier 2 and 2 tier 1B viruses. Whereas PGT121 neutralized 50 of these isolates, GLHL121 and MutHGLL121 Igs neutralized 12 and 23 tier 2 HIV-1 isolates respectively (Figure S3). Thus, the serum Igs isolated from sequentially immunized mice showed strong cross-clade tier 2 neutralizing activity, but less breadth than PGT121.
To determine when neutralization activity emerged, we assayed intermediate time points in the sequential immunization series. In both GLHL121 and MutHGLL121 mice, neutralization was first detected after immunization with 5MUT (Figure 3B). However, this activity was less broad and less potent than that found after the complete immunization series (Figure 3A,B).
To determine whether repeated doses of a single antigen or a simplified protocol can also elicit bNAbs, we immunized repeatedly with the priming antigen 10MUT, or alternatively with 10MUT followed by BG505 separated by a period of time equivalent to the original sequential protocol. Analysis of the serum from the GLHL121 mice by ELISA showed no significant binding to native-like Env proteins (Figure S4A,B). Repeated immunization with 10MUT elicited some weak neutralization activity in 3/6 MutHGLL121 mice and little or no detectable activity in any of the GLHL121 mice (Figure 3C). Similarly, 10MUT followed by BG5050 was entirely ineffective in GLHL121 mice and produced only weak neutralizing activity in 1/3 MutHGLL121 mice (Figure 3D). We conclude that repeated immunizations with 10MUT, or the combination of 10MUT and BG505 in absence of intermediate antigens, is far less effective than sequential immunization with different antigens.
To determine whether an abbreviated immunization protocol using some of the same antigens would also elicit bNAbs, we primed with 7MUT, and boosted with 5MUT, followed by wtBG505 and VLCs (Figure S4C). Analysis of the serum by ELISA revealed that this protocol was less effective in eliciting antibodies with binding affinity to native-like Env proteins (Figure S4C). Consistent with the ELISA data, the abbreviated protocol produced less potent neutralizing activity, and in only 1/3 GLHL121 and the 3 MutHGLL121 mice (Figure S4D). Similarly, priming with 11MUTA, a higher affinity variant of 10MUT followed by 5MUT, wtBG505, and VLCs elicited only weak ELISA activity against native-like proteins and low levels of neutralizing antibodies in MutHGLL121 and none in GLHL121 mice (Figure S4E–F).
We conclude that sequential immunization guided by ELISA measurements is effective in inducing maturation of PGT121 germline antibodies to become bNAbs in Ig knock-in mice.
Antibody Sequences from sequentially immunized mice
To characterize the antibodies elicited by sequential immunization, we cloned them from purified single IgG+ memory B cells binding to a mixture of 3 different gp120 proteins but not to mutants of these proteins that do not bind to PGT121.
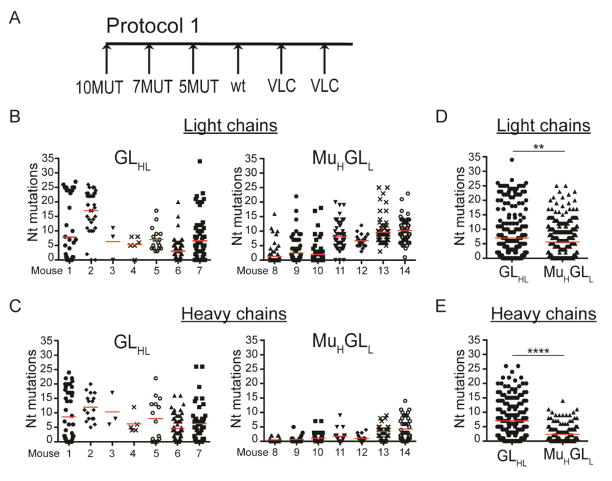
Sequential immunization with a series of related antigens induced remarkably high levels of somatic mutation in the IgL of the knock-in mice, reaching a maximum of 34 and 25 nucleotide, or 21 and 18 amino acid mutations in GLHL121 and MutHGLL121 B cells respectively (Figure 4A,B,D and Figure S5A,B,D). In contrast, high levels of IgH mutation were only found in GLHL121 and not in pre-mutated 121 IgH in MutHGLL121 mice (Figure 4C,E and Figure S5C,E). Thus, although the somatic mutation machinery was similarly active in the germinal center B cells of both knock-in mice, the MutH121 was less susceptible to additional mutation, possibly because some of the target sequences for activation induced cytidine deaminase (AID) had already been mutated (Figure S5F) (Dosenovic et al., 2015); (Pavri and Nussenzweig, 2011). Finally, despite the high level of mutation, we found no deletions or insertions in the IgH or IgL of either of the immunized mice (Kepler et al., 2014).
Figure 4.
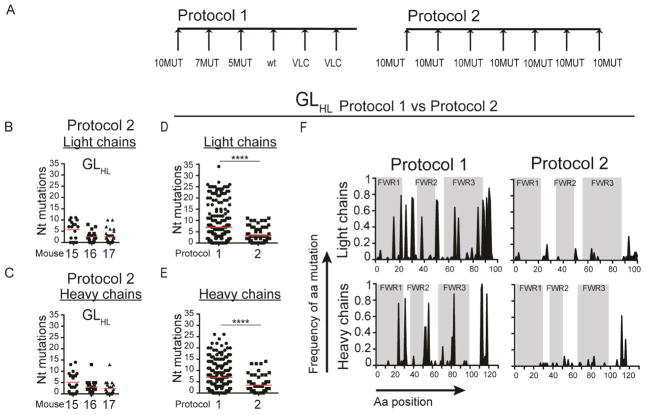
Somatic mutations in antibody sequences from immunized mice. (A) Diagram of the sequential immunization protocol (Protocol 1). (B) Number of nucleotide mutations in the IgL of antigen specific single B cells isolated from sequentially immunized GLHL121 (left) and MutHGLL121 (right) mice. Each column represents one mouse and each dot one B cell. (C) Number of nucleotide mutations in the IgH sequences of antigen specific single B cells isolated from sequentially immunized GLHL121 (left) and MutHGLL121 (right) mice. Each column represents one mouse and each dot one B cell. (D) Number of nucleotide mutations in the IgL obtained from sequentially immunized GLHL121 and MutHGLL121 mice. Data represent pooled sequences for each genotype. **p< 0.01. (E) Number of nucleotide mutations in the heavy chain sequences obtained from sequentially immunized GLHL121 and MutHGLL121 mice. Data represent pooled sequences for each genotype. ****p< 0.0001. Nt= Nucleotide. See also Figure S5 and S7.
To determine whether induction of high levels of mutation requires sequential immunization we isolated single antigen specific IgG+ memory B cells from mice immunized repeatedly with 10MUT or 10MUT followed by wtBG505. Multiple immunizations with 10MUT, or 10MUT followed by BG505 induced significantly lower levels of SHM at both the nucleotide and amino acid levels in GLHL121 and MutHGLL121 B cells than the sequential protocol (Figure 5A–F, Figure S5G,H,I). We conclude that repeated immunizations with one or two antigens fails to elicit the high levels of mutation found after sequential immunization.
Figure 5.
Somatic mutations after repeated or sequential immunization. (A) Diagram of the sequential or repeated immunization protocols (Protocol 1 and 2 respectively). (B) Number of nucleotide mutations in the IgL of single antigen specific B cells isolated from individual GLHL121 mice immunized repeatedly with 10mut. Each column represents one mouse and each dot one B cell. (C) Number of nucleotide mutations in the IgH of single antigen specific B cells isolated from individual GLHL121 mice immunized repeatedly with 10MUT. Each column represents one mouse and each dot one B cell. (D) Comparison of the number of nucleotide mutations in IgL obtained from GLHL121 mice immunized sequentially (7 mice pooled, protocol 1) or repeatedly with 10MUT (3 mice pooled, protocol 2) ****p< 0.0001. Each dot represents one B cell (E) Comparison of the number of nucleotide mutations in heavy chains obtained from GLHL121 mice immunized sequentially (7 mice pooled) or repeatedly (3 mice pooled) ****p< 0.0001. Each column represents one mouse and each dot one B cell. (F) Frequency of mutation per amino acid position in IgL (top) and IgH (bottom) of two representative mice immunized sequentially (M2, protocol 1) or repeatedly (M16, protocol 2). Framework regions (FWR) are shaded in gray and white areas correspond to complementarity determining regions (CDRs). Nt= Nucleotide; Aa= Amino acid. See also Figure S5 and S7.
Neutralization by monoclonal antibodies
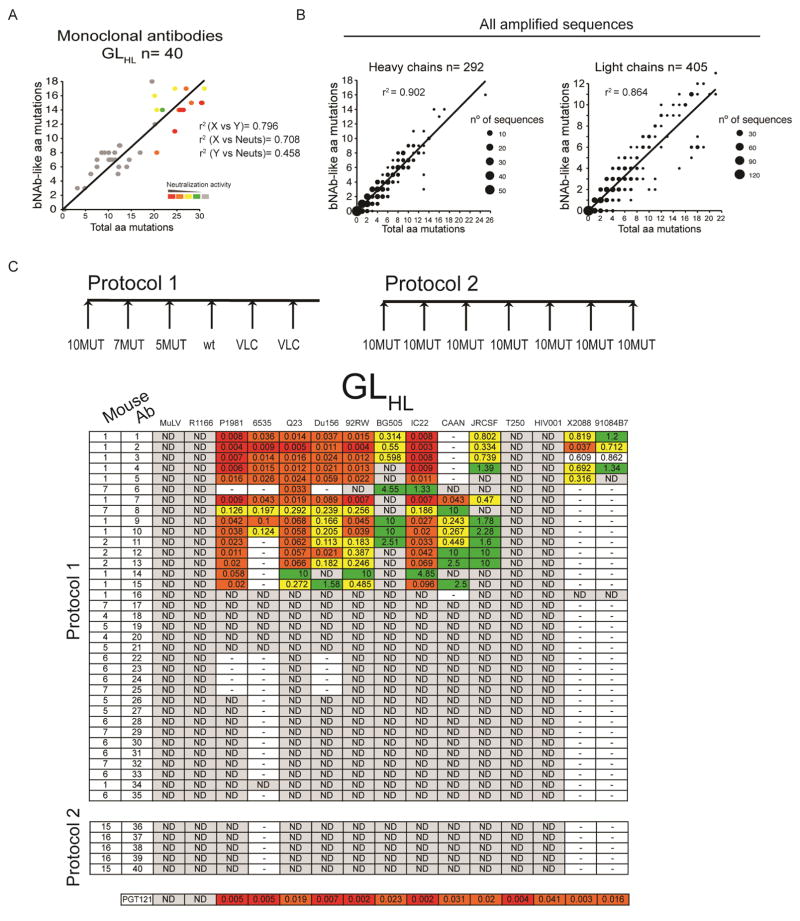
Antibodies from immunized mice showed somatic mutations that were in some cases identical or similar to mutations in PGT121, PGT122, PGT123 or 10-1074 (PGT121-like). Indeed there was a direct correlation between the total number of amino acid mutations and the number of amino acid substitutions that were similar to those found in the bNAbs (r2= 0.796, Fig. 6A and B). To further characterize individual antibodies we produced 77 monoclonal antibodies (40 from GLHL121 and 37 from MutHGLL121 mice), and tested them for neutralization against a panel of 12 tier 2 and 2 tier 1B viruses (Figure 6C, Figure 7 and Figure S6C). As a control we cloned 5 antibodies from GLHL121 mice and other 5 from MutHGLL121 mice immunized repeatedly with 10MUT (Figure 6, Figure 7 and Figure S6).
Figure 6.
Monoclonal antibody neutralizing activity. (A) 3D plot showing neutralization activity (color coded), total number of amino acid mutations (X axis) and the number of mutations that are identical or chemically equivalent to mutations in the human PGT121, PGT122, PGT123 or 10-1074 bNAbs (bNAb-like) (Y axis) for all monoclonal antibodies from GLHL121 mice immunized according to Protocol 1 and 2. Chemical equivalence is as in (B). Each antibody was assigned a neutralization score (see Materials and Methods). Red indicates higher neutralization scores. Correlation coefficients (r2) are indicated. r2 (X vs. Y) refers to the correlation between total amino acid mutations and the number of bNAb-like mutations; r2 (X vs. Neut) refers to the correlation between total amino acid mutations and neutralization score: r2 (Y vs. Neut) refers to the correlation between bNAb-like mutations and neutralization score. (B) Graphs show the total number of amino acid mutations (X axis) in IgH (left) and IgL (right) of all antibody sequences from GLHL121 mice immunized with protocols 1, 2 or 3 vs. the number of mutations that are identical or chemically equivalent to mutations in the human PGT121, PGT122, PGT123 or 10-1074 bNAbs (bNAb-like) (Y axis). 292 and 405 IgH and IgL sequences respectively were analyzed. The size of the dot is proportional to the number of sequences. Chemical equivalence was as follows: Group 1: G/A/V/L/I; Group 2: S/T; Group 3: C/M; Group 4: D/N/E/Q; Group 5: R/K/H; Group 6: F/Y/W; Group 7: P. Correlation coefficient (r2) is indicated. (C) Table shows the results of TZM-bl assays on monoclonal antibodies from GLHL121 mice immunized according to Protocol 1 and 2 as indicated in the diagrams on top. The protocol, mouse, antibody name, and the 14 tier 2/1B HIV-1 isolates are indicated at left and top respectively. Neutralization activity code: IC50 <0.01 in red; 0.01–0.1 in orange; 0.1–1 in yellow; >1 in green; not detectable (ND) in gray; not tested indicated by a dash. See also Figure S6.
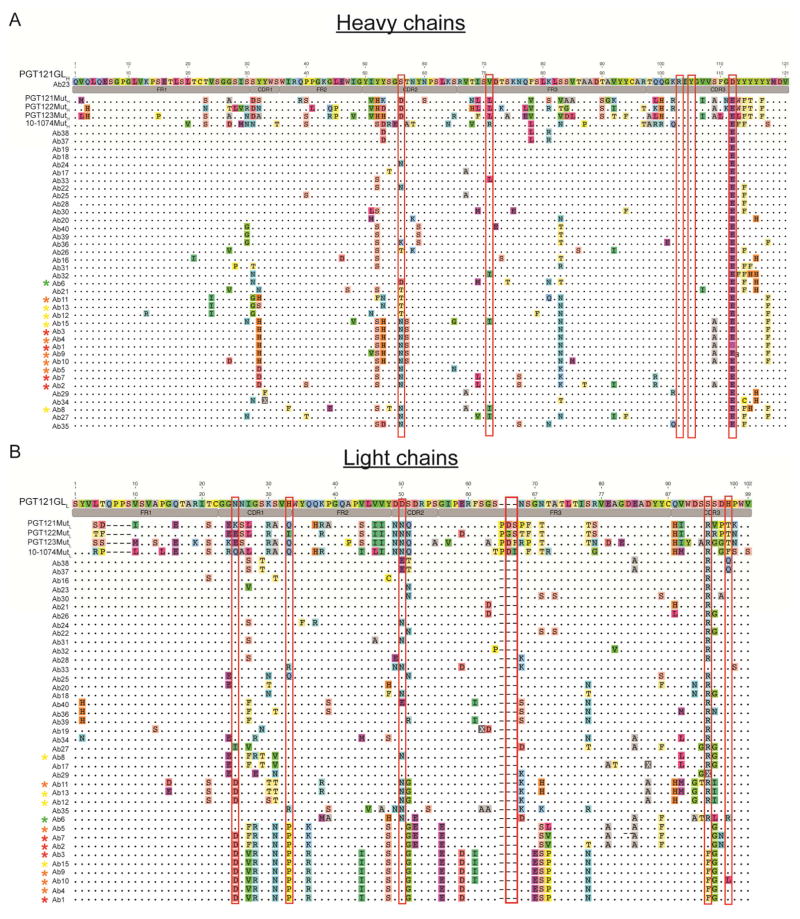
Figure 7.
Alignment of the monoclonal IgH and IgL to the inferred germline and mature mutated PGT121, PGT122, PGT123 and 10-1074 bNAbs. (A) Alignment of IgH sequences from the monoclonal antibodies in Figure 6. (B) Alignment of IgL sequences from monoclonal antibodies in Figure 6. The name of the antibody is indicated. Colors represent different amino acids. Dots represent identity to the germline reference sequence. Color coded asterisks next to the antibody name indicate neutralizing antibodies and their neutralization score (see Materials and Methods). Red indicates higher neutralization score. Amino acid substitutions that were previously identified as critical for binding to Env and neutralization of HIV-1 isolates are enclosed in a red rectangle. Antibody regions are indicated in gray. FR= framework region; CDR= complementarity determining region. Amino acid position is indicated with numbers on top of each reference sequence.
Among the 40 and 37 antibodies cloned from GLHL121 and MutHGLL121 mice, 15 and 17 respectively showed neutralizing activity against tier 2 HIV-1 isolates including antibodies Ab6 and Ab8 that were cloned from GLHL121 mouse M7 that had no detectable neutralizing activity in serum (Figures 3A and 6C). In contrast, none of the antibodies cloned from mice repeatedly immunized with 10MUT showed any neutralizing activity in TZM-bl assays (Figure 6 and Figure S6). Thus, the activity of the monoclonals generally reflected the neutralizing activity found in serum. However, bNAbs were also obtained from immunized mice with no detectable serologic activity (Figure 3 and 6).
Among the monoclonal antibodies tested, there was significant variation in neutralizing breadth and potency. As expected, the neutralizing activity of the monoclonal antibodies was directly related to their ability to bind to the native-like BG505 SOSIP protein in ELISA (Figure S6A,B) (Sanders et al., 2013). For example, antibodies Ab1, Ab2 and Ab3 showed high level binding in ELISA and neutralized 10 viral strains whereas antibodies Ab18 and Ab19 didn’t bind to BG505 and didn’t neutralize any of the strains in the panel (Figure 6 and Figure S6B). However, the potency of the autologous response to the BG505 T332N pseudovirus was unexpectedly low. Although additional analysis of these responses is required, the results suggest some structural differences between the epitopes on the immunogen and the pseudovirus.
Neutralizing activity was directly related to the number of somatic mutations (r2= 0.708) (Figure 6A). A direct correlation between the number of mutations and the number of PGT121-like mutations was also observed when all the amplified heavy chain and light chain sequences from all the mice immunized with the different protocols were analyzed (r2= 0.902 for heavy chains; r2= 0.864 for light chains)(Figure 6B). The antibodies with the best neutralizing activity carried amino acid substitutions at positions in IgH that make a critical contribution to Env binding, for example S56N contributes to the interaction with the N137 glycan and D100iE resides near the GDIR motif. (Garces et al., 2014; Sok et al., 2013). Moreover, CDRH3 residues found in the inferred germline that are also present in the mature PGT121 and involved in antigen-antibody interactions were retained (R103 and Y105 which make contacts with N332 glycan and the GDIR motif respectively). Similarly, the IgL in the neutralizing monoclonals showed amino acid substitutions at positions that contribute to the neutralizing activity of PGT121 (N25D, H33P, D50N (N332 glycan contact) and the S92, S93, S94 (GDIR recognition) (Mouquet et al., 2012; Sok et al., 2013) (Figure 7A,B). In addition, although none of the monoclonals carried the insertion in FWRL3 found in PGT121, the most potent antibodies had several amino acid mutations surrounding this region that may represent an alternative mechanism to enhance activity (Figure 7B).
We conclude that sequential immunization induces maturation of the predicted germline PGT121 antibody in a manner that in part recapitulates the maturation of the authentic monoclonal obtained from an HIV-1 infected individual.
Discussion
Antibodies hold great promise for HIV-1 prophylaxis because they can prevent infection in pre-clinical models even at low concentrations (Gautam et al., 2016; Gruell et al., 2013; Klein et al., 2012; Mascola et al., 2000; Moldt et al., 2012; Shibata et al., 1999; Shingai et al., 2013). Moreover, broad and potent antibodies to HIV-1 develop in a fraction of infected individuals suggesting that such antibodies might also be elicited by vaccination (Burton and Hangartner, 2016; Haynes and Montefiori, 2006; Hraber et al., 2014; Klein et al., 2013b; Mascola and Haynes, 2013; McCoy and Weiss, 2013; West et al., 2014). However, despite over 25 years and a very significant effort by numerous investigators, bNAbs have not been elicited by vaccination, and a preventative vaccine against HIV-1 has yet to be developed.
There are several reasons why bNAbs may be difficult to elicit. First and foremost, is that all the bNAbs isolated to date show unusually high levels of somatic mutation that are required for their activity (Klein et al., 2013a; Klein et al., 2013b; Mascola and Haynes, 2013; Mouquet et al., 2012; Mouquet et al., 2010; West et al., 2014). Hypermutation is a random process that introduces one nucleotide change per 103 bp per cell division in B cells dividing in the dark zone of the germinal center (Victora and Nussenzweig, 2012). B cell residence in germinal centers and selection by affinity is limited for any given antigen (Victora and Nussenzweig, 2012). Therefore, acquisition of the right combination of mutations to produce a bNAb is highly unlikely to occur by immunization with a monomorphic antigen. Based on these considerations, we and others proposed that sequential immunization would be required for bNAb development (Dimitrov, 2010; Haynes et al., 2012; Jardine et al., 2013; Klein et al., 2013a; Klein et al., 2013b; McGuire et al., 2013; Pancera et al., 2010; Scheid et al., 2009; Scheid et al., 2011; Zhou et al., 2010). This idea is strongly supported by data obtained from prospective studies of infected humans that showed that bNAbs develop in response to sequentially evolving variants of HIV-1, which are in turn selected by the developing antibodies (Bhiman et al., 2015; Doria-Rose et al., 2014; Liao et al., 2013; Wu et al., 2015; Wu et al., 2011).
An additional impediment to immunization to elicit broadly neutralizing responses is that few of the predicted germline precursors of bNAbs bind to native-like antigens (Doria- Rose et al., 2014; Hoot et al., 2013; Klein et al., 2013a; Liao et al., 2013; Mouquet et al., 2012; Mouquet et al., 2010; Scheid et al., 2011; Xiao et al., 2009). Consistent with these observations, specifically engineered Env-based immunogens or naturally occurring germline-binding Env (Andrabi et al., 2015; Doria-Rose et al., 2014; Gorman et al., 2016; Liao et al., 2013) appear to be necessary to initiate immune responses by B cells that express the germline precursors of CD4bs bNAbs (Dosenovic et al., 2015; Jardine et al., 2013; Jardine et al., 2015; McGuire et al., 2013). However, germline immunogens did not elicit neutralizing responses in mice that carry the germline precursors of CD4bs antibodies (Dosenovic et al., 2015; Jardine et al., 2015). Only a native-like antigen activated B cells expressing a synthetic intermediate antibody to produce tier-2 neutralizing antibodies (Dosenovic et al., 2015). These results provided the initial direct experimental support for the notion that a succession of different immunogens would be necessary to drive bNAb maturation (Dosenovic et al., 2015). Extension of these results to humans might be achieved by immunizing individuals that have modest levels of serologic activity with native-like antigens to see if this would increase potency and breadth (Schoofs et al., 2016).
Previous experiments with HIV-1 antigens in mice, rabbits, and macaques showed some increase in the breadth and potency of the response to Tier 1 viruses by sequential immunization. However, those experiments did not produce serologic responses or antibodies with broad neutralizing activity against tier 2 strains, nor were somatic mutations analyzed (Eda et al., 2006; Klinman et al., 1991; Malherbe et al., 2011). In addition, immunization with singular native-like envelope trimers elicits antibodies that neutralize the autologous virus but not heterologous strains (Sanders et al., 2015). None of those studies involved germline-targeting immunogens to prime bnAb precursors specifically. Our experiments demonstrate that immunization with a germline-targeting prime followed by ELISA-guided boosting with a sequence of directional immunogens designed to be progressively more native-like, with decreasing binding affinities to the inferred germline PGT121/10-1074 bNAb, induces high levels of somatic hypermutation. Similar to humans that develop bNAbs, the level of somatic mutation in knock-in mice was directly related to the acquisition of neutralizing activity. Finding that priming with a germline-targeting immunogen followed by a sequence of boosting immunogens partially reproduces the development of PGT121 antibodies in knock-in mice represents proof of concept for the idea that sequential immunization can elicit bNAbs.
The sequential boosting protocols that succeeded to elicit bNAbs and high levels of SHM were those in which the epitope structure changed gradually with each successive boost. In contrast, the protocols that failed to elicit bNAbs involved large changes in the epitope in at least one boost, such as 10MUT followed by BG505 (Figure S4B), or 11MUTA followed by 5MUT (Figure S4F). We have proposed that affinities of boosting immunogens for an intermediate mAb can be used to guide design of boosting protocols, that large affinity changes between successive immunogens corresponds to large changes in epitope structure, and that boosting protocols avoiding large affinity drops are more likely to succeed (Steichen et al., 2016). The observation that gradual protocols succeed here provides experimental evidence in support of that affinity drop analysis. This could allow future studies to prioritize experimental testing of sequential schemes (and optimize those schemes) based on similar affinity drop analyses.
Antibodies that target the glycan patch, including PGT121 are frequently found in infected individuals that develop bNAbs (Kong et al., 2013; Mouquet et al., 2012; Walker et al., 2011). These antibodies are highly mutated, but less so than some others in the VRC01 class of bNAbs, and are therefore particularly attractive targets for vaccine development (Gray et al., 2011; Mouquet et al., 2012; Scheid et al., 2009; Scheid et al., 2011; Walker et al., 2011; Walker et al., 2009; Wu et al., 2011; Zhou et al., 2010). Moreover, analysis of predicted intermediates in the PGT121 lineage showed that even antibodies with lower levels of mutation than PGT121, such as 3H+3L, have significant breadth and potency (Sok et al., 2013). Similarly, an antibody targeting the N332 supersite, BF520.1, showing relatively low levels of mutation was recently isolated from an infected infant (Simonich et al., 2016). However, this antibody is over 100 times less potent than PGT121. The antibodies that develop in GLHL121 knock-in mice appear to be far more potent than BF520.1 and closely resemble PGT121 intermediates.
The antibody responses elicited by our immunization scheme are suboptimal because they do not reach the full activity of PGT121. Additional variables should to be tested to further increase the potency and breadth of these antibody responses. These include the use of alternative adjuvants (Stills, 2005), timing between boosting, and immunogen dose (Gonzalez-Fernandez and Milstein, 1998). Moreover, inter-species differences in the Ig repertoire, B cell development, and FcR or Toll-like receptor expression should also be considered (Mestas and Hughes, 2004). Most importantly, our experiments are limited to knock-in mice and cannot be immediately translated to organisms such as humans with a diverse immune system where the germline precursor B cell frequency is limited. Additional immunogens with increased affinities for the predicted germline forms of bNAbs will likely be required to activate the rare bNAb precursors found in the normal naive B cell repertoire.
Despite these important caveats, our experiments establish the principle that a vaccination strategy based on sequential immunization with specifically engineered immunogens induces B cells expressing predicted germline precursors to develop bNAbs.
Methods and Resources
Contact for reagent and resource sharing
Further information and requests for reagents may be directed to the corresponding author Michel Nussenzweig (nussen@mail.rockefeller.edu)
Experimental model and subject details
Mice
Mice carrying the Ig V(D)J genes encoding the predicted germline or fully mature, mutated IgH and the predicted germline IgL corresponding to PGT121 (GLH121, MutH121, and GLL121 respectively) were produced by gene targeting Albino B6 (B6 (Cg)-Tyrc-2J/J) embryonic stem cells. The nucleotide sequence of the predicted germline PGT121 antibody was based on VH4-59*01, DH 3-3 and JH6*03 and the least mutated clonal relatives of PGT121/10-1074 like 10-996 (Mouquet et al., 2012). The GLL nucleotide sequence corresponds to germline VL3-21*02 and JL3*02 gene segments. The constant regions of IgH and IgL remain of mouse origin. The targeting vectors for IgH and IgL contained homologous regions flanking mouse D4-1 and J4. Recombination results in the deletion of the endogenous Js thereby minimizing rearrangement of the locus (Pelanda et al., 1997; Shih et al., 2002)
Mice were immunized every 2–4 weeks intraperitoneally (i.p.) with 10μg of protein, or a cocktail containing 3.3 μg of each VLC-SOSIP in Ribi (Sigma-Aldrich) or Alum (Thermo Scientific) adjuvant as specified. Serum samples were collected 2 weeks after immunization. Mice were maintained on regular chow. Male and female mice of 6–8 weeks of age were equally distributed in groups for knock-in mice characterization and immunization experiments. All animal procedures were performed in accordance to protocols approved by the Rockefeller University IACUC.
Methods details
Immunogens
Engineered Env proteins used as immunogens (11MUTA, 10MUT, 7MUT, 5MUT, 3MUT, BG505 (VLC2), VLC1, VLC3, VLC4 and VLC5 SOSIPs and 10MUT gp120) are described in (Steichen et al, submitted). These BG505-SOSIP.664 variants were modified as follows: 3MUT: T135A and T139I; 5MUT: V134Y, N136P, I138L, D140N; 6MUT: 5MUT+ T139I; 7MUT: 6MUT+ T135A; 9MUTA: 7MUT+ N137F+ Q328M; 9MUTB: 7MUT+ T135R+ Q328M; 10MUT: 7MUT+ N137F+ T320F+ Q328M; 11MUTA: 10MUT+ D141N. VLC (variable loop cocktail) contains 5 native-like SOSIPs differing in the variable loops (Steichen et al, submitted). Monomeric Env gp120s (11MUTA, 10MUT, 10MUT KO, 9MUTA, 9MUTB, 7MUT, 6MUT, 5MUT, 3MUT, 2MUT) and HIS tagged SOSIPs (BG505, VLCs) were used for ELISAs. AviTag-biotinylated Env gp120 proteins 10MUT, 5MUT, P1981_c5_3, CAAN5342.A2 and corresponding PGT121 epitope mutant (KO) versions were used for sorting. 10MUT gp120 KO and 5MUT gp120 KO have mutations T303A, H330A, and N332T; P1981_c5_3 and CAAN5342.A2 gp120s have mutations T303A, D325A, R327A, H330A and N332T.
ELISA
ELISA to the monomeric gp120 versions of 11MUTA,10MUT, 10MUT KO, 9 MUTB, 9MUTA, 7MUT, 6MUT, 5MUT, 3MUT and 2MUT were performed by coating of high bind 96 well plates (Corning #9018) with 100μl per well of a 2μg/ml protein solution in PBS overnight at 4°C. Plates were washed 6 times with washing buffer (1xPBS with 0.05% Tween 20 (Sigma-Aldrich)) and incubated in blocking buffer (1xPBS with 1% Milk) for 1 hour (h) at room temperature (RT). Immediately after blocking, monoclonal antibodies or serum samples were added in blocking buffer and incubated for 2 h at RT. Serum samples were assayed at a 1:100 starting dilution and seven additional 3 fold serial dilutions. Monoclonal antibodies were tested at 5 or 10μg/ml as specified in the Results section. Plates were washed 6 times with washing buffer and then incubated with anti-mouse or anti human IgG secondary antibody conjugated to horseradish peroxidase (HRP) (Jackson Laboratories) in washing buffer at a 1:5000 dilution. Plates were developed by addition of the HRP substrate, ABTS (Life Technologies) and absorbance was measured at 405nm with an ELISA microplate reader (FluoStar Omega, BMG Labtech). Alternatively, for ELISA to the wtBG505 SOSIP and VLC SOSIP, plates were pre coated with an anti-6X His tag antibody (Abcam) in 1xPBS overnight at 4°C. After overnight incubation, plates were washed six times and blocked for 1 h at RT using the same buffers as specified above. Immediately after blocking, the His tagged wtBG505 SOSIP and VLC SOSIP proteins were added at 2μg/ml in dilution buffer (1xPBS with 1% fetal bovine serum and 0.2% Tween20) to all the wells and incubated at RT for 1 h. Plates were then washed six times and blocked for 1 h at RT. Serum Ig or monoclonal antibodies were added in dilution buffer at the dilutions previously mentioned and incubated for 2 hours at 37°C. Plates were then washed six times and incubated with secondary antibody for 1.5 hours at 37°C. Finally plates were developed and measured as specified above.
Ig purification
Igs were purified from 200μl of mouse serum using Ab Spin Trap Protein G sepharose columns (GE Healthcare) following the manufacturers instructions. Igs were eluted in 2 fractions of 400μl or alternatively in 4 fractions of 200μl. The Ig containing fractions were buffer exchanged with PBS by overnight dialysis at 4°C (dialysis cassettes 20000 MWCO Thermo Scientific) and sterilized by filtration (0.22μM, Millex)
Neutralization assay
TZM-bl assays were performed as described (Montefiori, 2005). In brief, neutralization activity was calculated as a function of the reduction in Tat-induced luciferase expression in the TZM-bl reporter cell line after a single round of virus infection.
A total neutralization score was obtained by a sum of the individual scores assigned as (IC50 <0.01 score 4; 0.01–0.1 score 3; 0.1–1 score 2; >1 score 1; not detectable (ND) score 0) normalized by the number of viruses tested in the TZM-bl assay.
Flow cytometry
Cells from spleens were stained with cocktails of the following anti-mouse antibodies: anti-CD4 APC-eFluor 780, anti-CD8 APC-eFluor 780, anti-Ly-6G (Gr1) APC-eFluor 780, anti- F4/80 APC-eFluor 780, anti- B220 FITC, anti-CD38 Alexa Fluor 700, anti- IgM PerCP-eFluor 710, anti-CD21/CD35 eFluor 450 and anti-IgG1 BV421 (BD Biosciences); anti-CD23 PE (BioLegend); anti IgD PE/Cy7 and anti Ig light chain λ (Biolegend). Dead cells were excluded by staining with the Live-dead aqua stain (Life Technologies). Stained cell samples were analyzed in a BD LSR Fortessa analyzer.
Single B cell sorting
B cells were enriched from cell suspensions of spleen and lymph nodes by negative selection using magnetic anti-CD43 micro beads (Miltenyi Biotec) and LS MACS separation columns (Miltenyi Biotec) as specified by the manufacturer. CD43− cells were stained with a cocktail of the following antibodies: anti-CD4 APC-eFluor 780, anti-CD8 APC-eFluor 780, anti-Gr1 APC-eFluor 780, anti- F4/80 APC-eFluor 780, anti- B220 FITC, anti-CD38 Alexa Fluor 700, anti-IgM PerCP-eFluor 710, anti-IgG1 BV421 (BD Biosciences) and Live-dead aqua stain (Life Technologies). For immunized mice, we used combinations of the following biotinylated proteins 10MUT, 5MUT, P1981_c5_3, CAAN5342.A2 and corresponding PGT121 epitope mutant (KO) versions as baits at a concentration of 5μg/ml. Streptavidin-conjugated PE or APC was used to label the wild type and the KO proteins respectively. Naïve B cells were obtained by sorting on Live-dead− CD4−CD8−Gr1−F4/80−CD38+B220+IgM+. B cells from immunized mice were obtained by sorting on Live-dead−CD4−CD8−Gr1−F4/80−CD38+B220+IgM−IgG+bait+KO bait−. Single cells were sorted into individual wells of a 96-well plate containing 4μl of lysis buffer (RNASin (Promega) 40 U/ml (0,3 ml), 10xPBS (Dulbecco) (0.2 ml), DTT (Invitrogen) 100 mM (0.4 ml) and nuclease-free water (3.1 ml)) using a FACS Aria III (Becton Dickinson). Plates were immediately frozen on dry ice and stored at −80°C or processed for cDNA synthesis. cDNA from single cells was obtained by reverse transcription (Superscript III, Invitrogen) and used for amplification by nested PCR using the sequencing or cloning primers listed in (Figure S7) (von Boehmer et al, in press).PCR protocols (annealing (°C)/elongation (sec)/number of cycles): 1st PCR (IgG IgH): 52/55/50; 2nd PCR (IgG IgH): 54/55/50; 1st PCR (Igκ): 46/55/50; 2nd PCR (Igκ): 50/50/50; 1st PCR (IgM IgH): 52/55/50; 2nd PCR (IgM IgH): 54/55/50.
Antibody cloning and production
Amplified heavy chain and light chain cDNAs were individually cloned into expression vectors containing the corresponding mouse antibody constant regions by using the sequence and ligation-independent cloning (SLIC) methodology. (Li and Elledge, 2007) (von Boehmer et al, in press). DNA sequences that are complementary to the destination vector were added to the amplified IgH and IgL cDNAs by PCR using the cloning primers listed in (Figure S7). PCR protocol (annealing (°C)/elongation (sec)/number of cycles): 68/55/50. Amplified cDNA was purified with QIAquick 96 PCR Purification Kit following manufacturer instructions. The linearized vector (30–50ng) and the insert (4 μl of purified product) were ligated at 25°C for 2.5 min. Ligation was either stored at 4°C or transformed in DH5α competent bacteria. Next day, bacterial colonies were analyzed by PCR using the primers indicated in Figure S7. PCR protocol (annealing (°C)/elongation (sec)/number of cycles): 57/55/35.
Antibodies were produced by transient transfection of HEK293T cells with equal amounts of immunoglobulin heavy and light chain expression vectors. After 7 days, the supernatant was harvested and antibodies were concentrated by ammonium sulfate precipitation. IgG was purified with Protein G–Sepharose 4 Fast Flow (Klein et al., 2014).
Analysis software
Geneious 9.0.4 and MacVector 14.0.3 were used for sequence analysis and graphs were created using R language. Flow cytometry data was processed using FlowJo 10.0.7. GraphPad Prism 6.0f was used for data analysis.
Quantification and statistical analysis
Statistical information including n, mean and statistical significance values are indicated in the text or the figure legends. GraphPad Prism 6.0f was used to calculate Pearson correlation coefficients and for statistical analysis by one-way ANOVA and Tukey multi comparison test or unpaired T-Test. Data were considered statistically significant at *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 and ****p ≤ 0.0001.
Supplementary Material
Immunization elicits anti-HIV-1 broadly neutralizing antibodies in knock-in mice
ELISA-guided sequential immunization induces high levels of somatic mutation
New engineered HIV-1 envelope antigens
Mouse antibodies elicited by sequential immunization resemble human antibodies
Acknowledgments
We thank S. Hinklein and T. Eisenreich for help with mouse colony management, Neena Thomas for single cell FACS sorting, Harald Hartweger for help with experiments, Zoran Jankovic for laboratory support, and all members of the Nussenzweig Laboratory for helpful discussion and advice. We thank Chingwen Yang and Rada Norinsky for knock-in mice and Pamela J. Bjorkman for providing proteins. This work was supported by the following grants: Collaboration for AIDS Vaccine Discovery Grant OPP1033115 and OPP1124068 (M.C.N.); NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663 (M.C.N, W.R.S., D.R.B.); National Institute Of Allergy and Infectious Diseases of the NIH Grants AI100148 and AI109632 (M.C.N.); the International AIDS Vaccine Initiative Neutralizing Antibody Consortium and Center (W.R.S., D.R.B.); CAVD funding for the IAVI NAC Center (W.R.S., D.R.B.); the Ragon Institute of MGH, MIT and Harvard (W.R.S, D.R.B.); The Robertson Foundation and the Rockefeller University. A.E. was supported by an international postdoctoral fellowship from the Clarin COFUND- Marie Curie program (PCTI-FICYT). P.D. was supported by an international postdoctoral fellowship from the Swedish Research Council. A.D.G. was supported by MSTP grant T32GM07739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. M.C.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
Author contributions
A.E. planned and performed experiments, analyzed data and wrote the manuscript. J.S. planned experiments, provided immunogens, analyzed data and edited the manuscript. P.D. produced knock-in mice, performed experiments, and analysed data. D.K. provided immunogens, analyzed data and edited the manuscript. J. G. and A.G. produced antibodies, N.F and A.D.G. produced knock-in mice. T.O. analysed data. K.H., T.A., S.L., S. C., J.H. performed experiments. E.G. purified immunogen proteins. D. S. and K.L.S. planned and analyzed neutralization experiments. D.B. critically read and contributed to the manuscript preparation. W.R.S planned and analysed experiments and helped write the manuscript. M.C.N. planned and supervised experiments, analysed data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrabi R, Voss JE, Liang CH, Briney B, McCoy LE, Wu CY, Wong CH, Poignard P, Burton DR. Identification of Common Features in Prototype Broadly Neutralizing Antibodies to HIV Envelope V2 Apex to Facilitate Vaccine Design. Immunity. 2015;43:959–973. doi: 10.1016/j.immuni.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Preite S, Reboldi A, Ronchi F, Ansel KM, Lanzavecchia A, Sallusto F. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 2013;38:596–605. doi: 10.1016/j.immuni.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Bhiman JN, Anthony C, Doria-Rose NA, Karimanzira O, Schramm CA, Khoza T, Kitchin D, Botha G, Gorman J, Garrett NJ, et al. Viral variants that initiate and drive maturation of V1V2-directed HIV-1 broadly neutralizing antibodies. Nat Med. 2015;21:1332–1336. doi: 10.1038/nm.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol. 2004;5:233–236. doi: 10.1038/ni0304-233. [DOI] [PubMed] [Google Scholar]
- Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen YZ, Dolin R. Novel HIV vaccine strategies: overview and perspective. Ther Adv Vaccines. 2013;1:99–112. doi: 10.1177/2051013613494535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov DS. Therapeutic antibodies, vaccines and antibodyomes. MAbs. 2010;2:347–356. doi: 10.4161/mabs.2.3.11779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenovic P, von Boehmer L, Escolano A, Jardine J, Freund NT, Gitlin AD, McGuire AT, Kulp DW, Oliveira T, Scharf L, et al. Immunization for HIV- 1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell. 2015;161:1505–1515. doi: 10.1016/j.cell.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda Y, Takizawa M, Murakami T, Maeda H, Kimachi K, Yonemura H, Koyanagi S, Shiosaki K, Higuchi H, Makizumi K, et al. Sequential immunization with V3 peptides from primary human immunodeficiency virus type 1 produces cross-neutralizing antibodies against primary isolates with a matching narrow-neutralization sequence motif. J Virol. 2006;80:5552–5562. doi: 10.1128/JVI.02094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote J, Eisen HN. Kinetic and affinity limits on antibodies produced during immune responses. Proc Natl Acad Sci U S A. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Lee JH, de Val N, de la Pena AT, Kong L, Puchades C, Hua Y, Stanfield RL, Burton DR, Moore JP, et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces F, Sok D, Kong L, McBride R, Kim HJ, Saye-Francisco KF, Julien JP, Hua Y, Cupo A, Moore JP, et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam R, Nishimura Y, Pegu A, Nason MC, Klein F, Gazumyan A, Golijanin J, Buckler-White A, Sadjadpour R, Wang K, et al. A single injection of anti-HIV-1 antibodies protects against repeated SHIV challenges. Nature. 2016;533:105–109. doi: 10.1038/nature17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Fernandez A, Milstein C. Low antigen dose favours selection of somatic mutants with hallmarks of antibody affinity maturation. Immunology. 1998;93:149–153. doi: 10.1046/j.1365-2567.1998.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodnow CC, Vinuesa CG, Randall KL, Mackay F, Brink R. Control systems and decision making for antibody production. Nat Immunol. 2010;11:681–688. doi: 10.1038/ni.1900. [DOI] [PubMed] [Google Scholar]
- Gorman J, Soto C, Yang MM, Davenport TM, Guttman M, Bailer RT, Chambers M, Chuang GY, DeKosky BJ, Doria-Rose NA, et al. Structures of HIV-1 Env V1V2 with broadly neutralizing antibodies reveal commonalities that enable vaccine design. Nat Struct Mol Biol. 2016;23:81–90. doi: 10.1038/nsmb.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H, Bournazos S, Ravetch JV, Ploss A, Nussenzweig MC, Pietzsch J. Antibody and antiretroviral preexposure prophylaxis prevent cervicovaginal HIV-1 infection in a transgenic mouse model. J Virol. 2013;87:8535–8544. doi: 10.1128/JVI.00868-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nat Biotechnol. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines. 2006;5:347–363. doi: 10.1586/14760584.5.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoot S, McGuire AT, Cohen KW, Strong RK, Hangartner L, Klein F, Diskin R, Scheid JF, Sather DN, Burton DR, et al. Recombinant HIV envelope proteins fail to engage germline versions of anti-CD4bs bNAbs. PLoS Pathog. 2013;9:e1003106. doi: 10.1371/journal.ppat.1003106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Kulp DW, Havenar-Daughton C, Sarkar A, Briney B, Sok D, Sesterhenn F, Ereno-Orbea J, Kalyuzhniy O, Deresa I, et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science. 2016;351:1458–1463. doi: 10.1126/science.aad9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepler TB, Liao HX, Alam SM, Bhaskarabhatla R, Zhang R, Yandava C, Stewart S, Anasti K, Kelsoe G, Parks R, et al. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe. 2014;16:304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013a;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013b;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Nogueira L, Nishimura Y, Phad G, West AP, Jr, Halper-Stromberg A, Horwitz JA, Gazumyan A, Liu C, Eisenreich TR, et al. Enhanced HIV-1 immunotherapy by commonly arising antibodies that target virus escape variants. J Exp Med. 2014;211:2361–2372. doi: 10.1084/jem.20141050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinman DM, Higgins KW, Conover J. Sequential immunizations with rgp120s from independent isolates of human immunodeficiency virus type 1 induce the preferential expansion of broadly crossreactive B cells. J Exp Med. 1991;173:881–887. doi: 10.1084/jem.173.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR, Nabel GJ. Broadly neutralizing antibodies and the search for an HIV-1 vaccine: the end of the beginning. Nat Rev Immunol. 2013;13:693–701. doi: 10.1038/nri3516. [DOI] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malherbe DC, Doria-Rose NA, Misher L, Beckett T, Puryear WB, Schuman JT, Kraft Z, O’Malley J, Mori M, Srivastava I, et al. Sequential immunization with a subtype B HIV-1 envelope quasispecies partially mimics the in vivo development of neutralizing antibodies. J Virol. 2011;85:5262–5274. doi: 10.1128/JVI.02419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- McCoy LE, Weiss RA. Neutralizing antibodies to HIV-1 induced by immunization. J Exp Med. 2013;210:209–223. doi: 10.1084/jem.20121827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Hoot S, Dreyer AM, Lippy A, Stuart A, Cohen KW, Jardine J, Menis S, Scheid JF, West AP, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestas J, Hughes CCW. Of Mice and Not Men: Differences between Mouse and Human Immunology. The Journal of Immunology. 2004;172:2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- Moldt B, Rakasz EG, Schultz N, Chan-Hui PY, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, et al. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;Chapter 12(Unit 12):11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature. 2010;467:591–595. doi: 10.1038/nature09385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don’t know, what we should know. Nat Med. 2004;10:806–810. doi: 10.1038/nm0804-806. [DOI] [PubMed] [Google Scholar]
- Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Pelanda R, Schwers S, Sonoda E, Torres RM, Nemazee D, Rajewsky K. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity. 1997;7:765–775. doi: 10.1016/s1074-7613(00)80395-7. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science. 2015;349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffner T, Sattentau QJ, Dorrell L. Development of prophylactic vaccines against HIV-1. Retrovirology. 2013;10:72. doi: 10.1186/1742-4690-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs T, Klein F, Braunschweig M, Kreider EF, Feldmann A, Nogueira L, Oliveira T, Lorenzi JC, Parrish EH, Learn GH, et al. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science. 2016;352:997–1001. doi: 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho MW, Martin MA. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- Shih TA, Roederer M, Nussenzweig MC. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nat Immunol. 2002;3:399–406. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Jr, Lifson JD, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonich CA, Williams KL, Verkerke HP, Williams JA, Nduati R, Lee KK, Overbaugh J. HIV-1 Neutralizing Antibodies with Limited Hypermutation from an Infant. Cell. 2016;166:77–87. doi: 10.1016/j.cell.2016.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Doores KJ, Briney B, Le KM, Saye-Francisco KL, Ramos A, Kulp DW, Julien JP, Menis S, Wickramasinghe L, et al. Promiscuous glycan site recognition by antibodies to the high-mannose patch of gp120 broadens neutralization of HIV. Sci Transl Med. 2014;6:236ra263. doi: 10.1126/scitranslmed.3008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, Laserson U, Laserson J, Liu Y, Vigneault F, Julien JP, Briney B, Ramos A, Saye KF, Le K, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen J, et al. submitted. [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stills HF., Jr Adjuvants and antibody production: dispelling the myths associated with Freund’s complete and other adjuvants. ILAR J. 2005;46:280–293. doi: 10.1093/ilar.46.3.280. [DOI] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Scheid JF, Klein F, Bjorkman PJ, Nussenzweig MC. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell. 2014;156:633–648. doi: 10.1016/j.cell.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhang Z, Schramm CA, Joyce MG, Kwon YD, Zhou T, Sheng Z, Zhang B, O’Dell S, McKee K, et al. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Chen W, Feng Y, Zhu Z, Prabakaran P, Wang Y, Zhang MY, Longo NS, Dimitrov DS. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem Biophys Res Commun. 2009;390:404–409. doi: 10.1016/j.bbrc.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.