Summary
The motor of the membrane‐anchored archaeal motility structure, the archaellum, contains FlaX, FlaI and FlaH. FlaX forms a 30 nm ring structure that acts as a scaffold protein and was shown to interact with the bifunctional ATPase FlaI and FlaH. However, the structure and function of FlaH has been enigmatic. Here we present structural and functional analyses of isolated FlaH and archaellum motor subcomplexes. The FlaH crystal structure reveals a RecA/Rad51 family fold with an ATP bound on a conserved and exposed surface, which presumably forms an oligomerization interface. FlaH does not hydrolyze ATP in vitro, but ATP binding to FlaH is essential for its interaction with FlaI and for archaellum assembly. FlaH interacts with the C‐terminus of FlaX, which was earlier shown to be essential for FlaX ring formation and to mediate interaction with FlaI. Electron microscopy reveals that FlaH assembles as a second ring inside the FlaX ring in vitro. Collectively these data reveal central structural mechanisms for FlaH interactions in mediating archaellar assembly: FlaH binding within the FlaX ring and nucleotide‐regulated FlaH binding to FlaI form the archaellar basal body core.
Introduction
Motility is a critical feature of many prokaryotes (Jarrell and McBride, 2008). The archaeal motility structure, the archaellum, is a unique structure: its subunits share homologies with the components of the assembly systems of type IV pili (T4P) (Fig. 1A), which facilitate movement by extension and retraction (Craig et al., 2004; Berry and Pelicic, 2015). Remarkably, the archaella rotate to propel the cell forward, similar to the structurally unrelated bacterial flagella (Jarrell and Albers, 2012). Archaella are composed of only 7–13 proteins, and the genes encoding the subunits of the archaella are usually found clustered in the genome. Deletion of these genes leads to non‐archaellated strains, as demonstrated in Methanococcales sp., Sulfolobales sp. and Halobacteriaceae sp. (Patenge et al., 2001; Thomas et al., 2001; Chaban et al., 2007; Tripepi et al., 2010). Although archaella were discovered four decades ago and are widely found in archaea, it only recently became possible to study them using a combined genetic, biochemical and structural approach. Most of the biochemical and structural data are available from the crenarchaeon Sulfolobus acidocaldarius making this an exemplary system for archaella structural biochemistry and genetics.
Figure 1.
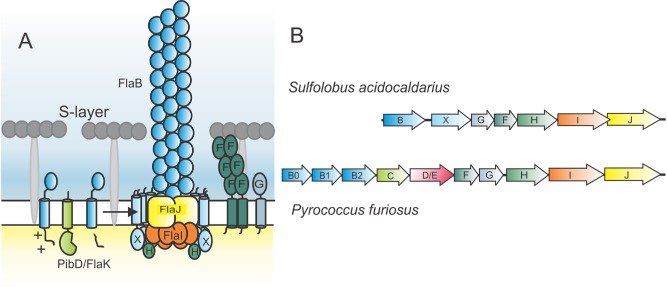
Schematic model of the S . acidocaldarius archaellum.
A. The pre‐archaellin FlaB is processed by the peptidase PibD/FlaK and then assembled into the archaellum filament, which is a three start helical filament. The motor complex is formed by the ring‐forming scaffold protein FlaX in which FlaH and FlaI probably interact with the integral membrane protein FlaJ. The dimeric soluble domain of FlaF interacts with the S‐layer. FlaG most probably has a similar function as FlaF as its soluble domain has homologies to FlaF.
B. Operons encoding the archaellum in S . acidocaldarius and P . furiosus. Genes have the same color as the respective proteins in (A).
In S. acidocaldarius, seven different proteins build the archaellum complex and are essential for archaellation (Fig. 1) (Lassak et al., 2012). FlaF and FlaG are essential for the formation of archaella and predicted to be monotopic membrane proteins. FlaF is a stator component anchoring the archaellum to the cell envelope by binding to the S‐layer protein, the sole cell wall protein of S. acidocaldarius (Banerjee and Albers, 2015). Like the pilin subunits of the T4P systems, the archaellin FlaB contains an N‐terminal class III signal peptide that is processed by a membrane‐bound aspartic acid protease called PibD (Albers et al., 2003). It is so far unknown how processed archaellins are assembled into the filament. However, it is believed that the polytopic membrane protein of the archaella assembly system FlaJ and the P‐loop ATPase FlaI (homologue of the bacterial T4P inner‐membrane protein (e.g. PilC of Myxococcus xanthus) and the assembly ATPase (e.g. PilB of M. xanthus)) play a major role in this process (Peabody et al., 2003; Ghosh and Albers, 2011).
The FlaI ATPase from S. acidocaldarius forms hexamers upon binding to ATP, and its activity is stimulated by tetraether lipids (Ghosh et al., 2011). It is composed of a flexible N‐terminal domain and a C‐terminal domain that contains the ATP binding site. Deletion of the first 29 amino acids of FlaI leads to an S. acidocaldarius cell that still formed archaella but was no longer motile, demonstrating a bifunctional role of FlaI in both assembly and rotation of the archaellum (Reindl et al., 2013). The S. acidocaldarius FlaX cytoplasmic domain forms ring‐like oligomeric structures of 30 nm diameter (Banerjee et al., 2012). Deletion of three predicted alpha‐helices on the C‐terminus of FlaX abolishes formation of the ring and interaction with FlaI in vitro (Banerjee et al., 2012). FlaX also interacts with FlaH.
Notably, the formation of a complex containing FlaX, FlaH and FlaI was confirmed in vitro, suggesting that these three proteins form the core cytoplasmic motor complex of the archaellum (Banerjee et al., 2013); the role of FlaH, however, has been enigmatic. FlaH is a putative ATP‐binding protein with a possible regulatory function toward FlaI (Albers and Jarrell, 2015), as FlaH has a predicted Walker A motif but a non‐canonical Walker B motif. Although crystals were obtained from the Methanocaldococcus jannaschii FlaH (Meshcheryakov et al., 2014), the only homologous structures are from PH0284 and PH0186 (Yokoyama et al., 2000; Kang et al., 2009), two RecA fold proteins from Pyrococcus horikoshii (2DR3.pdb and 2ZTS.pdb, respectively), which form hexameric rings. However, these proteins have not been characterized otherwise.
To better understand the biological roles of FlaH, we solved its crystallographic structure and coupled structural analysis to biochemical assays. We then combined this new structural and biochemical information with electron microscopy, mutational analyses of FlaH and studies of its interactions with FlaI and FlaX. Strains expressing FlaH mutants that no longer bound nucleotides were immotile and unable to assemble archaella, indicating that ATP‐binding by FlaH is essential for its function in the archaellum motor. Together with the interaction studies of the FlaH/FlaI and FlaH/FlaX archaellum sub‐complexes, our results uncover the core structure of the archaellum motor needed for archaeal motility.
Results
The crystal structure of SaFlaH
FlaH is conserved in all archaellated archaea (Jarrell and Albers, 2012), essential for archaella assembly and rotation (Patenge et al., 2001; Thomas et al., 2001; Chaban et al., 2007; Lassak et al., 2012) and interacts with FlaI (Banerjee et al., 2013). To better understand the role of FlaH in the archaellum, we determined the crystal structure of FlaH from S. acidocaldarius (SaFlaH). Diffracting crystals of SaFlaH were obtained in the presence of ATP and Mg2+, and the structure was solved by molecular replacement using the structure of the RecA superfamily ATPase PH0284 of P. horikoshii OT3 (2DR3.pdb) as search model (Yokoyama et al., 2000). The final model was refined to a resolution of 2.3 Å with an Rfree and Rwork of 24.8% and 19.3%, respectively (4YDS.pdb; see Table 1). Contrary to the structural homologues PH0284 and PH0186 (Yokoyama et al., 2000; Kang et al., 2009) of P. horikoshii, which crystallized as hexamers, SaFlaH crystallized as monomer with one molecule of SaFlaH in the asymmetric unit. The final model encompasses residues 1–226, only missing the last two residues and the C‐terminal His‐tag (see Fig. 2A). SaFlaH contains a typical RecA/RAD51 superfamily ATPase domain with a large twisted central β‐sheet, sandwiched by α‐helices on both sides. The central β‐sheet consists of nine strands and no additional domains besides the classical RecA/RAD51 ATPase fold. Consistent with the high affinity for ATP and the lack of ATPase activity (Fig. 3A), the structure contains an Mg2+‐ion and a clearly resolved ATP (Fig. 2B). Importantly, ATP binds on an exposed and conserved surface of the FlaH subunit (Fig. 2D and E) where it would be expected to influence oligomerization, as seen in Rad51 archaeal homologues (Shin et al., 2003; Wu et al., 2004).
Table 1.
Crystallographic data collection and refinement statistics
| Data collection | |
|---|---|
| Space group | C 2 |
| Cell dimensions (Å/°) | a = 103.4; b = 52.9; c = 73.6 |
| 90; 130.8; 90 | |
| Wavelength (Å) | 1.0 |
| Resolution range (Å) | 50–2.3 |
| Completeness (%)a | 85.5/43.2 |
| Observed reflections | 28 367 |
| Unique reflections | 11 612 |
| Multiplicitya | 2.4/1.9 |
| Rsym (%)a | 2.8/33.3 |
| mean I/σa | 19.3/2.4 |
| Refinement statistics | |
|---|---|
| Resolution range (Å) | 42.9–2.3 |
| Rwork (%)/Rfree (%)b | 19.3/24.8 |
| Average B‐factor (Åb) | 60.1 |
| Protein | 59.8 |
| Solvent | 64.6 |
| Number of atoms | 1 878 |
| Protein | 1 765 |
| Ligand | 32 |
| Water molecules | 81 |
| RMSD bond lengths | 0.005 |
| RMSD bond angles | 0.98 |
| Ramachandran favored (%) | 95 |
| Ramachandran allowed (%) | 0.45 |
| PDB ID | 4YDS.pdb |
Overall/last shell.
Rfree is the cross‐validation R factor for 10% of reflections against which the model was not refined.
Figure 2.
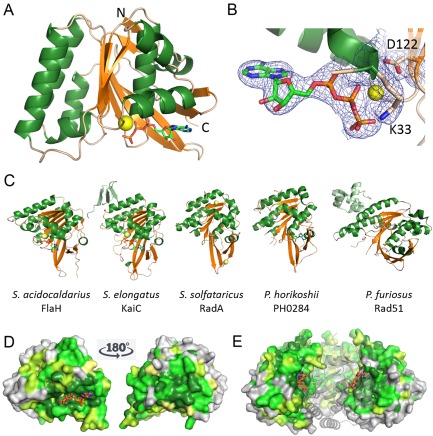
Crystal structure of FlaH with ATP showing fold, ATP binding and conserved surface regions.
A. Ribbon representation of the S . acidocaldarius FlaH monomer showing fold, secondary structure and ATP binding site. ATP (sticks) bind to the active site and Mg2+ ion (yellow sphere). N‐ and C‐termini are indicated.
B. Active site with electron density (2|Fo|‐|Fc| map at 1σ) for the bound ATP and Mg2+‐ion. The Walker A and Walker B residues K33 and D122 are indicated.
C. FlaH compared with other RecA superfamily ATPases. Similar structures revealed by a DALI search for structural homologues are shown. Additional subunits besides the RecA/Rad51 domain are shown in light green. Nucleotides or phosphate bound to the active sites are shown in sticks, ions as spheres.
D. Conservation of residues is depicted on the SaFlaH surface. Dark green and light green indicate identical and similar residues respectively. FlaH monomers are in the same orientation as in A and turned 180° and shown as surface representation. ATP is shown as orange sticks. Mg2+ ion as a yellow sphere.
E. FlaH interface within the hexamer modeled by superposition on the PH0284 hexamer: one monomer shown as cartoon to visualize the highly conserved interfaces. ATP is shown as orange sticks. Mg2+ ion as yellow sphere.
Figure 3.
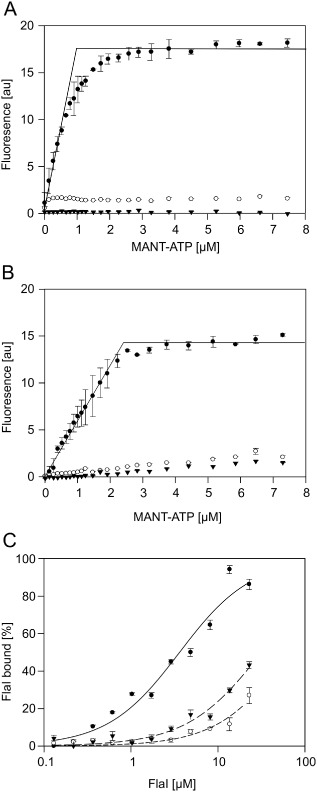
Mutations in the FlaH Walker A and B Motifs strongly reduce nucleotide binding and interaction with FlaI. The increase in fluorescence upon MANT‐ATP binding to 1 μM FlaH was determined for increasing MANT‐ATP concentrations for (A) WT SaFlaI (closed circles), SaFlaH K33A (open circles) and SaFlaH D122N (triangles) and (B) WT PfFlaH (closed circles) PfFlaH K53A (open circles) and PfFlaH D140N (triangles). (C) The binding of PfFlaH to PfFlaI was studied by microscale thermophoresis. The 34 nM labeled WT PfFlaH, PfFlaH K53A or PfFlaH D140N were mixed with increasing concentrations (14 nM–29 μM) of unlabeled PfFlaI, and thermophoresis was performed. Curves were obtained from at least two independent experiments. Binding curves were fitted to the Hill equation, and binding is depicted as the fraction bound.
The DALI search (Holm and Rosenström, 2010) for structural homologues of SaFlaH identified other members of the RadA/Rad51/RecA family (Fig. 2C). The closest hit is the RecA domain of the circadian clock protein kinase KaiC (3S1A.pdb). The next closest hits were the archaeal RadA Sso2452 paralogue with antirecombinase activity (2W0M.pdb), the archaeal RecA superfamily ATPase PH0284 (2DR3.pdb) that was used for molecular replacement as input model and the archaeal Rad51 protein (1PZN.pdb) followed by other members of the RecA superfamily of ATPases (Fig. 2C). An additional domain that is present in some of those ATPases (shown in light green in Fig. 2C) is missing in the SaFlaH structure, which consists of only the RecA domain. Mapping the conservation of different FlaH homologues on the structure of SaFlaH identified two regions of conservation (Fig. 2C). Modeling the monomeric SaFlaH structure on the hexameric PH0284 structure reveals that the two regions of conservation correspond to the interfaces of monomers in the hexameric PH0284 structure (Fig. 2D and E). This strongly suggests that SaFlaH is also able to form a hexamer.
FlaH walker motifs are essential for ATP binding and FlaI interaction
Examining the FlaH structure coupled to alignment of sequences of several FlaH homologues from euryarchaeota and crenarchaeota showed a high sequence and structural homology between FlaH homologues (see Supplementary Fig. S1). FlaH sequences contain the conserved Walker A motif, GXXXXGK(T/S), which is essential for nucleotide binding in P‐loop ATPases and RecA family proteins (Hanson and Whiteheart, 2005). In FlaHs, the aspartate residue in the Walker B motif, hhhhDE, which functions in chelating the divalent cation, is conserved, whereas the glutamate is replaced by a serine or threonine in all homologues (Fig. S1). Mutation of this glutamate in other ATPases often results in proteins that bind but do not hydrolyze nucleotides (Hanson and Whiteheart, 2005). We therefore generated mutants in the lysine of the Walker A (SaFlaHK33A) and the aspartic acid in the Walker B (SaFlaHD122N) motifs. WT SaFlaH and both mutants were expressed in Escherichia coli and purified. Purified SaFlaH is stabilized by the presence of nucleotides and lower yields were observed for the mutants compared with WT SaFlaH. Both mutants were concentrated to obtain similar concentrations as WT SaFlaH (see Supplementary Figure S2A). Determination of the OD260/280 ratio demonstrated that purification in the absence of nucleotides or ammonium sulfate precipitation removed the nucleotide from WT SaFlaH but resulted in unstable protein.
ATP binding assays were performed with SaFlaH using the fluorescent ATP analogue MANT‐ATP. This nucleotide analog shows an increase in fluorescence quantum yield upon binding to proteins. Indeed an increase in fluorescence was observed when increasing concentrations of MANT‐ATP were incubated with 5 μM of SaFlaH (Fig. 3A). However, only 20% of the purified SaFlaH bound MANT‐ATP suggesting that most of the purified SaFlaH is in a nucleotide‐bound state. Similar experiments with the two Walker mutants showed that binding of MANT‐ATP was abolished in both mutants (Fig. 3A). Thus, WT SaFlaH bound MANT‐ATP with an affinity higher than 1 μM, but SaFlaHK33A and SaFlaHD122N only bound MANT‐ATP with strongly reduced affinity. Although different conditions and metal‐cofactors were tested, no ATP hydrolysis could be detected either for WT SaFlaH or for the two mutants.
SaFlaH and SaFlaI interact with high affinity (KD = 35 nM) (Banerjee et al., 2013). ATP was present in these experiments, as it was co‐purified with SaFlaH, but no extra ATP was added. Experiments to study whether the interaction between SaFlaH and SaFlaI depends on nucleotide binding to FlaH were complicated by the fact that WT SaFlaH, SaFlaHK33A and SaFlaHD122N were unstable in the nucleotide‐free form. Therefore, we set out to perform these experiments with the homologous proteins from P. furiosus. WT PfFlaH, and the Walker A (PfFlaHK53A) and Walker B mutants (PfFlaHD140N) were expressed in E. coli. PfFlaH, PfFlaHK53A and PfFlaHD140N were purified to homogeneity (see Supplementary Fig. S2B). Determination of the OD260/280 ratio of purified WT PfFlaH showed that the purified protein still contained nucleotides, but for PfFlaH, the nucleotide could be removed with ammonium sulfate precipitation. WT PfFlaH, PfFlaHK53A and PfFlaHD140N were much more stable in the absence of nucleotides than the proteins derived from S. acidocaldarius. MANT‐ATP binding assays with PfFlaH showed that WT PfFlaH bound MANT‐ATP with a 1:1 stoichiometry and an affinity higher than 1 μM, whereas the PfFlaHK53A and PfFlaHD140A mutants bound MANT‐ATP only with much lower affinity (Fig. 3B). To enable the study of the interaction with P. furiosus FlaI (PfFlaI), PfFlaI was also expressed in E. coli and purified to homogeneity (see Fig. S2A) and shown to be highly active (6 μmoles freed phospate/mg protein/min). To test whether PfFlaI and PfFlaH interact, PfFlaH was fluorescently labeled, and microscale thermophoresis (MST) was performed using increasing concentrations of PfFlaI. For these assays, the nucleotide‐bound form of PfFlaH was used. PfFlaI bound to nucleotide‐bound PfFlaH with a KD of around 1 μM (Fig. 3C). Interestingly, PfFlaI bound to the PfFlaHK53A and PfFlaHD140A mutants with strongly reduced affinities, suggesting that nucleotide binding by PfFlaH is important for its interaction with PfFlaI (Fig. 3C). Thus, mutagenesis of the Walker A lysine and the Walker B aspartic acid in the motifs of SaFlaH and PfFlaH strongly reduced nucleotide binding, and in the case of PfFlaH also the interaction with PfFlaI.
ATP binding to SaFlaH is essential for archaellum assembly
To test whether nucleotide binding to SaFlaH is important for assembly of the archaellum, the SaflaH K33A and SaflaH D122N genes as well as the wild‐type flaH were expressed in a ΔaapFΔflaH deletion strain. Strains with a deletion in the aapF gene no longer produce aap pili. Sulfolobus strains that do not express aap pili are more motile in motility plate assays, and the archaellum is more easily identified by electron microscopy (Henche et al., 2012). The ΔaapF and ΔaapFΔflaH deletion strains as well as the ΔaapFΔflaH complemented strains were tested on motility plates (Fig. 4A and B). Deletion of flaH rendered the hypermotile ΔaapF strain non‐motile (Fig. 4A), but the in trans complementation of the ΔaapFΔflaH strain with wild‐type flaH restored motility (Fig. 4A, most right panel). To be able to detect FlaH expression levels, wild‐type FlaH as well as SaflaH K33A and SaflaH D122N were expressed with HA‐tags in the ΔaapFΔflaH strain and tested for motility (Fig. 4B). The HA‐tagged wild‐type FlaH restored motility in the ΔaapFΔflaH strain, although the motility radius was slightly smaller than in the strain complemented with the wild‐type FlaH without the HA‐tag, probably because the tag has an influence on the optimal assembly of the motor complex. But more importantly, the expression of the SaflaH K33A and SaflaH D122N resulted in non‐motile cells (Fig. 4B), although both proteins were expressed at the same levels as the wild‐type protein (Fig. 4C). It was described that in a flaH deletion mutant, the motor scaffold protein FlaX is unstable, suggesting that FlaX is directly or indirectly stabilized by FlaH (Lassak et al., 2012)(see Fig. 4C, lower blot). However, in the complemented ΔaapFΔflaH strains FlaX was stabilized by the presence of the expressed wild type FlaH and FlaH mutant proteins (Fig. 4C, lower blot). These data were confirmed by an analogous dataset in which the FlaH mutations were introduced into the genome of the ΔaapFΔflaH strain (Fig. S3). In the SaflaH K33A and SaflaH D122N expressing strains archaella were not detected by electron microscopy (Fig. S3C). These data suggest that the interaction between FlaH and FlaI depends on the nucleotide bound state of FlaH and that mutations that result in loss of nucleotide binding by FlaH abolish the assembly of the archaellum.
Figure 4.
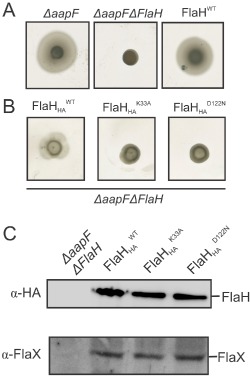
Motility assays and Western Blot analysis of in trans complemented ΔaapFΔflaH strains using wild‐type FlaH and HA tagged FlaH and its Walker mutants.
A. Motility assays of the ΔaapF, ΔaapFΔflaH strain and the in trans with flaH complemented ΔaapFΔflaH strain. Motility plates shown are representative results of three repetitions.
B. Motility assays of the ΔaapFΔflaH strain expressing either HA ‐tagged flaH, flaH K33A or flaH D122N. Motility plates shown are representative results of three repetitions.
C. Whole cell pellets from the ΔaapFΔflaH strain and the in trans complemented strains from (B) were lysed, separated on SDS‐PAGE and blotted on PVDF membranes. Western blot analysis was performed using anti‐HA antibodies and anti‐FlaX antibodies confirming expression of HA‐tagged FlaH wild‐type protein, FlaH Walker mutants and the presence of FlaX.
FlaH interacts with the FlaX C‐terminal domain
The above described experiments supported the earlier notion that the interaction of FlaX with FlaH stabilizes the motor complex formation in vivo. The formation of a ternary complex of SaFlaH, SaFlaI and SaFlaX without its N‐terminal trans‐membrane domain was shown in vitro (Banerjee et al., 2013). FlaX contains a triple helix coiled‐coil region which separates the N‐ and C‐terminal domains (Banerjee et al., 2012). FlaXc, which lacks the first 37 amino acid residues and therefore its transmembrane domain (Fig. 5A), interacted with the N‐terminal as well as with the C‐terminal part of SaFlaI, suggesting that FlaXc builds a molecular scaffold around the FlaI hexamer during archaellum assembly (Banerjee et al., 2013). Moreover, it was demonstrated that the presence of the coiled‐coil region in FlaX was not important for the FlaX‐FlaI interaction. To map the interaction domains of FlaX and FlaH, His6‐FlaXc and StrepII‐FlaH were mixed and incubated with Streptactin beads. As shown in Fig. 5B, FlaXc was only present in the elution fraction when it was incubated in the presence of FlaH but did not bind to the Streptactin beads itself. Therefore, it could be confirmed that FlaXc and FlaH interact in vitro.
Figure 5.
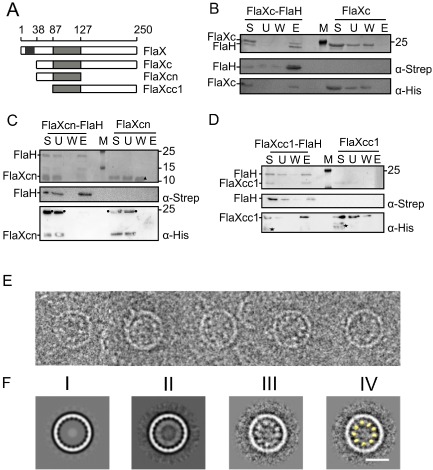
SaFlaH interacts with SaFlax and forms substructures inside the SaFlaX ring in vitro.
A. A schematic of SaFlaX truncates used in this study. The black box indicates the membrane domain (residues 10–32), and the light gray box is the coiled coil region (residues 87–127) of SaFlaX.
B. StrepII‐tagged SaFlaH and His6‐tagged SaFlaXc were mixed (S, starting material) and incubated with streptactin beads. Unbound proteins were removed (U, unbound fraction), the beads were washed (W, wash fraction) and proteins were eluted with 5 mM d‐desthiobiotin (E, elution). As a control, a similar experiment was performed in the absence of SaFlaH. Fractions were separated on a SDS‐PAGE and visualized by Coomassie staining. Fractions were also transferred after SDS‐PAGE to PVDF membranes, and proteins were detected using anti‐Strep and anti‐His antibodies. M depicts the PAGE size standard. An analogous interaction experiment was performed with His6‐tagged SaFlaXcn (C), where no interaction with FlaH was observed. In contrast SaFlaXcc1 (D) showed interaction with FlaH. These results suggest that the C‐terminal domain of FlaX is essential for the interaction with FlaH. Consistent with Banerjee et al. (2012), an asterisk (*) depicts degraded protein in the case of FlaXcc1 in D, a circle (●) depicts an oligomeric forms of the FlaXcn as visualized in the anti‐His blot, a filled triangle (Δ) depict monomers of FlaXcn visualized on SDS‐PAGE.
E. Cryo‐EM images of FlaX/FlaH complexes: selected rings belonging to the class with 20‐fold symmetry. Discrete particles are visible inside the rings, probably representing SaFlaH.
F. Class averages of ring images. (I) SaFlaX ring with 20‐fold symmetry (Banerjee et al., 2012); (II) 20‐fold symmetric SaFlaX/SaFlaH complexes aligned to the SaFlaX ring; (III) Particles belonging to the class average in II aligned to the inner part of the ring. Discrete densities are visible; these densities were smeared out in II resulting in a continuous inner ring; (IV) The densities inside the SaFlaX rings correspond to the size and shape of the monomer structure of SaFlaH (yellow, filtered to 40 Å resolution). Scale bar = 20 nm.
To identify whether the coiled‐coil region of FlaXc is important in the FlaXc–FlaH interaction, we purified N terminal His6 tagged FlaXcn (a deletion of the C‐terminal domain: residue 128–250) and FlaXcc1 (a deletion of the N‐terminal domain: residue 38–86) that both contained the coiled coil region (Fig. 5A) (Banerjee et al., 2012). When both FlaX variants were incubated with StrepII tagged FlaH and Streptactin beads, the results indicated that FlaXcn did not interact with FlaH (Fig. 5C), whereas FlaXcc1 formed a complex with FlaH (Fig. 5D). This result confirmed that the C terminal domain of FlaX (residues 128–250) independent of its coiled‐coil domain interacts with FlaH.
Single particle electron microscopy analysis of the SaFlaX/FlaH complex
To visualize the interaction between SaFlaX and SaFlaH, we mixed the purified proteins in vitro in an equimolar ratio and analyzed them by cryo‐EM. As in the structure of FlaXc alone (Banerjee et al., 2012), we observed hollow rings of varying diameter with symmetries ranging from 15 to 23 fold. Remarkably, almost every SaFlaXc ring contained a ring of extra density on the inside (Fig. 5E; Supplementary Figure S4), demonstrating that FlaH binds inside the FlaX ring. Although individual particles are recognizable in the single images (Fig. 5E), no interpretable substructure was visible in the class averages (Fig. 5F–II), indicating a symmetry mismatch between the SaFlaXc ring and the inner ring. In order to solve the symmetry mismatch, the SaFlaXc rings were first sorted into classes that displayed the same symmetry, then a further alignment and classification against the internal density was performed for each class (Fig. 5F–III). This method gave the best results for the ring with 20‐fold symmetry where nine or 10 discrete particles were observed inside the ring (Fig. 5F–III) correlating in size and shape with the crystal structure of SaFlaH (Fig. 5F–IV) and similar to the densities in the raw data (Fig. 5E, Supplementary Fig. S4A). This suggests that the substructures in the SaFlaX ring with 20‐fold symmetry are formed by monomeric SaFlaH. Thus, SaFlaH binds to the inside of the ring formed by SaFlaX, most likely to the C‐terminal domain of SaFlaX.
Discussion
Archaea are present in most biological environments including e.g. the human gut. All motile archaea use the archaellum for motility so the structural biochemistry and activities of this machinery is of great interest. The flaH, flaI and flaJ genes are conserved in all fla operons among archaea, and the proteins encoded by these genes are proposed to form the archaellum motor core complex (Ghosh and Albers, 2011; Banerjee et al., 2013). To better understand the archaellum assembly, we characterized FlaH, a component whose role was unknown. Our analysis of crenarchaeal and euryarchaeal FlaHs showed that FlaH binds ATP with high affinity but lacks detectable in vitro ATPase activity. The crystal structure of ATP‐bound SaFlaH shows the expected positioning of the residues of the ATP‐binding Walker A and Walker B motifs and that ATP is positioned at a conserved FlaH surface where it most likely is involved in protein–protein interactions (Fig. 2D and E). The closest structural homologue of SaFlaH, PH0284 and PH0186 of P. horikoshii, crystallized as a hexamer (Yokoyama et al., 2000; Kang et al., 2009). Superposition of the monomeric SaFlaH structure on the hexameric PH0284 structure shows that the conserved surfaces surrounding the bound ATP molecule (see Fig. 2E) form the interface between two monomers in the hexamer. It is therefore likely that upon binding to ATP, SaFlaH has the ability to hexamerize as we also showed for PfFlaH using cross‐linking (Fig. 6A).
Figure 6.
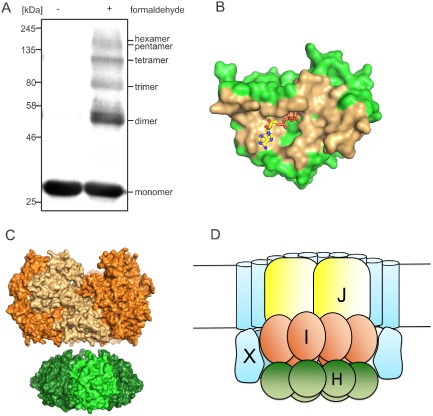
Proposed model of FlaH binding within the FlaX ring and nucleotide‐regulated FlaH binding to FlaI to form the archaellar basal body core.
A. FlaH oligomerization by cross‐linking. FlaH was incubated with or without formaldehyde, separated on SDS‐PAGE and detected by anti His‐tag antibody.
B. FlaH residues involved in interface formation are colored orange for the FlaH‐FlaH interaction modeled by comparison with the PH0284 hexamer.
C. Side view of the SaFlaH hexamer (green) modeled based on the hexameric PH0284 of P . horikoshii with SaFlaI (orange) illustrate that the outer diameters of both protein oligomers are of comparable size.
D. Cartoon of our proposed quaternary structure of the archaellum motor complex taking into account previous data, the cross‐linking result and the FlaH‐FlaX localization and interaction data from Fig. 5.
Mutations in the Walker A and Walker B motifs resulted in proteins that bound nucleotides with a strongly reduced affinity (Fig. 3). Strains expressing these SaFlaH mutants were unable to assemble archaella and were no longer motile, indicating that ATP‐binding by SaFlaH is important for its function in the archaellum motor (Fig. 4). Most importantly, thermophoresis showed that the FlaH–FlaI interaction depends on nucleotide binding by FlaH. FlaI and FlaH can both be present in at least the monomeric and hexameric state. In the interaction studies, the only nucleotides present were derived from FlaH, which was added in the nucleotide‐bound state. Thus, nucleotide‐bound FlaH most likely binds to a monomeric nucleotide‐free FlaI. How these interactions are influenced by the different oligomeric states of FlaH and FlaI is focus of further studies as it is important to determine where the interaction site of FlaH and FlaI is.
We further demonstrated that the C‐terminal domain of SaFlaX is crucial for the interaction with FlaH. EM analysis of in vitro mixed SaFlaXc‐SaFlaH samples showed that SaFlaH is located inside the SaFlaXc ring. Rings of widely different diameters were observed with 15‐ to 23‐fold symmetry, probably as an artifact of the in vitro assembly. Image analysis of the rings with 20‐fold symmetry indicated 9–10 particles of the size and shape of the SaFlaH monomers on the inner side of the SaFlaXc rings. Thus, SaFlaH can interact as a monomer with the SaFlaX ring. The size of the FlaX ring in the in vivo archaellum complex is currently still unknown, and therefore we do not know the exact stoichiometry, or exact numbers of FlaX and FlaH in the rings. The observed hexameric structures of both FlaI and FlaH structural homologues make it possible to speculate that FlaH forms a hexamer in vivo, however (Fig. 6A and B). Interestingly, euryarchaeota like P. furiosus do not encode a homologue of FlaX but have three additional proteins in their archaella operons: FlaC/D/E (Fig. 1). These have probably evolved during the acquisition of the chemotaxis system in these archaea. Although FlaC/D/E do not contain membrane domains like FlaX, they do have coiled‐coil domains and therefore we propose that these take over the role of FlaX in the euryarchaeal archaellum motor complex (Kira Makarova, personal communication).
Thus, these new data along with other existing information indicate that SaFlaX, SaFlaH and SaFlaI interact with each other and together form the archaellar basal body core (Banerjee et al., 2013), FlaI and FlaH can be present in monomeric and hexameric forms, and the interaction between FlaI and FlaH is dependent on nucleotide binding to FlaH. FlaX forms large rings inside which FlaH can bind. FlaI interacts with both FlaX and FlaH, thus placing FlaI inside the FlaX ring (Fig. 6D). Also the inner membrane protein FlaJ is expected to be located inside this FlaX ring. Taken together, the emerging picture indicates a central role for FlaH interactions in the assembly and rotation of the archaellum that is most likely driven by the interactions among FlaH, FlaI, FlaJ and FlaX. In particular, nucleotide‐binding by FlaH promotes FlaI interaction within the FlaX ring (see Fig. 6D).
This model is still speculative but can be used to help design experiments to address three of the next important questions. How do FlaH and FlaI work together in the core complex of the archaellum in both archaellum assembly and rotation? How and when does the switch between assembly and rotation occur, and how are FlaH and FlaI proteins involved in this switch? Likely comparative examination of archaellum among different organisms will play key roles in addressing these and other emerging questions. However, the many strengths of the S. acidocaldarius system including powerful genetics and structures for FlaH, FlaI and FlaF and their assemblies suggest S. acidocaldarius will continue to be one of the strong platforms for these investigations.
Material and methods
Protein expression and purification
Overexpression constructs were transformed into E. coli codon plus cells and grown as preculture overnight at 37°C in LB medium containing ampicillin (50 μg ml−1) and chloramphenicol (34 μg ml−1). Two liters of fresh medium containing antibiotic was inoculated with 2 ml preculture and grown at 37°C to an OD600 of 0.6–0.8. Subsequently, the cultures were chilled on ice for 30 min and induced with 0.5 mM of isopropyl β‐D‐thiogalactopyranoside. Growth was continued for 16 h at 17°C. Cells were collected by centrifugation and frozen in liquid nitrogen and stored at −80°C.
Cells expressing SaFlaH, PfFlaH or mutants of these proteins were thawed on ice, resuspended in 50 ml lysis buffer (50 mM HEPES‐NaOH, pH 7.3, 150 mM NaCl) containing Complete EDTA‐free protease inhibitor cocktail (Roche) and lysed by sonication. Cell debris was removed by centrifugation at 4°C for 30 min at 4600 × g and for 30 min at 20 000 × g. The isolated supernatant was then incubated with DNase I for 1 h on ice followed by precipitation with 20% (w/v) ammonium sulfate. The pellet was collected by centrifugation for 1 h at 4500 × g at 4°C, resuspended in 10 ml lysis buffer and dialyzed overnight against 1 l dialysis buffer (50 mM HEPES pH 7.3, 300 mM NaCl). Further Ni‐NTA (Sigma Aldrich, Seelze, Germany) or Streptactin affinity (IBA GmbH, Göttingen, Germany) based purification were performed using columns equilibrated in purification buffer (50 mM MES, 500 mM NaCl, pH 6.0), and proteins were eluted in purification buffer containing 500 mM imidazol or 2.5 mM d‐desthiobiotin. Further purification of FlaH was achieved by incubation at 50°C for 20 min, followed by centrifugation for 15 min at 10 000 × g, and the supernatant was dialyzed overnight against 1 l purification buffer. The protein aliquots were frozen in liquid nitrogen and stored at −80°C until further use. His6‐tagged PfFlaI was purified and precipitated using 80% (w/v) ammonium sulfate and resuspended in a buffer containing 50 mM Tris‐HCl pH 8, 150 mM NaCl. SaFlaI and SaFlaXc and its variants were purified as described (Ghosh et al., 2011; Banerjee et al., 2012).
Crystallization and structure determination of SaFlaH
For crystallization of SaFlaH, purified SaFlaH was concentrated in purification buffer supplemented with 2 mM ATP and 5 mM MgCl2 using Amicon centrifugal filter devices with a cutoff of 10 kDa. Concentrated SaFlaH was loaded on a Superose 6 column equilibrated in 20 mM Tris pH 7.0, 500 mM NaCl and 5% glycerol. Peak fractions corresponding to momomeric SaFlaH were pooled, concentrated to 6 mg ml−1 and crystallized in 100 mM HEPES pH 7.0, 32.5% PEG 3350 and 150 mM NaSCN. Crystals were flash‐frozen in liquid nitrogen using 1,4‐butanediol as cryo‐protectant. Diffraction data (Otwinowski and Minor, 1997) were collected at the ALS SIBYLS beamline (Berkeley, CA) (Classen et al., 2013) and processed with HKL2000 (Table S1). The FlaH structure was solved with molecular replacement using Phaser (McCoy et al., 2007) and the RecA superfamily ATPase PH0284 from P. horikoshii (2DR3.pdb) as search model. The structure was refined using PHENIX (Adams et al., 2010) and manually fit using COOT (Emsley and Cowtan, 2004). Refinement statistics are shown in Table S1. The structure was deposited at the PDB database as 4YDS.pdb.
Molecular cloning and mutagenesis
To construct expression plasmids for SaFlaH and PfFlaH, the SaflaH or PfflaH genes were amplified from pSVA284 (carrying a codon optimized flaH gene) or genomic DNA of P. furiosus DSM3638, using primer pairs 771/772, 3612/3613, 786/2168 or 3681/3682 respectively. His6 or StrepII tags were included in the primers when necessary. PCR products were cloned as NdeI/XhoI, EcoRI/HindIII, and BamHI/NotI digested fragments into pETDuet1, yielding plasmids pSVA293, pSVA2100, pSVA2108 and pSVA2167. SaFlaH or PfFlaH point mutations were constructed using site directed mutagenesis via round PCR on pSVA2100 or pSVA2167. SaFlaHK33A, SaFlaHD122N, PfFlaHK53A and PfFlaHD140N mutants were created using primer pairs 3632/3633, 3634/3635, 3699/6000 and 6003/6004 yielding plasmids pSVA2130, pSVA2131, pSVA2176 and pSVA2178 respectively. Strains, plasmids and primers used in this study are summarized in Tables S1–S3.
Generation of FlaH in vivo point mutations
In vivo point mutations in S. acidocaldarius were generated via a modification of the protocol described by Lassak et al. (2012). The flaH gene with about 700 bp up‐ and down‐stream regions was amplified by PCR from genomic DNA of S. acidocaldarius DSM639 with primers 3614/3617 and cloned as ApaI/PstI fragment into pSVA406, yielding pSVA2126. Site directed mutagenesis of SaflaH was performed via round PCR on pSVA2126 using primer pairs 3615/3616 and 3627/3628 to obtain pSVA2113 and pSVA2124 respectively. The constructs were methylated in E. coli strain ER1821 and transformed into S. acidocaldarius MW455 (ΔaapF/ΔflaH) to express the protein in trans. All vectors constructed and primers used are shown in Supplementary Tables S2–S4.
MANT‐ATP binding assay
Binding of MANT‐ATP (2′‐/3′‐O‐(N′‐methylanthraniloyl) adenosine‐5′‐O‐triphosphate) was monitored by fluorescence resonance energy transfer (FRET) from excited tryptophanes to the bound MANT‐ATP using a temperature controlled ISS PC1 spectrofluorometer. To reduce background fluorescence by only exciting bound MANT‐nucleotide, the MANT‐ATP was excited via excitation of the tryptophans resulting in Fluorescence Resonance Energy Transfer (FRET). The excitation wavelength was set to 285 nm and the emission detected at 450 nm. Slit widths for excitation and emission were set to 1 nm. Binding was measured by titration of 1 μM SaFlaH or PfFlaH with increasing MANT‐ATP concentrations (0–7 μM) at 25°C in binding buffer (20 mM MES, 500 mM NaCl, 5 mM MnCl2, pH 6.2). Fluorescence was corrected for MANT‐ATP fluorescence in the absence of protein.
ATP hydrolysis assays
ATP hydrolysis was determined for SaFlaH, PfFlaH and mutants of these proteins using the malachite green assay in a buffer containing 20 mM Tris pH 7.0, 500 mM NaCl, 5 % glycerol, 1 mM ATP and 5 mM MgCl2 as described previously (Lanzetta et al., 1979).
Microscale thermophoresis (MST)
For MST measurements, PfFlaH, PfFlaHK53A and PfFlaHD140N were fluorescently labeled using the amine‐reactive Monolith NTTM protein labeling kit NT‐647 (NanoTemper Technologies GmbH) according to the manufacturer's instructions. To determine the binding affinity of PfFlaI, 33.3 nM labeled PfFlaH in 20 mM Tris‐HCl pH 8, 200 mM NaCl and 0.05 % (v/v) Tween‐20 was titrated with increasing concentrations of PfFlaI. The measurements were performed at 25°C, 80% LED power and infrared‐laser power, resulting in a temperature jump of ∼ 2°C. Samples were incubated for 15 min before measurements. The laser on and off times were adjusted to 30 and 5 s respectively. Measurements were performed on a NanoTemper Monolith NT.115 instrument in hydrophilic‐treated capillaries, and analyzed using NT analysis software version 1.4.27 (NanoTemper Technologies GmbH) by fitting the normalized fluorescence (Fnorm) to the Hill equation (Fnorm = (Fnorm,max − Fnorm,min) × [FlaI]n/([FlaI]n + Kdn), where Fnorm,max = maximal normalized fluorescence, Fnorm,min = minimal normalized fluorescence, [FlaI] = concentration of FlaI, Kd = dissociation constant and n = hill coefficient. The percentage bound PfFlaI was determined using the Fnorm,max and Fnorm,min values of the curve fit.
Motility assays and electron microscopy
Swimming assay on semi solid plates and transmission electron microscopy of S. acidocaldarius cells were performed as described previously (Lassak et al., 2012).
In vitro binary complex formation
In vitro protein–protein complex formation was performed essentially as described previously (Banerjee et al., 2013). His6‐tagged FlaH was incubated with either FlaHcc, FlaHcn or FlaHcc1 in a buffer containing 50 mm Tris‐HCl pH 8.0 and 150 mM NaCl, and then applied to Ni‐NTA affinity beads (Ni‐NTA; Sigma Aldrich, Seelze, Germany). Beads were washed, and proteins were eluted in the same buffer containing 500 mM imidazole. As a control FlaHcc, FlaHcn or FlaHcc1 were bound to a column in the absence of His6‐tagged FlaH. Fractions were separated on SDS‐PAGE and either stained by Coomassie or transferred to PVDF membranes and analyzed using α‐His or α‐Strep antibodies.
Cryo EM/single particle analysis
Purified SaFlaX and SaFlaH were mixed in a stoichiometric manner. For initial analysis, FlaX/FlaH complexes were diluted to a final concentration of 0.05 mg ml−1 and negatively stained with 1% (w/v) uranyl acetate. Electron micrographs were collected using a Philips CM120 at 120 kV at a magnification of 44 000× on a Gatan 2k × 2k CCD camera.
For cryo‐EM, FlaX/FlaH samples at a protein concentration of 2.5 mg ml−1 were applied to glow‐discharged Quantifoil grids (Quantifoil MicroTools) and vitrified by injection into liquid ethane using a Vitrobot plunge‐freezing device (FEI). Images were recorded using a back‐thinned FEI Falcon II direct electron detector. Images were recorded manually at a nominal magnification of 39 000× yielding a pixel size at the specimen of 2.74 Å at a defocus of 2–6 μm. Particles were selected using the boxer module from EMAN (Ludtke et al., 1999). The CTF was determined using EMAN2 (Tang et al., 2007). A data set of 16 000 particles was processed with Imagic V (van Heel et al., 1996). The images were subjected to multireference alignment and multivariate statistical analysis. The images were classified and images assigned to the same class were averaged.
Cross‐linking experiment
The oligomerization behavior of FlaH was assessed using chemical cross‐linking as described earlier in (Banerjee et al., 2012) and formaldehyde was used as chemical cross‐linker. 37% formaldehyde was diluted to a final concentration of 5% (v/v) in 20 mM HEPES, pH 7.2, 150 mM NaCl buffer to prepare the fresh working stock. 0.5 mg ml−1 of protein was incubated with 0.1% (v/v) formaldehyde, and the mixture was incubated at room temperature for 5 min, and the cross‐linking reaction was stopped using 125 mM glycine‐NaOH, pH 10.
Finally, the cross‐linked product was precipitated with trichloroacetic acid (TCA). One volume of 50% TCA stock was mixed with two volumes of cross‐linked sample and incubated for 30 min on ice. Samples were centrifuged at 14 000 × g for 5 min. The supernatant was removed, and the white fluffy pellet was washed three times with 200 μl of ice‐cold 100% acetone for the complete removal of the TCA. The pellet was then dried in a heat block at 95°C to remove residual acetone and resuspended in 20 mM HEPES, pH 7.2, 150 mM NaCl buffer. The sample was mixed with Laemmli buffer, and the cross‐linked oligomers of FlaH were visualized on 11% SDS‐PAGE with Coomassie blue staining and anti‐His western blot.
Authors contributions
PC, TN and AB purified protein, performed activity and interaction assays, in vivo experiments and designed experiments, wrote manuscript and made figures. EDI performed cryo‐EM and single particle analysis and interpretation in cooperation with JV; they both wrote the part of the paper discussing the cryo‐EM results. SR crystallized FlaH, collected diffraction data and built the structure. AG set up protein purification and designed experiments. ASA helped to solve the FlaH structure. DJM assisted with cryo‐EM sample preparation and data collection. JAT directed the structural experiments. CvdD, JAT, JV and SVA wrote and revised the manuscript and designed experiments.
Supporting information
Supporting information
Acknowledgements
TN and PC were supported by an ERC starting grant (Nr. 311523, Archaellum). AG, AB und SVA were supported by intramural funds of the Max Planck Society. We are grateful to Werner Kühlbrandt for his support. We thank Juan Castillo‐ Hernández for computer support. This work was supported by the National Institutes of Health grant MINOS (Macromolecular Insights on Nucleic Acids Optimized by Scattering) GM105404 (MH/JAT). SIBYLS beamline efforts to combine structural methods at the Advanced Light Source of Lawrence Berkeley National Laboratory are supported in part by United States Department of Energy program Integrated Diffraction Analysis Technologies (IDAT). The authors declare no conflict of interests.
References
- Adams, P.D. , Afonine, P.V. , Bunkóczi, G. , Chen, V.B. , Davis, I.W. , Echols, N. , et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers, S.‐V. , and Jarrell, K.F. (2015) The archaellum: how Archaea swim. Front Microbiol 6: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers, S.V. , Szabo, Z. , and Driessen, A.J. (2003) Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol 185: 3918–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A. , and Albers, S.‐V. (2015) FlaF is a β‐sandwich protein that anchors the archaellum in the archaeal cell envelope by binding the S‐layer protein. Structure 23: 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A. , Ghosh, A. , Mills, D.J. , Kahnt, J. , Vonck, J. , and Albers, S.‐V. (2012) FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring‐shaped structures and interacts with the motor ATPase FlaI. J Biol Chem 287: 43322–43330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A. , Neiner, T. , Tripp, P. , and Albers, S.‐V. (2013) Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS J 280: 6141–6149. [DOI] [PubMed] [Google Scholar]
- Berry, J.‐L. , and Pelicic, V. (2015) Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol Rev 39: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban, B. , Ng, S.Y. , Kanbe, M. , Saltzman, I. , Nimmo, G. , Aizawa, S. , and Jarrell, K.F. (2007) Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis . Mol Microbiol 66: 596–609. [DOI] [PubMed] [Google Scholar]
- Classen, S. , Hura, G.L. , Holton, J.M. , Rambo, R.P. , Rodic, I. , McGuire, P.J. , et al (2013) Implementation and performance of SIBYLS: a dual endstation small‐angle X‐ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J Appl Crystallogr 46: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, L. , Pique, M.E. , and Tainer, J.A. (2004) Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol 2: 363–378. [DOI] [PubMed] [Google Scholar]
- Emsley, P. , and Cowtan, K. (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Ghosh, A. , and Albers, S.‐V. (2011) Assembly and function of the archaeal flagellum. Biochem Soc Trans 39: 64–69. [DOI] [PubMed] [Google Scholar]
- Ghosh, A. , Hartung, S. , Does, C. , van der Tainer, J.A. , and Albers, S.‐V.V. (2011) Archaeal flagellar ATPase motor shows ATP‐dependent hexameric assembly and activity stimulation by specific lipid binding. Biochem J 437: 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, P.I. , and Whiteheart, S.W. (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6: 519–529. [DOI] [PubMed] [Google Scholar]
- van Heel, M. , Harauz, G. , Orlova, E.V. , Schmidt, R. , and Schatz, M. (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24. [DOI] [PubMed] [Google Scholar]
- Henche, A.‐L. , Ghosh, A. , Yu, X. , Jeske, T. , Egelman, E. , and Albers, S.‐V. (2012) Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius . Environ Microbiol 14: 3188–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm, L. , and Rosenström, P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res 38: W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell, K.F. , and Albers, S.‐V. (2012) The Archaellum: an old structure with a new name. Trends Microbiol 20: 307–312. [DOI] [PubMed] [Google Scholar]
- Jarrell, K.F. , and McBride, M.J. (2008) The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6: 466–476. [DOI] [PubMed] [Google Scholar]
- Kang, H.‐J. , Kubota, K. , Ming, H. , Miyazono, K. , and Tanokura, M. (2009) Crystal structure of KaiC‐like protein PH0186 from hyperthermophilic archaea Pyrococcus horikoshii OT3. Proteins Struct Funct Bioinforma 75: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Lanzetta, P.A. , Alvarez, L.J. , Reinach, P.S. , and Candia, O.A. (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem 100: 95–97. [DOI] [PubMed] [Google Scholar]
- Lassak, K. , Ghosh, A. , and Albers, S.‐V. (2012) Diversity, assembly and regulation of archaeal type IV pili‐like and non‐type‐IV pili‐like surface structures. Res Microbiol 163: 630–644. [DOI] [PubMed] [Google Scholar]
- Lassak, K. , Neiner, T. , Ghosh, A. , Klingl, A. , Wirth, R. , and Albers, S.‐V. (2012) Molecular analysis of the crenarchaeal flagellum. Mol Microbiol 83: 110–124. [DOI] [PubMed] [Google Scholar]
- Ludtke, S.J. , Baldwin, P.R. , and Chiu, W. (1999) EMAN: semiautomated software for high‐resolution single‐particle reconstructions. J Struct Biol 128: 82–97. [DOI] [PubMed] [Google Scholar]
- McCoy, A.J. , Grosse‐Kunstleve, R.W. , Adams, P.D. , Winn, M.D. , Storoni, L.C. , and Read, R.J. (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshcheryakov, V.A. , Yoon, Y.‐H. , Matsunami, H. , and Wolf, M. (2014) Purification, crystallization and preliminary X‐ray crystallographic analysis of the flagellar accessory protein FlaH from the methanogenic archaeon Methanocaldococcus jannaschii . Acta Crystallogr Sect F Struct Biol Commun 70: 1543–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. , and Minor, W. (1997) Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Patenge, N. , Berendes, A. , Engelhardt, H. , Schuster, S.C. , and Oesterhelt, D. (2001) The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum . Mol Microbiol 41: 653–663. [DOI] [PubMed] [Google Scholar]
- Peabody, C.R. , Chung, Y.J. , Yen, M.R. , Vidal‐Ingigliardi, D. , Pugsley, A.P. , and Saier, M.H., Jr (2003) Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149: 3051–3072. [DOI] [PubMed] [Google Scholar]
- Reindl, S. , Ghosh, A. , Williams, G.J. , Lassak, K. , Neiner, T. , Henche, A.‐L. , et al (2013) Insights into FlaI functions in archaeal motor assembly and motility from structures, conformations, and genetics. Mol Cell 49: 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, D.S. , Pellegrini, L. , Daniels, D.S. , Yelent, B. , Craig, L. , Bates, D. , et al (2003) Full‐length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J 22: 4566–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G. , Peng, L. , Baldwin, P.R. , Mann, D.S. , Jiang, W. , Rees, I. , and Ludtke, S.J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157: 38–46. [DOI] [PubMed] [Google Scholar]
- Thomas, N.A. , Pawson, C.T. , and Jarrell, K.F. (2001) Insertional inactivation of the flaH gene in the archaeon Methanococcus voltae results in non‐flagellated cells. Mol Genet Genomics 265: 596–603. [DOI] [PubMed] [Google Scholar]
- Tripepi, M. , Imam, S. , Pohlschröder, M. , and Pohlschroder, M. (2010) Haloferax volcanii flagella are required for motility but are not involved in PibD‐dependent surface adhesion. J Bacteriol 192: 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , He, Y. , Moya, I.A. , Qian, X. , and Luo, Y. (2004) Crystal structure of archaeal recombinase RADA: a snapshot of its extended conformation. Mol Cell 15: 423–435. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S. , Hirota, H. , Kigawa, T. , Yabuki, T. , Shirouzu, M. , Terada, T. , et al (2000) Structural genomics projects in Japan. Nat Struct Biol 7 (Suppl.): 943–945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


