Abstract
Inositol 1,4,5-trisphosphate receptors (IP3R) and ryanodine receptors are the channels responsible for Ca2+ release from the endoplasmic and sarcoplasmic reticulum. High-resolution structures of these enormous channels are beginning to reveal the inter-domain re-arrangements that link binding of their key regulators (IP3 and/or Ca2+) to opening of the Ca2+-permeable pore. Activation of IP3R is initiated when IP3 causes its clam-like binding domain to close, thereby disrupting interactions with other domains that may then communicate with the pore. All four IP3-binding sites within the tetrameric IP3R must bind IP3 before the channel can open. This has important consequences for the distribution of both IP3 and IP3R activity within cells, and it may contribute to the complex spatial organization of intracellular Ca2+ signals.
The endoplasmic reticulum (ER), which ramifies throughout the cell making contacts with many other intracellular membranes, is well placed to deliver Ca2+ signals rapidly to specific targets via its Ca2+-permeable channels. Opening of these channels allows Ca2+ to flow into the cytosol, causing the increase in cytosolic Ca2+ concentration that regulates many cellular processes. Pumps within the ER membrane eventually return Ca2+ to the ER lumen. Ca2+ fluxes across the plasma membrane can also generate Ca2+ signals, but cycling of Ca2+ across ER membranes is more economical, and it allows more targeted delivery to intracellular destinations. Within the ER, two related families of Ca2+ channels, inositol 1,4,5-trisphosphate receptors (IP3R) and ryanodine receptors (RyR), mediate most Ca2+ signalling. Both IP3R and RyR are stimulated by Ca2+, allowing them to propagate Ca2+ signals regeneratively as Ca2+ released by one channel ignites the activity of another (1). For IP3Rs, IP3 sets the gain on this potentially explosive Ca2+ release because IP3Rs must bind IP3 before they can respond to Ca2+. However, for IP3R the IP3-binding core (IBC) and pore lie at opposite ends of the primary sequence and far apart within the quaternary structure (Fig. 1), and the location of the stimulatory Ca2+-binding site is unknown (2). So how does IP3, via its effects on Ca2+ binding, lead to opening of a distant pore? Structural analyses and a clever manipulation of IP3R subunits have now revealed how IP3 binding initiates IP3R activation.
Fig. 1. Structure of the IP3R.
(A) The essential 4- and 5-phosphate groups of IP3 bind to opposite sides (β- and α-domains) of the clam-like IP3-binding core (IBC) causing the clam to close and re-position the suppressor domain (SD), which moves with the α-domain of the IBC (Protein Data Bank, PDB, 3U4J) (5). Locations of key domains within the primary sequence are shown below. (B) View from the cytosol showing the four N-terminal regions (SD and IBC) and the C-terminal domains (CTD) from each subunit (the central splayed α-helices). The coloured subunits (orange and green) show how the CTD from one subunit contacts the N-terminal region (the β-IBC) of another. The N- (blue) and C-termini (red) of each subunit, through which Alzayady et al. linked subunits to form concatamers (16), are highlighted; the termini of one subunit are enclosed in the yellow box. Within each IBC, the position where IP3 would bind is shown by a yellow circle (PDB, 3JAV) (11). (C) View across the ER membrane, with 2 subunits highlighted in shades of orange and green. The transmembrane region with its 24 α-helices, six from each subunit, are shown (PDB, 3JAV) (11). (D) One of the likely inter-subunit interactions through which IP3R binding to the IBC is communicated to the pore. The β-domain of the IBC of one subunit contacts the C-terminal domain (CTD) of another. The CTD extends as an almost unbroken α-helix through the centre of the protein to helix S6, which lines the pore. For the pore to open and allow Ca2+ to pass, S6 must move to disrupt the hydrophobic constriction that occludes the closed pore (red arrow).
IP3R and RyR are bigger than any other ion channel. Each channel is tetrameric, invariably a homo-tetramer for RyR, but IP3R form homo- and heterotrameric structures from three related subunits (IP3R1-3) and their splice variants (3). The enormous size of these channels and the diversity of IP3R tetramers have hampered structural analyses, although crystal structures of receptor fragments have provided valuable insight. High-resolution structures of the N-terminus of IP3R, for example, showed how IP3 is recognized and initiates IP3R activation by closing the clam-like IBC, thereby displacing the N-terminal suppressor domain (SD) (4, 5). The SD is essential for channel gating (Fig. 1A). Similar analyses of RyR showed that the inactive N-terminal structures of IP3R and RyR are similar (6). Indeed, the structures of the N-terminal and pore regions of the two receptors are sufficiently similar to allow chimeric IP3R/RyR channels to function (5). Improvements to single-particle electron cryomicroscopy, notably use of direct electron capture cameras and improved image reconstruction algorithms, is transforming structural biology (7). Among the many beneficiaries of this ‘resolution revolution’ (7) are intracellular Ca2+ channels, with near-atomic resolution structures of RyR1 (8–10) and IP3R1 (11) published in 2015. Each channel forms a mushroom-like structure anchored in the ER by its stalk. The N-terminal domains of each subunit, which include the IBC of the IP3R, are arranged like the four blades of a propeller around a hole in the middle of the domed cytoplasmic cap of the mushroom (Fig. 1B). The stalk forms the ion-conducting pore, the architecture of which is broadly similar to that of voltage-gated cation channels. The pore is lined by the tilted S6 helices from each subunit, and together the helices form a hydrophobic constriction towards the cytosolic end of the ER membrane. In the open channel, this constriction must dilate to allow Ca2+ to pass (10). A selectivity filter at the luminal entrance of the pore (12) and a ring of acidic residues around the cytosolic vestibule (11) contribute to the cation-selectivity of the open pore. The S6 helix of the intracellular Ca2+ channels is unusual in reaching well beyond the ER membrane and into the cytosol. Within IP3R, each extended S6 helix is linked to the α-helical C-terminal domain (CTD) by a pair of short orthogonal helices. Together these domains form an almost continuous α-helical structure extending from the pore, through the core of the protein, to the cytosolic surface, where the CTD contacts the IBC of a neighbouring subunit (Fig. 1D). This arrangement, which had been presciently predicted from analyses of the effects of IP3 and Ca2+ on the accessibility of C-terminal residues (2), is intriguing because it suggests a direct link between IP3-evoked closure of the clam-like IBC and the distant pore. Other inter-subunit interfaces, notably those involving the essential SD and the IBC (5) or ARM3 domain of an adjacent subunit (11) may also facilitate communication between IP3 binding and the pore. The idea that IP3R activation is associated with movements at domain interfaces aligns with evidence from RyRs showing that most disease-causing mutations, which tend to cause channels to open too willingly, are located at the boundaries between domains (6). The structures beautifully illustrate how IP3 binds to evoke conformational changes around the IBC, they reveal the structure of the pore, and they suggest how the initial conformational changes might be transmitted to the pore (Fig. 1). But where does Ca2+ binding sit within this sequence? The answer to that is likely to come from structures of the IP3R in its active state.
Steep concentration-effect relationships for IP3-evoked Ca2+ release suggest that IP3 must bind to several of the four IP3-binding sites before the channel can open (13), but it has never been clear whether every site must bind IP3. A recent report resolves the issue by demonstrating that within a tetrameric concatamer of IP3R subunits, each must have a functional IP3-binding site for IP3 to evoke Ca2+ release. Tying protein subunits together with flexible linkers to provide concatenated structures of defined composition has been used for other ion channels (14), but it is challenging for subunits as large as the IP3R, and daunting because residues near the N- and C-termini of IP3Rs control gating. The task is made a little easier by the proximity of the N- and C-termini of adjacent subunits within a tetrameric IP3R (Fig. 1B), allowing them to be joined with a 14-residue linker (15). Alzayady et al. (16) used these methods to produce a concatenated IP3R tetramer that behaves normally, but it opens only if all four subunits can bind IP3. Furthermore, the need for all four IP3-binding sites could not be circumvented by high concentrations of the co-agonist, Ca2+, or by an allosteric regulator of gating, ATP. IP3R can open only when all four IP3-binding sites are occupied, and seemingly no amount of help from co-regulators can surmount that need. This is an important observation because it suggests that dissociation of any one of four bound IP3 is enough to close an IP3R, and it implies that regenerative propagation of Ca2+ signals by Ca2+-induced Ca2+ release can proceed only via IP3R in which all four IP3-binding sites have bound IP3. That conclusion has some interesting consequences.
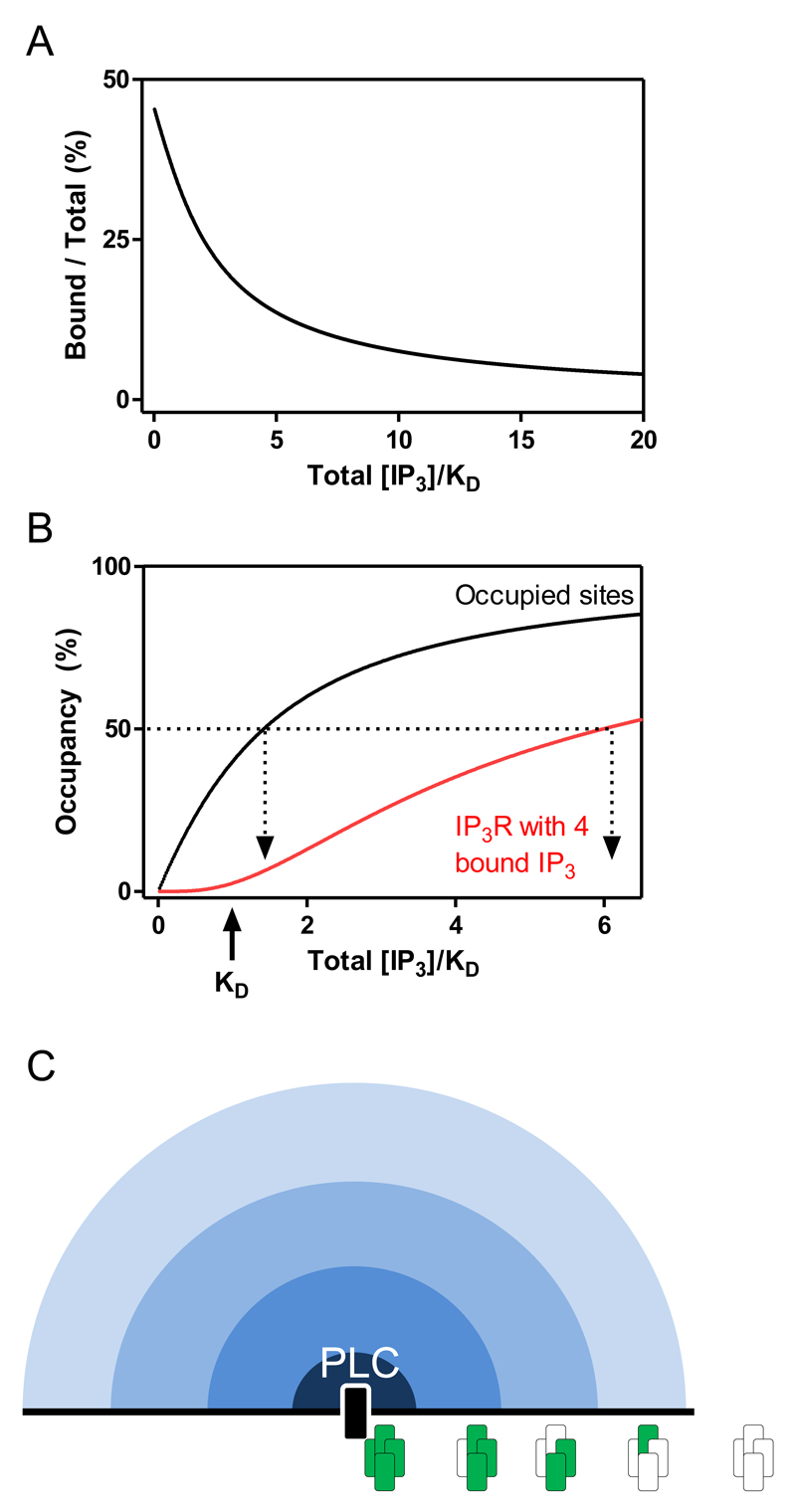
In a typical cell, expressing ~7200 tetrameric IP3Rs (17), the cytosolic concentration of IP3-binding sites is ~100nM (18). That concentration is similar to the affinity of IP3R for IP3 (equilibrium dissociation constant, KD = 119nM) measured under conditions that match those used for functional assays (19). Hence, across the range of IP3 concentrations that achieve graded occupancy of IP3Rs, the IP3Rs are themselves significant buffers of the free IP3 concentration (Fig. 2A). Many years ago, an influential analysis of Ca2+ and IP3 diffusion coefficients in cytoplasm concluded that whereas Ca2+ was heavily buffered, IP3 diffused freely (20), leading to a view that only Ca2+ functions as a local messenger. However, that analysis used cytoplasm from Xenopus oocytes, where the concentration of IP3Rs is likely to be substantially lower than in more typical cells. Since IP3Rs must bind four IP3 molecules to become active, binding events at low concentrations of IP3 are mostly ‘unproductive’ because the IP3 is too thinly spread across IP3Rs for individual receptors to acquire the four IP3 molecules they need. With a free concentration of IP3 (~40nM) that occupies 25% of the IP3-binding sites, for example, only 0.4% of IP3Rs will have four IP3 bound (25%4), just 28 IP3Rs in our typical cell. But at this concentration of IP3, equating to 12,000 IP3 molecules per cell, some 7200 IP3 would be bound to IP3R (Fig. 2A). The total concentration of IP3 would therefore need to increase to 64nM to provide a free concentration of 40nM. Hence, buffering of IP3 by IP3R and the need for IP3R to bind four IP3 conspire to ensure that substantial increases in IP3 concentration are required to achieve appreciable IP3R activity. En route to that activity most IP3Rs will have bound some IP3 (Fig. 2B). This distribution of IP3 across IP3Rs may allow local signalling to IP3Rs from phospholipase C (PLC), and perhaps thereby explain how different PLC-coupled receptors can evoke Ca2+ release from different IP3-sensitive stores within a single cell (21). We can envisage a steeply declining IP3 concentration radiating outwards from PLC as IP3 encounters IP3Rs (Fig. 2C). Close to PLC, IP3 concentrations may be sufficient to achieve IP3R activation, but beyond that there will be larger zones within which many IP3Rs have bound some IP3. Although these partially occupied IP3Rs may not open, IP3 binding may modify their behaviour in other ways. We, for example, have suggested that partially liganded IP3Rs may cluster and so prime IP3R to respond to Ca2+ released by their neighbours (22), although that idea has been contentious (1). Another possibility is that IP3 might trigger dissociation of IRBIT (IP3R-binding protein released by IP3), an endogenous antagonist of IP3Rs (23), from a neighbouring subunit and so prime it to bind IP3. The key point is that under physiological conditions, opening of IP3Rs is preceded by a wave of IP3 binding to very many more IP3Rs, and that may play some part in better preparing them to respond when they acquire additional IP3.
Fig. 2. IP3Rs bind and buffer IP3.
(A) Within a typical cell, the concentration of IP3-binding sites and their affinity (KD) for IP3 are similar (~100nM). An inescapable consequence is that within the range of IP3 concentrations that achieve graded occupancy of the binding sites, a substantial fraction of the IP3 is bound, thereby depleting the cytosol of free IP3 (18). (B) Buffering of cytosolic IP3 by IP3R shifts the relationship between IP3 concentration and IP3 binding to higher concentrations of IP3: a total concentration that exceeds the KD is now required to occupy 50% of the binding sites. Because 4 sites must be occupied to activate an IP3R, the IP3 concentrations needed to activate 50% of IP3Rs are much higher than those needed to occupy 50% of the binding sites. In this simple scheme, a concentration of IP3 that occupied 50% of all binding sites, would allow only 6% of all IP3Rs, about 450 IP3Rs in a typical cell, to bind the 4 IP3 needed for activity. (C) IP3 produced by PLC is captured by IP3Rs as it diffuses into the cytosol creating a radial concentration gradient. Within the gradient, IP3Rs very close to PLC may acquire the 4 IP3 molecules needed for activity (green), but beyond that there is an extensive penumbra of IP3Rs that bind IP3 to fewer of their subunits.
The Ca2+ signals detected in intact cells after photolysis of caged IP3, where IP3 is uniformly delivered to the cytosol, are remarkable for two reasons that become more so in light of the evidence that IP3Rs can open only after they bind four IP3 molecules (16). First, IP3 evokes local Ca2+ signals that reflect the almost simultaneous opening of a handful of IP3Rs within a small cluster. Second, these Ca2+ release events are sparse, just a few per cell, and yet they often revisit the same site (1). Ca2+ is proposed to mediate the rapid recruitment of IP3Rs within these clusters, but the new evidence from Alzayady et al. (16) tells us that this can occur only if the neighbours have four IP3 bound. How, when so few IP3Rs open (just a handful from the 7000 in a cell), does the IP3 binding become so concentrated into a few small clusters of IP3Rs?
After more than 30 years of waiting, cryo-electron microscopy has provided our first glimpse of the IP3R in sufficient detail (11) to begin to see how IP3 binding to each of its four subunits (16) leads to opening of the Ca2+ pore that initiates cytosolic Ca2+ signalling. That breakthrough comes as gene-editing and high-resolution measurements of Ca2+ signals in intact cells provide the techniques needed to explore the structural determinants of IP3R behaviour in living cells.
Acknowledgments
Funding: Work from the authors’ laboratory is supported by the Wellcome Trust.
Footnotes
Competing Interests: The authors declare that they have no competing interests.
References and Notes
- 1.Smith IF, Wiltgen SM, Shuai J, Parker I. Ca2+ puffs originate from preestablished stable clusters of inositol trisphosphate receptors. Sci Signal. 2009;2:ra77. doi: 10.1126/scisignal.2000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anyatonwu G, Khan MT, Schug ZT, da Fonseca PC, Morris EP, Joseph SK. Calcium-dependent conformational changes in inositol trisphosphate receptors. J Biol Chem. 2010;285:25085–25903. doi: 10.1074/jbc.M110.123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor CW, Tovey SC. IP3 receptors: toward understanding their activation. Cold Spring Harb Persp Biol. 2010;2 doi: 10.1101/cshperspect.a004010. a004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosanac I, Alattia J-R, Mal TK, Chan J, Talarico S, Tong FK, Tong KI, Yoshikawa F, Furuichi T, Iwai M, Michikawa T, et al. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature. 2002;420:696–700. doi: 10.1038/nature01268. [DOI] [PubMed] [Google Scholar]
- 5.Seo M-D, Velamakanni S, Ishiyama N, Stathopulos PB, Rossi AM, Khan SA, Dale P, Li C, Ames JB, Ikura M, Taylor CW. Structural and functional conservation of key domains in InsP3 and ryanodine receptors. Nature. 2012;483:108–112. doi: 10.1038/nature10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Petegem F. Ryanodine receptors: structure and function. J Biol Chem. 2012;287:31624–31632. doi: 10.1074/jbc.R112.349068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henderson R. Overview and future of single particle electron cryomicroscopy. Arch Biochem Biophys. 2015;581:19–24. doi: 10.1016/j.abb.2015.02.036. [DOI] [PubMed] [Google Scholar]
- 8.Yan Z, Bai XC, Yan C, Wu J, Li Z, Xie T, Peng W, Yin CC, Li X, Scheres SH, Shi Y, et al. Structure of the rabbit ryanodine receptor RyR1 at near-atomic resolution. Nature. 2015;517:50–55. doi: 10.1038/nature14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zalk R, Clarke OB, Georges AD, Grassucci RA, Reiken S, Mancia F, Hendrickson WA, Frank J, Marks AR. Structure of a mammalian ryanodine receptor. Nature. 2015;517:44–49. doi: 10.1038/nature13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efremov RG, Leitner A, Aebersold R, Raunser S. Architecture and conformational switch mechanism of the ryanodine receptor. Nature. 2015;517:39–43. doi: 10.1038/nature13916. [DOI] [PubMed] [Google Scholar]
- 11.Fan G, Baker ML, Wang Z, Baker MR, Sinyagovskiy PA, Chiu W, Ludtke SJ. Serysheva, II, Gating machinery of InsP3R channels revealed by electron cryomicroscopy. Nature. 2015;527:336–341. doi: 10.1038/nature15249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schug ZT, da Fonseca PC, Bhanumathy CD, Wagner L, 2nd, Zhang X, Bailey B, Morris EP, Yule DI, Joseph SK. Molecular characterization of the inositol 1,4,5-trisphosphate receptor pore-forming segment. J Biol Chem. 2008;283:2939–2948. doi: 10.1074/jbc.M706645200. [DOI] [PubMed] [Google Scholar]
- 13.Meyer T, Holowka D, Stryer L. Highly cooperative opening of calcium channels by inositol 1,4,5-trisphosphate. Science. 1988;240:653–656. doi: 10.1126/science.2452482. [DOI] [PubMed] [Google Scholar]
- 14.Sack JT, Shamotienko O, Dolly JO. How to validate a heteromeric ion channel drug target: assessing proper expression of concatenated subunits. J Gen Physiol. 2008;131:415–420. doi: 10.1085/jgp.200709939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alzayady KJ, Wagner LE, 2nd, Chandrasekhar R, Monteagudo A, Godiska R, Tall GG, Joseph SK, Yule DI. Functional inositol 1,4,5-trisphosphate receptors assembled from concatenated homo- and heteromeric subunits. J Biol Chem. 2013;288:29772–29784. doi: 10.1074/jbc.M113.502203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alzayady KJ, Wang L, Chandrasekhar R, Wagner LE, Van Petegem F, Yule DI. Defining the stoichiometry of inositol 1,4,5-trisphosphate binding required to initiate Ca2+ release. Sci Signal. 2016 doi: 10.1126/scisignal.aad6281. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tovey SC, Dedos SG, Taylor EJA, Church JE, Taylor CW. Selective coupling of type 6 adenylyl cyclase with type 2 IP3 receptors mediates a direct sensitization of IP3 receptors by cAMP. J Cell Biol. 2008;183:297–311. doi: 10.1083/jcb.200803172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A HEK cell has a total volume of ~1pL, with ~50% of the cytoplasm occupied by organelles. With 28,800 IP3-binding sites/cell, this suggests that within the cytosolic volume into which IP3 can distribute, the concentration of IP3-binding sites is ~100nM
- 19.Ding Z, Rossi AM, Riley AM, Rahman T, Potter BVL, Taylor CW. Binding of inositol 1,4,5-trisphosphate (IP3) and adenophostin A to the N-terminal region of the IP3 receptor: thermodynamic analysis using fluorescence polarization with a novel IP3 receptor ligand. Mol Pharmacol. 2010;77:995–1004. doi: 10.1124/mol.109.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allbritton NL, Meyer T, Stryer L. Range of messenger action of calcium ion and inositol 1,4,5-trisphosphate. Science. 1992;258:1812–1815. doi: 10.1126/science.1465619. [DOI] [PubMed] [Google Scholar]
- 21.Short AD, Winston GP, Taylor CW. Different receptors use inositol trisphosphate to mobilize Ca2+ different intracellular pools. Biochem J. 2000;351:683–686. [PMC free article] [PubMed] [Google Scholar]
- 22.Rahman TU, Skupin A, Falcke M, Taylor CW. Clustering of IP3 receptors by IP3 retunes their regulation by IP3 and Ca2+ Nature. 2009;458:655–659. doi: 10.1038/nature07763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ando H, Kawaai K, Mikoshiba K. IRBIT: A regulator of ion channels and ion transporters. Biochim Biophys Acta. 2014;1843:2195–2204. doi: 10.1016/j.bbamcr.2014.01.031. [DOI] [PubMed] [Google Scholar]