Abstract
Sodium/proton exchangers (NHX) are key players in plant responses to salinity and have a central role in establishing ion homeostasis. NHXs can be localized in tonoplast or plasma membranes, where they exchange sodium ions for protons, resulting in the removal of ions from the cytosol into vacuole or extracellular spaces. In the present study, the expression pattern of the gene encoding Na+/H+ antiporter in the vacuolarmembrane (NHX1 gene) in Leptochloa fusca (Kallar grass) was measured by a semi- quantitative RT-PCR method under different treatments of NaCl and CdCl2. Results indicated that NaCl positively affected expression levels of LfNHX1, and that the amount of LfNHX1 mRNA increased in conjunction with the rise of salinity pressure, This finding suggests that vacuolar Na+/H+ antiporter might play an important role in the salt tolerance ability of kallar grass. The results also showed that cadmium exposure significantly modulated the mRNA expression of the LfNHX1 gene, suggesting that cadmium exposure disturbed Na+ homeostasis across the tonoplast and decreased the salt tolerance ability of kallar grass.
Key Words: Kallar grass, Salt stress, Cadmium, NHX1, Semi-quantitative RT-PCR
INTRODUCTION
Soil salinization has become a pervasive environmental concern and a worldwide agricultural productivity issue. Most crop plants are sensitive to salinity caused by high concentrations of soil salts [1]. More than 800 million ha of land throughout the world are salt-affected, accounting for about 6% of the Earth’s total land area. Out of 1500 million ha of land farmed by dryland agriculture, 32 million ha (2%) are affected by varying degrees of salinity, and out of the current 230 million ha of irrigated land, 45 million ha (20%) are affected by salt [2]. Unfortunately, saline soil is still rapidly expanding because of irrigation with salty water, improper drainage, entry of seawater in coastal areas, and salt accumulation in arid and semiarid regions [3].
Salt stress is mainly caused by sodium ions, whose high concentrations can be toxic to most organisms. Besides ion toxicity, osmotic stress and K+/Na+ ionic imbalances can cause aberrant metabolism such as the inhibition of cytosolic enzyme activities [4].
Plants use two strategies to maintain low Na+ concentrations: sodium exclusion and sodium compartmentation. Plasma membrane-bound Na+/H+ antiporters operate to transport sodium out of the cell [5]. These antiporters are ubiquitous membrane proteins which catalyze the exchange of Na+ for H+ across membranes, thereby playing an important role in cellular pH and Na+ homeostasis throughout the biological kingdom [6].
Salt toxicity can be mainly attributed to Na+-specific damage to the cytoplasm of the cell [7]. Na+/H+ antiporter activities, which are involved in transporting Na+ out of the cytosol into the vacuole and the apoplast, have been detected in both plasma membrane and tonoplast vesicle preparations of different plant species [6]. Physiological studies suggest the role of these Na+/H+ antiporters in plant salt tolerance [8].
The vacuolar Na+/H+ antiporter (NHX1) has long been proposed to play importantroles in salt tolerance. The compartmentalization of Na+ into the vacuole might reduce the deleterious effects of excess Na+ in the cytosol and maintain osmotic balance using Na+ as a cheap osmoregulatory substance, thus enhancing water uptake and salt tolerance of plants [9]. While transporting Na+ into vacuoles, Na+/H+ antiporters use the pH gradient generated by vacuolar H+-translocating enzymes H+ ATPase and PPiase [10].
Blumwald and Poole [11] first reported the existence of Na+/H+ antiporters in tonoplast vesicles of red beet tap roots. Gaxiola et al., [12] found that AtNHX1, an Arabidopsis homologue of the yeast NHX1, was able to suppress some of the phenotypes of the yeast nhx1 mutant. They found that AtNHX1 mRNA level increased in plants treated with 250 mM NaCl or 250 mM KCl for 6 h [12]. Apse et al. [13] demonstrated AtNHX1 to be localized in tonoplasts, functioning as a Na+/H+ antiporterin Arabidopsis. Moreover, overexpressions of AtNHX1 were shown to improve salt tolerance in both Arabidopsis and tomato plants, further supporting the role of vacuolar Na+/H+ antiporters in salt tolerance [6].
In addition to Arabidopsis thaliana [5], vacuolar Na+/H+ antiporter genes have also been identified in various halophytic and salt-tolerant glycophytic species such as Oryza sativa, Atriplex gmelini, Suaeda salsa and Triticum aestivum [4, 14-16].
Leptochloa fusca L. Kunth, also known as Diplachne fusca, is a highly salt tolerant C4 perennial halophytic forage plant that grows well in coastal salt marshes [17]. Due to its characteristics as a typical euhalophyte with both accumulating and excreting properties, it is an attractive model plant for the study of salt tolerance mechanisms [18].
Despite rapid advances in the molecular identification and biochemical characterization of Na+/H+ antiporters in plants, the expression pattern and regulation of vacuolar Na+/H+ antiporter genes have not been vigorously investigated. As mentioned above, the Na+/H+ antiporter has a crucial function in the exchange of Na+ for H+ across membranes, which is important for the plant's salt tolerance due to the fact that it maintains cellular ion homeostases. However, little information is available about the NHX1 gene's response to cadmium stress [19]. Cadmium, an abundant and non- essential heavy metal, is a toxic trace pollutant for humans, animals and plants, added to the environment and transferred to the food chain mainly by industrial processes and phosphate fertilizers [20]. Cadmium can cause many adverse effects on plants including disturbance in metabolism [21], secondary water stress [22] and oxidative stress [23]. Since heavy metals are persistent environmental pollutants, it is necessary to indicate their toxicological effects on immobile organism such as plants [19]. The purpose of this study is to discover gene expression patterns of the L. fusca vacuolar Na+/H+ antiporter (NHX1) in response to salt and cadmium stresses.
MATERIALS AND METHODS
Plant material and growth conditions: Seeds of kallar grass (provided by the Department of Agronomy, Ferdowsi University of Mashhad, Iran) were sown in 15-cm pots filled with prewashed sand and irrigated daily with a half-strength Hoagland nutrient solution. Plants were grown in a greenhouse at 26±2 ◦C in a photoperiod of 16 h. Fertilization was performed twice a week with Hoagland solution. NaCl andcadmium treatments were performed on six-week-old seedlings. For the salinity treatment, seedlings were drenched with 150, 300, 450 and 600 mM sodium chloride solutions. For the cadmium treatment, three experimental groups of 50, 100 and 150 µM cadmium chloride were considered. Sampling was performed in a time course of 0, 6,12, 24, and 72 hours after treatment. All treatments started at 8 am to account for the potential diurnal rhythm in the gene expression patterns.
RNA extraction and cDNA synthesis: RNAs were extracted from the treated seedlings by Topazol plus Kit (TopazGene, Iran) according to the manufacturer's instructions. Using a RevertAid First Strand cDNA Synthesis Kit (Thermo SCIENTIFIC, # K1691), 1 μg RNA was reversely transcribed to its corresponding cDNA after the DNaseI treatment of the RNA samples. These were later used as templates for the semi-quantitative RT–PCR.
Semi-quantitative RT–PCR: To normalize cDNA amounts of each treatment at different time points,the PCR product intensity of actin was considered as the house- keeping gene. Primer pairs for NHX1 (accession number: JF933902) and actin (accession number: FJ603097) are given in Table 1. Each PCR was performed in a total volume of 25 µl containing 10 mM Tris-HCl pH 8.3, 50 mM KCl, 2 mM MgCl2, 200 μM dNTPs (50 µM of each), 0.3 μM of each primer and 1U Taq DNA polymerase (Thermo SCIENTIFIC). For amplification, the reaction mixtures were denatured at 94°C for 3 min, followed by 35 cycles at 94°C for 30 sec (denaturation), 37°C for 60 sec (annealing) and 72°C for 60 sec (extension), with an additional extension period of 8 min at 72°C. Amplification was carried out in a thermocycler (Eppendorf, Germany) with a heated lid. PCR reaction products were mixed with a fifth of the volume of gel loading buffer (SinaClone, Iran) and separated by electrophoresis in a 1.5% agarose gel, using a Tris-borate-EDTA (TBE) system (0.5 x TBE = 45 mM Tris-base, 45 mM boric acid, and 1 mM EDTA). Electrophoresis was carried out at room temperature at 75 V for 1.5 h, after which the gels were stained with EB solution (0.015%) in distilled water. The gel images were then visualized with a UV Transilluminator. Experiments were repeated at least twice to verify the results.
Table 1.
The sequences of primers used to amplify the genes encoding a vacuolar Na+/H+ antiporter(target gene) and actin as reference gene in Semi-Quantitative RT-PCR.
| Genes | Primers | Sequences (5'-3') | Amplicon Size (bp) |
|---|---|---|---|
| Vacuolar Na + /H + antiporter | NHX1-F | TTCTGGATTGCTCAGTGCTT | 200 |
| NHX1-R | CAGCCAGCATGTAAGAGAGG | ||
| Actin | actin-F | CGTACAACTCCATCATGAAG | 199 |
| actin-R | AGTGTTTGGATTGGTGGCTC |
Statistical analysis: Amplified products were quantified by Total Lab TL120 software (Nonlinear Dynamics Ltd). DNA band intensities amplified from the vacuolar Na+/H+ antiporter gene were normalized compared to the actin gene by dividing the former to the latter. Using SPSS (Version 16.0), a one-way ANOVA was performed followed by Duncan's multiple range test (DMRT) with a critical value of P ≤ 0.01, to analyze statistical differences among the data from different salt and cadmium concentrations at each time point.
RESULTS AND DISCUSSION
Previous studies have identified salt tolerance determinants in organisms ranging from cyanobacteria to fungi and from algae to higher plants. However, a complete understanding of these factors in halophytic species requires more investigation. To the best of our knowledge, little information is available regarding salt stress responses of kallar grass at the molecular level. To further examine the key genes that respond to salinity stress and tolerance in this species, the authors of the present study have previously focused on its isolation, characterization and gene expression pattern analyses [24]. In the present study, to characterize the engagement of NHX1 in kallar grass responses to saline conditions, its expression pattern was checked 6, 12, 24 and 72 hours after treatment with 0, 150, 300, 450 and 600 mM NaCl and 0, 50 100 and 150µM cadmium chloride.
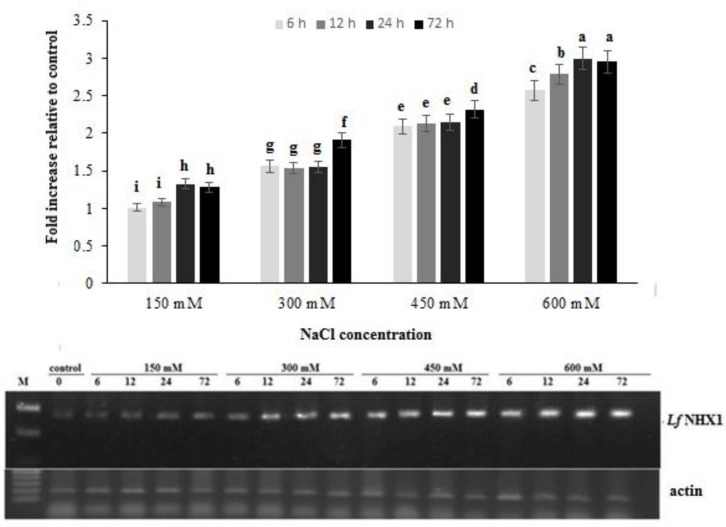
To assess the effect of salt on the expression pattern of the LfNHX1 gene, total RNA from shoots of NaCl treated plants were isolated. Expression levels of LfNHX1 nwere evaluated by semi-quantitative RT-PCR. A basal transcript level was found in non-stressed plants which increased significantly with salt treatments. Quantification of NHX1 transcripts in kallar grass shoot parts showed that salinization affected NHX1 levels positively. In other words, the higher the salinity, the higher the NHX1 levels. In comparison with the control, the relative amounts of mRNA of NHX1 were 1.28, 1.9, 2.3 and 3 fold higher in 150 mM, 300 mM, 450 mM and 600 mM stressed plants respectively after 72 h of exposure to salt (Fig. 1).
Figure 1.
Relative expression levels of LfNHX1 gene after exposure to different concentrations of NaCl. actin gene was used as an internal reference to evaluate the comparative expression level of LfNHX1. Different letters above each column denoted significant difference between them (P ≤ 0.01).
NaCl treatment is known to up-regulate vacuolar Na+/H+ antiporter activities and enhance Na+ compartmentation into vacuoles. This up-regulation is either due to the activation of existing proteins and/or the increase of gene transcript levels [24]. Similar results have been reported in Aeluropus littoralis [25], AtNHX1 of A. thaliana [26], HvNHX1 of Hordeum vulgare [27] AgNHX1 of Atriplex gmelini [4], AhNHX1 of Arachis hypogaea [28] and CmNHX1 of Cucumis melo [29]. In wheat, TaNHX1transcripts also showed increased expression levels in the seedlings after salt treatments [30]. Similarly, the SbNHX1 gene of Salicornia brachiata showed very a high expression level with NaCl treatments [31].
Besides its function under salt stress, the role of vacuolar Na+/H+ under normal growth conditions remains to be examined. In the present study, the relatively high basal expression level of V-NHX indicated the important physiological function of LfNHX1 in kallar grass, even in the absence of stress. A shift from purple buds to blue open flowers correlated with the activity of an NHX1 homologue in the Japanese morning glory (Ipomoea nil) [32]. A T-DNA insertion mutant of AtNHX1 showed a much lower Na+/H+ and K+/H+ exchange activity. The mutant plants exhibited altered leaf development and reduced leaf area compared with their wild-type counterparts [33]. These results suggest the Na+/H+ antiporter to be important not only for salt tolerance, but also for the vacuolar pH regulation and the developmental processes of leaves. LfNHX1 was expressed at a high level in the shoots and gradually increased with NaCl stress. After 72 h of exposure to 600 mM salinity, the relative expression of the kallar grass vacuolar Na+/H+ antiporter in the shoots was 3 times larger than that of the controls (Fig. 1). The higher NHX1 expression in the leaves was a prompt response to NaCl treatment which could have helped decrease the Na+ content in the cytoplasm and maintain water concentrations [34]. Such high expression may indicate the sequestration of more Na+ into the vacuoles, hence protecting the cytoplasm against the toxic effects of Na+. This is in line with the fact that Na+ compartmentalization into the vacuoles of leaf cells is an important salt tolerance mechanism [35].
Previous transgenic studies have revealed that the overexpression of the NHX1 gene significantly enhanced plant salt tolerance abilities. In transgenic Arabidopsis overexpressing AtNHX1, higher activities of the vacuolar Na+/H+ antiporter were observed and enabling it to grow in the presence of 200 mM NaCl [13]. The overexpression of AtNHX1 in tomatos resulted in the growth of transgenic plants,flowers and set fruits at high salt concentrations [36].
The role of sodium compartmentalization in salt tolerance has been further demonstrated in transgenic Brassica napus. The transcription level of BnNHX1, a vacuolar Na+/H+ antiporter from Brassica napus, increased upon treatment with 200 mM NaCl [37]. The overexpression of the cotton Na+/H+ antiporter gene GhNHX1 in tobacco improved salt tolerance in comparison with wild-type plants [38].
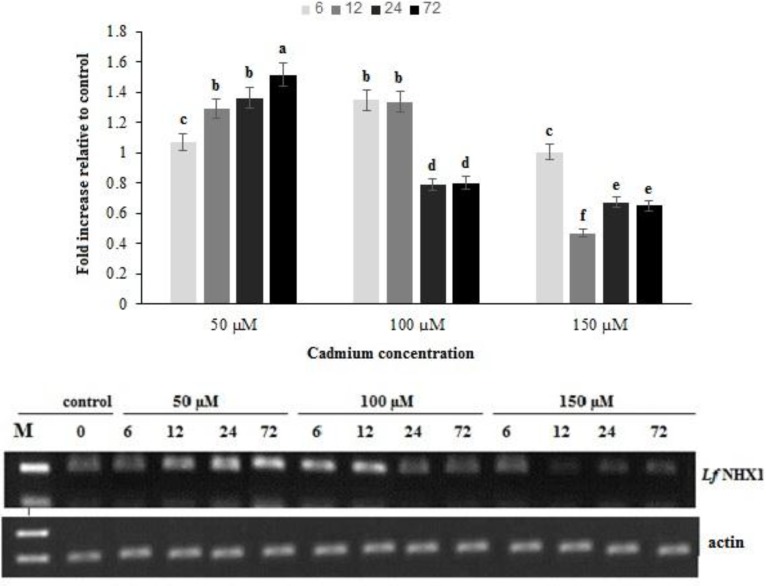
The of V-NHX mRNA expression variation profile in response to cadmium is shown in Figure 2, in which no consistent results were discovered with regard to cadmium levels or time durations. In low cadmium concentrations (50 µM group), the the NHX1 gene transcript amount was up-regulated in all sampling times. As time elapsed, increments of the NHX1 gene expression were observed. The increase ofcadmium concentrations in the medium, the transcript levels of the NHX1 gene, gradually decreased and returned to a state which was even lower than basal expression levels of the control group (Fig. 2).
Figure 2.
Relative expression levels of LfNHX1 gene after exposure to different concentrations of cadmium. actin gene was used as an internal reference to evaluate the comparative expression level of LfNHX1. Different letters above each column denoted significant difference between them (P ≤ 0.01
Na+/H+ antiporter is an important membrane protein responsible for pumping Na+ into the vacuole to reduce Na+ toxicity and alleviate the adverse effects of salt stress [38]. In the present study, the investigation of the NHX1 gene mRNA expression in response to cadmium helped understand whether cadmium exposure affected kallar grass salt-tolerance ability. The findings suggest that treatment with 50 µM cadmium enhanced the transcript amount of the NHX1 gene during measured time-courses. Nevertheles, with increasing cadmium concentrations, NHX1gene expression levels and salt tolerance ability decreased in kallar grass.
When cadmium concentration was low in the medium, significant NHX1 transcript enhancements occurred in kallar grass shoots. However, in the medium and high concentration (100 and 150 µM) groups, gene expression levels and salt tolerance ability gradually decreased. This implies that exposure to low cadmium concentrations (50 µM) induces higher salt-tolerance ability in kallar grass. Nevertheless, this effect disappeared with increasing cadmium concentrations and exposure time. Exposure to100 and 150 µM cadmium concentrations impaired the normal homeostasis of Na+ around the membrane and decreased the salt-tolerance ability of kallar grass.
In conclusion, the results suggest that tonoplast LfNHX1 plays important roles in the compartmentation of Na+ into vacuoles, and that antiporter activity is probably one of the most important factors determining salt tolerance in kallar grass. Other results showed that kallar grass Na+ homeostasis and salt tolerance ability were also affected by cadmium exposure but exhibited dose-response over time. However, for more insight into the function of this gene further studies will be needed on the interaction between salt and cadmium.
Acknowledgements: This research was financially supported by Shahid Bahonar
University, Kerman.
Conflict of Interest: The authors declare that they have no competing interest.
References
- 1.Wang H, Tang X, Shao C, Shao H, Wang L. Molecular cloning and bioinformatics analysis of a new plasma membrane Na+/H+ antiporter gene from the halophyte Kosteletzkya virginica. Sci World J. 2014 doi: 10.1155/2014/141675. Article ID 141675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- 3.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 4.Hamada A, Shono M, Xia T, Ohta M, Hayashi Y, Tanaka A, Hayakawa T. Isolation and characterization of a Na+/H+ antiporter gene from the halophyte Atriplex gmelini. Plant Mol Biol. 2001;46:35–42. doi: 10.1023/a:1010603222673. [DOI] [PubMed] [Google Scholar]
- 5.Aharon GS, Apse MP, Duan S, Hual X, Blumwald E. Characterization of a family of vacuolar Na+/H+ antiporters in Arabidopsis thaliana. Plant Soil. 2003;253:245–256. [Google Scholar]
- 6.Shi H, Zhu JK. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol Biol. 2002;50:543–550. doi: 10.1023/a:1019859319617. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Zhang S, Dong L, Chu J. Incorporation of Na+/H+ antiporter gene from Aeluropus littoralis confer salt tolerance in soybean (Glycine max L.) Indian J Biochem Bio. 2014;51:58–65. [PubMed] [Google Scholar]
- 8.Ji H, Pardo JM, Batelli G, Oosten MJV, Bressan RA, Li X. The salt overly sensitive (SOS) pathway: established and engineering roles. Mol Plant. 2013;279:812–819. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- 9.Bhaskaran S, Savithramma DL. Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J Exp Bot. 2011;62:5561–5570. doi: 10.1093/jxb/err237. [DOI] [PubMed] [Google Scholar]
- 10.Saqib M, Zorb C, Rengel Z, Schubert S. The expression of the endogenous vacuolar Na+/H+ antiporters in roots and shoots correlates positively with the salt resistance of wheat (Triticum aestivum L.) Plant Sci. 2005;169:959–965. [Google Scholar]
- 11.Blumwald E, Poole RJ. Na+/H+ antiporter in isolated tonoplast vesicles from storage tissue of Beta vulgaris. Plant Physiol. 1985;78:163–167. doi: 10.1104/pp.78.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda A, Nakamura A, Tanaka Y. Molecular cloning and expression of the Na+/H+exchanger gene in Oryza sativa. Biochim Biophys Acta. 1999;1446:149–155. doi: 10.1016/s0167-4781(99)00065-2. [DOI] [PubMed] [Google Scholar]
- 15.Ma XL, Zhang Q, Shi HZ, Zhu JK, Zhao YX, Ma CL, Zhang H. Molecular cloning and different expression of a vacuolar Na+/H+ antiporter gene in Suaeda salsa under salt stress. Biol Plantarum. 2004;48:219–225. [Google Scholar]
- 16.Yu JN, Huang J, Wang ZN, Zhang JS, Chen SY. An Na+/H+ antiporter gene from wheat plays an important role in stress tolerance. J Biosciences. 2007;32:1153–1161. doi: 10.1007/s12038-007-0117-x. [DOI] [PubMed] [Google Scholar]
- 17.Qureshi RH, Salim M, Abdullah M, Pitman MG. Diplachne fusca: An Australian salt tolerant grass used in Pakistan agriculture. J Aust Inst Agri Sci. 1982;48:195–199. [Google Scholar]
- 18.Abdullah M, Akram M, Khan A, Dqureshi RH. Internal water resources management by plants under various root environment stresses with special reference to kallar grass Leptochloa fusca. Proceedings of National Seminar on Water Resources Development and its Management in Arid Areas. 6-8 Oct 1990, Quetta, Pakistan. :145–157. [Google Scholar]
- 19.Cong M, Lv J, Liu X, Zhao J, Wu H. Gene expression in Suaeda salsa after cadmium exposure. Springer Plus. 2013;2:232. doi: 10.1186/2193-1801-2-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandalio LM, Dalurzo HC, Gomez M, Romero-Puetras MC, Del Rio LA. Cadmium- induced changes in the growth and oxidative metabolism of pea plants. J Exp Bot. 2001;52:2115–2126. doi: 10.1093/jexbot/52.364.2115. [DOI] [PubMed] [Google Scholar]
- 21.Poschenrieder C, Gunse B, Barcelo J. Influence of cadmium on water relations, stomatal-resistance, and abscisic-acid content in expanding bean leaves. Plant Physiol. 1989;90:1365–1371. doi: 10.1104/pp.90.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nedjimi B, Daoud Y. Cadmium accumulation in Atriplex halimus subsp. schweinfurthii and its influence on growth, proline, root hydraulic conductivity and nutrient uptake. Flora. 2009;204:316–324. [Google Scholar]
- 23.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Taherinia B, Kavousi HR, Dehghan S. Isolation and characterization of plasma membrane Na+/H+ antiporter (SOS1) gene during salinity stress in kallar grass (Leptochloa fusca) Eurasia J Biosci. 2015;9:12–20. [Google Scholar]
- 25.Zhang GH, Su Q, An LJ, Wu S. Characterization and expression of a vacuolar Na+/H+ antiporter gene from the monocot halophyte Aeluropus littoralis. Plant Physiol Bioch. 2008;46:117–126. doi: 10.1016/j.plaphy.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 26.Yokoi S, Quintero FJ, Cubero B, Ruiz MT, Bressan RA, Hasegawa PM, Pardo JM. Differential expression and function of Arabidopsis thaliana NHX Na+/H+antiporters in the salt stress response. Plant J. 2002;30:529–539. doi: 10.1046/j.1365-313x.2002.01309.x. [DOI] [PubMed] [Google Scholar]
- 27.Fukuda A, Chiba K, Maeda M, Nakamura A, Maeshima M, Tanaka Y. Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+- pyrophosphatase, H+ ATPase subunit A, and Na+/H+ antiporter from barley. J Exp Bot. 2004;55:585–594. doi: 10.1093/jxb/erh070. [DOI] [PubMed] [Google Scholar]
- 28.Xing J, Wang B, Jia K, Wan S, Meng J, Guo F, Xionguo L. Isolation of Arachis hypogaea Na+/H+ antiporter and its expression analysis under salt stress. Afr J Biotechnol. 2001;10:14302–14310. [Google Scholar]
- 29.Wang S, Zhang YD, Perez PG, Deng YW, Li ZZ, Huang DF. Isolation and characterization of a vacuolar Na+/H+ antiporter gene from Cucumis melo L. Afr J Biotechnol. 2011;10:1752–1759. [Google Scholar]
- 30.Wang ZN, Zhang JS, Guo BH, He S, Tian AG, Chen SY. Cloning and characterization of the Na+/H+ antiporter genes from Triticum aestivum. Acta Bot Sin. 2002;44:1203–1208. [Google Scholar]
- 31.Jha A, Joshi M, Yadav NS, Agrawal PK, Jha B. Cloning and characterization of the Salicornia brachiata Na+/H+ antiporter gene SbNHX1 and its expression by abiotic stress. Mole Biol Rep. 2011;38:1965–1973. doi: 10.1007/s11033-010-0318-5. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi T, Fukada-Tanaka S, Inagaki Y, Saito N, Yonekura-Sakakibara K, Tanaka Y, Kusumi T, Iida S. Genes encoding the vacuolar Na+/H+ exchanger and flower coloration. Plant Cell Physiol. 2001;42:451–461. doi: 10.1093/pcp/pce080. [DOI] [PubMed] [Google Scholar]
- 33.Apse MP, Sottosanto JB, Blumwald E. Vacuolar cation/H+ exchange, ion homeostasis, and leaf development are altered in a T-DNA insertional mutant of AtNHX1, the Arabidopsis vacuolar Na+/H+ antiporter. Plant J. 2003;36:229–239. doi: 10.1046/j.1365-313x.2003.01871.x. [DOI] [PubMed] [Google Scholar]
- 34.Khedr AHA, Serag MS, Nemat-Alla MM, El-Naga AZA, Nada RM, Quick WP, Abogadallah GM. Growth stimulation and inhibition by salt in relation to Na+ manipulating genes in xero-halophyte Atriplex halimus L. Acta Physiol Plant. 2011;33:1769–1784. [Google Scholar]
- 35.Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- 36.Zhang HX, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol. 2001;19:765–768. doi: 10.1038/90824. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Zuo K, Wu W, Song J, Sun X, Lin J, Li X, Tang K. Molecular cloning and characterization of a new Na+/H+ antiporter gene from Brassica napus. DNA Sequence. 2003;14:351–358. doi: 10.1080/10855660310001596211. [DOI] [PubMed] [Google Scholar]
- 38.Wu CA, Yang GD, Meng QW, Zheng CC. The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol. 2004;45:600–607. doi: 10.1093/pcp/pch071. [DOI] [PubMed] [Google Scholar]