Summary
In Arabidopsis roots, the transcription factor MYB72 plays a dual role in the onset of rhizobacteria‐induced systemic resistance (ISR) and plant survival under conditions of limited iron availability. Previously, it was shown that MYB72 coordinates the expression of a gene module that promotes synthesis and excretion of iron‐mobilizing phenolic compounds in the rhizosphere, a process that is involved in both iron acquisition and ISR signaling. Here, we show that volatile organic compounds (VOCs) from ISR‐inducing Pseudomonas bacteria are important elicitors of MYB72. In response to VOC treatment, MYB72 is co‐expressed with the iron uptake‐related genes FERRIC REDUCTION OXIDASE 2 (FRO2) and IRON‐REGULATED TRANSPORTER 1 (IRT1) in a manner that is dependent on FER‐LIKE IRON DEFICIENCY TRANSCRIPTION FACTOR (FIT), indicating that MYB72 is an intrinsic part of the plant's iron‐acquisition response that is typically activated upon iron starvation. However, VOC ‐induced MYB72 expression is activated independently of iron availability in the root vicinity. Moreover, rhizobacterial VOC‐mediated induction of MYB72 requires photosynthesis‐related signals, while iron deficiency in the rhizosphere activates MYB72 in the absence of shoot‐derived signals. Together, these results show that the ISR‐ and iron acquisition‐related transcription factor MYB72 in Arabidopsis roots is activated by rhizobacterial volatiles and photosynthesis‐related signals, and enhances the iron‐acquisition capacity of roots independently of the iron availability in the rhizosphere. This work highlights the role of MYB72 in plant processes by which root microbiota simultaneously stimulate systemic immunity and activate the iron‐uptake machinery in their host plants.
Keywords: induced resistance, iron homeostasis, MYB transcription factor, volatile organic compounds, Arabidopsis thaliana, plant growth‐promoting rhizobacteria
Significance Statement
Plant roots intimately interact with plant growth‐promoting rhizobacteria that prime the plant immune system and aid in iron uptake two functions facilitated by the root‐specific transcription factor MYB72. Here we show how MYB72 and iron uptake responses are systemically activated by photosynthesis‐related signals and volatiles produced by plant growth‐promoting rhizobacteria, highlighting the important role of beneficial root microbiota in supporting plant growth and health.
Introduction
Plant roots host an immense number of bacteria at the root–soil interface and within the root compartment (Mendes et al., 2011; Bulgarelli et al., 2012; Lundberg et al., 2012). These so‐called root microbiota provide important services to the plant as they improve plant nutrition and provide protection against root pathogens (Berendsen et al., 2012; Bulgarelli et al., 2013). Selected plant growth‐promoting rhizobacteria from the root microbiota confer systemic immunity against a broad spectrum of foliar pathogens, a phenomenon that is called induced systemic resistance (ISR) (Pieterse et al., 2014). In the roots of the model plant Arabidopsis (Arabidopsis thaliana), initiation of ISR by root‐colonizing Pseudomonas fluorescens WCS417 bacteria (recently renamed Pseudomonas simiae WCS417; Berendsen et al., 2015) is dependent on the root‐specific transcription factor MYB72 (Van der Ent et al., 2008; Segarra et al., 2009). In addition to its role in ISR, MYB72 expression is induced in roots under iron limitation and growth conditions that distort iron uptake, such as high zinc concentrations (Colangelo and Guerinot, 2004; De Mortel et al., 2008; Buckhout et al., 2009). Recently, MYB72 and its closest paralog MYB10 were shown to be required for plant survival in alkaline soils where iron availability is greatly restricted (Palmer et al., 2013). Hence, MYB72 is emerging as a node of convergence in immune and iron‐deficiency signaling pathways.
Iron is an essential element for plant and animal life. Although iron is one of the most abundant elements on Earth, its low solubility makes it poorly available. To cope with iron‐limited conditions, plants have evolved sophisticated iron‐uptake mechanisms (Connolly and Guerinot, 2002). In Arabidopsis and other dicotyledonous plants, iron limitation induces a set of coordinated responses, collectively referred to as strategy I, which foster iron mobilization and uptake by the roots. Strategy I includes three main steps that take place in the epidermal cells of the root: (i) rhizosphere acidification via proton extrusion by plasma membrane‐localized H+‐ATPases, resulting in enhanced solubility of ferric iron (Fe3+) in the soil, (ii) reduction of ferric iron to ferrous iron (Fe2+) via the plasma membrane protein FRO2 (ferric reduction oxidase 2), and (iii) transport of ferrous ion from the soil environment to the root interior via the high‐affinity iron transporter IRT1 (iron‐regulated transporter 1) (Walker and Connolly, 2008).
Plants reprogram their transcriptome in roots under conditions of iron deficiency (Colangelo and Guerinot, 2004; Dinneny et al., 2008; Buckhout et al., 2009). In the roots of Arabidopsis, responses to iron limitation are regulated by the basic helix‐loop‐helix (bHLH) transcription factor FER‐like iron deficiency induced transcription factor (FIT) (Colangelo and Guerinot, 2004; Jakoby et al., 2004; Yuan et al., 2005; Bauer et al., 2007). FIT forms heterodimers with members of the Ib sub‐group of the bHLH gene family (bHLH38/39/100/101), resulting in up‐regulation of FRO2 and IRT1 (Yuan et al., 2008; Wang et al., 2013). Recent studies demonstrated additional regulation of FIT activity at the protein level through mechanisms that involve ethylene and nitric oxide (NO) signaling (Lingam et al., 2011; Meiser et al., 2011; Sivitz et al., 2011). The transcription factor gene MYB72, whose expression is rapidly induced in roots upon iron starvation (Buckhout et al., 2009), has been identified as one of the iron deficiency‐induced genes that are activated in a FIT‐dependent manner (Colangelo and Guerinot, 2004; Sivitz et al., 2012).
In an attempt to understand the dual role of the root‐specific transcription factor MYB72 in ISR and the iron‐deficiency response, we recently demonstrated that MYB72 controls a gene regulatory module that involves synthesis of iron‐mobilizing phenolic metabolites and their release into the rhizosphere through activity of the β‐glucosidase BGLU42 (Zamioudis et al., 2014). Interestingly, both MYB72 and BGLU42 are also required for the onset of ISR, suggesting a mechanistic link between the induction of MYB72‐dependent iron‐uptake responses in the roots and the development of rhizobacteria‐mediated ISR in the leaves (Zamioudis et al., 2014). In this study, we focused on the transcriptional regulation of MYB72 in Arabidopsis roots exposed to ISR‐inducing rhizobacteria or conditions of iron deficiency. First, we show that volatile compounds from ISR‐inducing rhizobacteria are important elicitors of MYB72. Detailed analysis of the spatial expression pattern of MYB72 revealed that rhizobacteria and local iron deficiency in the root vicinity differentially affect MYB72 expression in the Arabidopsis root. Moreover, rhizobacteria‐induced MYB72 expression appears to fully depend on photosynthesis‐related signal(s), whereas local iron deficiency‐induced MYB72 expression only partially depends on such signals. Collectively, this work shows how volatiles of rhizosphere bacteria enhance the iron‐acquisition capacity of the root system independently of the iron availability in the root vicinity, and highlights the sophisticated strategies that root microbiota have evolved to simultaneously benefit their host plants in terms of host immunity and nutrient uptake.
Results
ISR‐inducing rhizobacteria trigger an iron‐deficiency response in Arabidopsis roots
Whole‐genome transcript profiling of Arabidopsis roots in response to ISR‐inducing P. simiae WCS417 bacteria revealed a total of 672 up‐regulated and 799 down‐regulated genes (P < 0.05 and >1.5‐fold change; Table S1; a subset of these have been described in Zamioudis et al., 2014). Because of our particular interest in the root‐specific ISR regulator MYB72, we noticed that MYB72 was activated in concert with many iron deficiency‐induced genes, including the well‐established marker IRT1 (Vert et al., 2002). Classification of the WCS417 up‐regulated genes into biological categories using AmiGO Term Enrichment software (Carbon et al., 2009) revealed a significant over‐representation of the Gene Ontology category ‘cellular response to iron ion’ (Zamioudis, 2012). By comparing the WCS417‐induced gene set with the iron‐deficiency root transcriptome described by Dinneny et al. (2008), we found that 20% of all WCS417‐induced genes are also activated in response to iron deprivation (Figure 1a), indicating that colonization by WCS417 bacteria activates an iron‐deficiency response in Arabidopsis roots.
Figure 1.
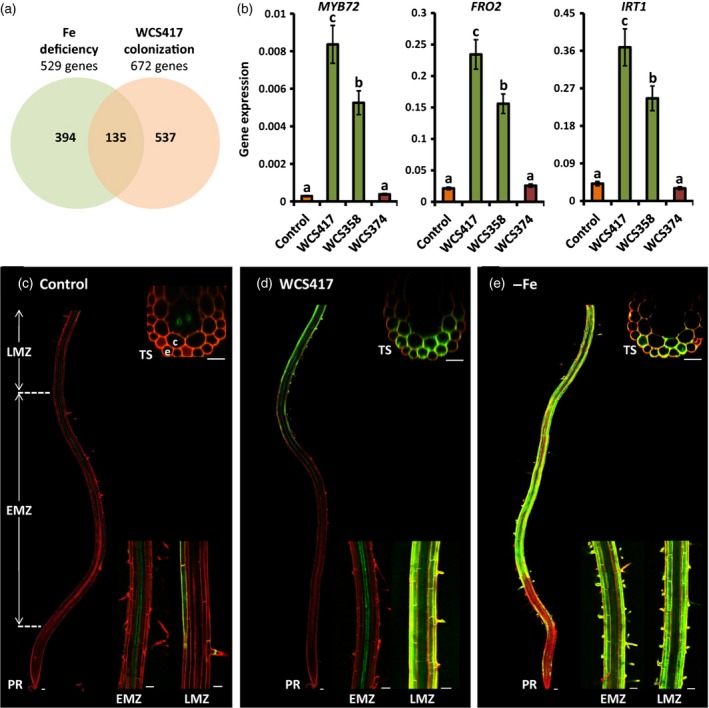
Beneficial rhizosphere bacteria stimulate an iron‐deficiency response in Arabidopsis roots.
(a) Venn diagram indicating the overlap between genes that are up‐regulated in Arabidopsis roots by WCS417 (Table S1) and iron deficiency (Dinneny et al., 2008).
(b) Quantitative RT‐PCR analysis of MYB72,FRO2 and IRT1 transcript levels in the roots of Arabidopsis Col‐0 seedlings treated with 10 mm MgSO 4 (control) or the indicated rhizobacterial Pseudomonas strains. Gene expression was normalized to that of ACTIN7. Values are means ± SD of three replicates. Different letters indicate statistically significant differences (Tukey's HSD test; P < 0.05).
(c–e) Representative confocal images of pMYB72:GFP‐GUS roots that were mock‐treated (c), treated with WCS417 bacteria (d), or subjected to iron deficiency (e). Seedlings were treated at 12 days old, and images were taken 2 days after treatment. PR, primary root; EMZ, early maturation zone; LMZ, late maturation zone; TS, transverse optical section; e, epidermis; c, cortex. Scale bars = 50 μm (longitudinal sections) and 25 μm (transverse sections).
To investigate whether the ability of rhizosphere bacteria to induce ISR correlates with their ability to activate iron deficiency‐inducible mechanisms, we tested three well‐characterized root‐associated Pseudomonas strains for their ability to up‐regulate expression of MYB72 and that of the iron‐deficiency marker genes FRO2 and IRT1 (Robinson et al., 1999; Vert et al., 2002). Colonization of Arabidopsis roots by WCS417 or Pseudomonas putida WCS358 (recently renamed Pseudomonas capeferrum WCS358; Berendsen et al., 2015), which both trigger ISR in Arabidopsis (Van Wees et al., 1997), strongly activated MYB72, FRO2 and IRT1 gene expression (Figure 1b). However, the rhizobacterial strain Pseudomonas fluorescens WCS374 (recently renamed Pseudomonas defensor WCS374; Berendsen et al., 2015), which is not capable of inducing ISR in Arabidopsis (Van Wees et al., 1997), did not induce expression of these genes (Figure 1b). Likewise, ferric chelate reductase activity was enhanced in WCS417‐ and WCS358‐treated roots, but remained at basal levels in WCS374‐colonized roots (Figure S1). Together, these results suggest that the ability of rhizosphere bacteria to induce ISR correlates with their ability to activate the iron‐deficiency response.
Tissue‐specific expression pattern of MYB72
To investigate the tissue‐specific expression pattern of MYB72 upon root colonization by WCS417 or iron limitation, we generated stable transgenic lines expressing a GFP–GUS fusion protein under the control of the 1.7 kb MYB72 promoter region (pMYB72:GFP‐GUS). Confocal imaging revealed that, under control conditions (no bacteria, sufficient iron), MYB72 is expressed at a low basal level in the xylem parenchyma cells in the early maturation zone of the roots, and sporadically in the epidermis and cortex in the late maturation zone (Figure 1c). Upon colonization of the roots by WCS417, we observed massive accumulation of the GFP fluorophore in the epidermal and cortical cells of WCS417‐colonized roots (Figure 1d). Similar spatial expression patterns were observed in roots subjected to iron deficiency (Figure 1e). However, in contrast to WCS417‐treated roots in which expression of MYB72 was induced in the late maturation zone, conditions of iron deficiency also stimulated MYB72 gene expression in the elongation and early maturation zone (Figure 1d,e). Interestingly, both under iron limitation and upon root colonization, MYB72 expression in the epidermis was most pronounced in trichoblasts and root hair cells. In line with the quantitative RT‐PCR data on bacterized roots shown in Figure 1(b), only the ISR‐inducing strains WCS417 and WCS358 were capable of enhancing MYB72 promoter activity in colonized roots, and WCS374 was not (Figure S2).
Bacterial volatiles stimulate MYB72 expression independently of the iron status in the root vicinity
Several bacterial molecules have been proposed to function as ISR determinants in Arabidopsis and other plant species, including iron‐chelating siderophores, microbe‐associated molecular patterns such as flagellin and chitin, and volatile organic compounds (VOCs) (Ryu et al., 2004; Van Loon et al., 2008; De Vleesschauwer and Höfte, 2009; Millet et al., 2010). We reasoned that iron‐chelating siderophores of ISR‐inducing rhizobacteria may deplete iron, potentially resulting in activation of MYB72 and the iron‐deficiency response. However, two siderophore‐defective Tn5 transposon‐insertion mutants of WCS417 (Duijff et al., 1993) activated MYB72 expression to the same extent as the wild‐type WCS417 strain, suggesting that iron chelation by microbe‐secreted siderophores is probably not the reason for induction of MYB72 in WCS417‐colonized roots (Figure S3a). Likewise, neither flg22 nor chitosan, two well‐studied microbial elicitors that trigger defense responses in Arabidopsis roots (Millet et al., 2010), were capable of up‐regulating MYB72 or FRO2 expression (Figure S3b). Previously, VOCs from the ISR‐inducing bacterium Bacillus subtilis GB03 was shown to stimulate iron‐uptake mechanisms in Arabidopsis, thereby improving plant iron content (Zhang et al., 2009). Bacterial VOCs have also been implicated in ISR (Ryu et al., 2004). Thus, we investigated whether VOCs of the ISR‐inducing strain WCS417 are capable of inducing the expression of MYB72, FRO2 and IRT1. To this end, 12‐day‐old Arabidopsis seedlings growing in a two‐compartment plate were exposed to WCS417 VOCs for 2 days, after which root samples were collected for gene expression analysis. WCS417 VOCs strongly up‐regulated MYB72 expression, as well as expression of the strategy I markers FRO2 and IRT1 (Figure 2), suggesting that VOCs of WCS417 are major determinants for elicitation of the iron‐deficiency response in Arabidopsis roots.
Figure 2.
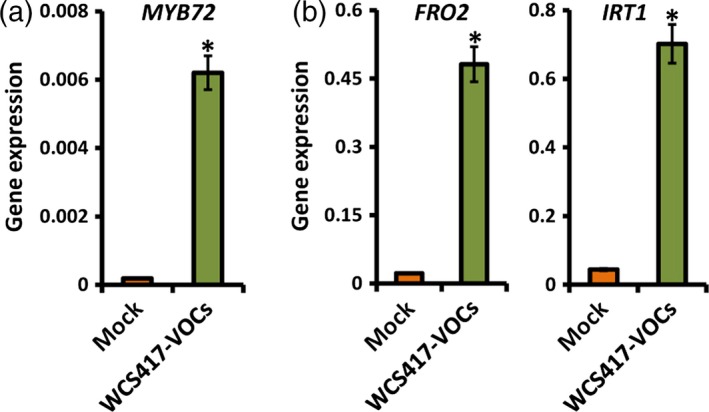
Bacterial VOCs induce the expression of strategy I marker genes.
Quantitative RT‐PCR analysis of (a) MYB72 and (b) FRO2 and IRT1 transcripts in the roots of Arabidopsis Col‐0 seedlings upon exposure to P. simiae WCS417 VOCs. Gene expression was normalized to that of ACTIN7. Twelve‐day‐old seedlings were exposed to WCS417 VOCs in a split‐plate assay, and samples were collected for gene expression analysis 2 days after treatment. Values are means ± SD of three replicates. Asterisks indicate statistically significant differences compared with mock‐treated roots (Student's t test; P < 0.05).
To obtain further insight into the mechanisms by which WCS417 VOCs activate MYB72, we first investigated whether different levels of iron in the root vicinity affect the ability of the VOCs to up‐regulate this gene. Supplementing the plant growth substrate with increasing iron concentrations proportionally reduced the level of MYB72 expression, but did not impair the ability of the WCS417 VOCs to enhance MYB72 transcript levels by approximately 20‐fold (Figure S4). Similar results were obtained for FRO2, suggesting that WCS417 VOCs up‐regulate MYB72 and related strategy I iron‐deficiency responses independently of the iron availability. This suggests that bacterial VOCs interact with plant processes that increase endogenous iron needs. To further investigate this, we measured the transcript levels of the iron storage protein gene FER1, which reflects metabolic iron needs and serves as a robust molecular marker for the cellular iron status (Gaymard et al., 1996). As shown in Figure 3(a), FRO2 and IRT1 expression were strongly up‐regulated in VOC‐treated seedlings during the first 2 days of VOC exposure. Subsequently, their expression levels decreased to levels that were significantly below those of mock‐treated roots on day 3, after which they returned to basal levels on day 4. Interestingly, we found that MYB72 was co‐expressed with the iron‐uptake genes FRO2 and IRT1, both under control and VOC‐stimulated conditions, confirming previous findings showing that MYB72 regulates processes that are directly linked to strategy I (Zamioudis et al., 2014). Activation of strategy I on the first day of VOC application coincided with a significant decrease in FER1 transcripts in both roots and shoots (Figure 3b). This suggests that VOCs interact with cellular processes that rapidly deplete iron from cellular stores, consequently resulting in onset of the iron‐deficiency response. In line with the documented ability of B. subtilis GB03 VOCs to improve iron nutrition (Zhang et al., 2009), FER1 transcripts in WCS417 VOC‐treated seedlings stabilized to the levels of mock‐treated plants at 2 days after VOC treatment, after which FER1 transcription continued to significantly increase on days 3 and 4 in both roots and shoots of VOC‐treated plants (Figure 3b). Increased FER1 transcription coincided with a significant increase in chlorophyll content in iron‐deprived WCS417 VOC‐treated seedlings (Figure 3c), indicative of enhanced iron nutrition in the leaves (Briat et al., 2015). Further evidence for this effect of bacterial VOCs comes from the observation that treatment of iron‐deprived Arabidopsis seedlings with WCS417 VOCs resulted in darker green seedlings relative to iron‐deprived seedlings that were not treated with bacterial VOCs (Figure 3c). Collectively, these results indicate that WCS417 VOCs transiently stimulate iron‐uptake mechanisms.
Figure 3.
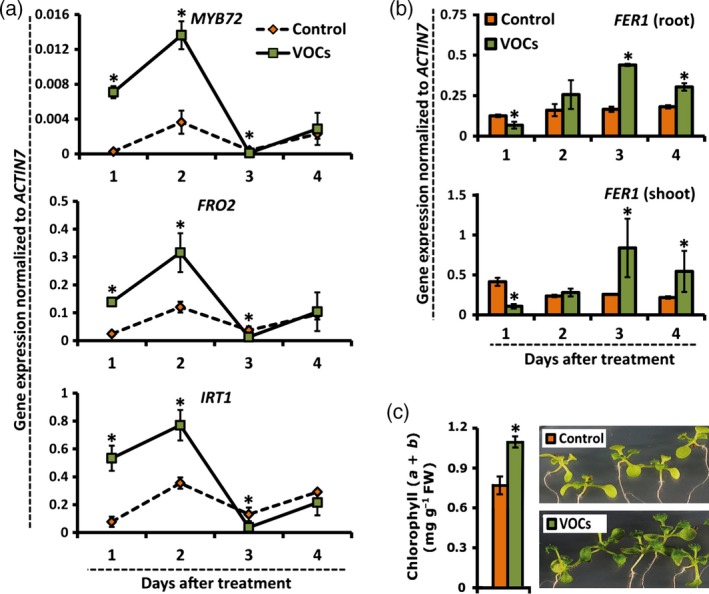
Bacterial VOCs stimulate expression of markers for improved iron nutrition.
(a,b) Kinetics of expression of MYB72,FRO2 and IRT1 (a) and FER1 (b) in the roots of mock‐treated (control) and P. simiae WCS417 VOC‐treated Arabidopsis Col‐0 seedlings as indicated by quantitative RT‐PCR analysis. Seedlings were treated in a split‐plate assay when 12 days old, and samples were collected 1, 2, 3 and 4 days after treatment. Values are means ± SD of three replicates. Asterisks indicate statistically significant differences compared with mock‐treated roots (Student's t test; P < 0.05).
(c) Chlorophyll (a + b) content and phenotypes of control and bacterial VOC‐treated seedlings grown under iron limitation (5 μm Fe(III)EDTA). Seedlings were transferred to low‐iron plates when 10 days old, and treated or not with bacterial VOCs for 9 days, after which chlorophyll content was measured. Photos were taken 5 days after VOC treatment. of three replicates alues are means ± SD of nine replicates. The asterisk indicates a statistically significant difference compared with mock‐treated seedlings (Student's t test; P < 0.05).
Natural root microbiota activate MYB72
To investigate whether bacterial members of the natural Arabidopsis root microbiota exert similar effects on the iron‐uptake response to those exerted by the model Pseudomonas spp. strain WCS417, we tested VOCs of 40 culturable root‐derived bacteria belonging to the phyla Proteobacteria, Actinobacteria, Firmicutes and Bacteroidetes, that were isolated from healthy Arabidopsis plants grown in natural soil (Bulgarelli et al., 2012; Y. Bai, N. Dombrowski, & P. Schulze‐Lefert unpublished data) based on their ability to activate the MYB72 promoter in the pMYB72:GFP‐GUS reporter line. Of 40 bacterial strains representing 40 operational taxonomic units, the VOCs of seven strains belonging to the genus Pseudomonas (Gammaproteobacteria) and the phylum Actinobacteria potently activated the MYB72 promoter to the same extent as WCS417 VOCs (Table S2). The VOCs of another 28 strains induced the MYB72 promoter to a lesser but still detectable extent. These results suggest that members of the natural root microbiota play an important role in enhancing the iron‐uptake capacity in plant roots.
Rhizobacteria‐induced expression of MYB72 requires a photosynthesis‐related signal
Previous work in Arabidopsis proposed that the high‐affinity iron‐uptake system in the roots is regulated by local and long‐distance signals (Vert et al., 2003). To investigate whether the bacterial VOC‐induced iron‐deficiency response is regulated locally in the roots or requires systemic signals from the shoot, we performed shoot decapitation experiments and monitored expression of the iron‐deficiency markers FRO2 and IRT1 in the roots. We first tested whether shoot decapitation affects the ability of plants to respond to iron‐limited conditions. Interestingly, we found that shoot decapitation did not compromise the ability of roots to respond to iron limitation (Figure 4a), indicating that iron limitation in the rhizosphere is autonomously registered by a root‐resident iron‐sensing system. By contrast, shoot decapitation at the time that WCS417 bacteria were applied to the two‐compartment plate completely abolished the ability of WCS417 VOCs to induce FRO2 and IRT1 expression in the roots (Figure 4b). Similar to FRO2 and IRT1, WCS417 VOC‐induced expression of MYB72 was blocked in roots of decapitated plants (Figure 4c). Interestingly, stalling photosynthesis by transferring seedlings to the dark also compromised the ability of WCS417 VOCs to induce MYB72, FRO2 and IRT1 expression (Figure 4c). Likewise, depriving the seedlings of carbon dioxide (CO2) by utilizing a KOH trap in the presence of light, or blocking photosynthesis by transferring seedlings into medium supplemented with the photosynthesis inhibitor norflurazon, completely blocked the ability of VOCs to activate MYB72 reporter gene expression (Figure 4d). Together, these data indicate that the bacterial VOC‐stimulated iron‐uptake system requires shoot‐derived, photosynthesis‐related signals that systemically up‐regulate the iron‐deficiency response in the roots.
Figure 4.
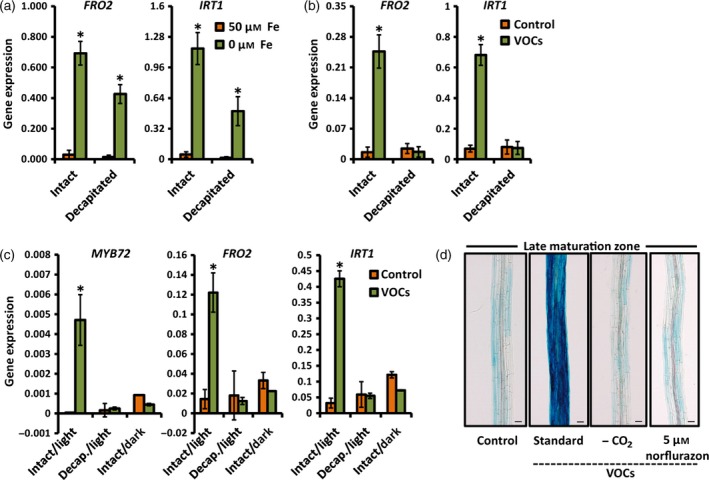
Bacterial VOC‐mediated induction of iron‐uptake responses in Arabidopsis roots requires shoot‐derived signals.
(a) Quantitative RT‐PCR analysis of FRO2 and IRT1 transcript levels in the roots of intact and decapitated 15‐day‐old Col‐0 seedlings that were grown under standard conditions (+Fe) or subjected to iron deficiency for 3 days (−Fe, 300 μm Ferrozine). Shoot decapitation was performed immediately prior to the start of the iron‐deficiency treatment.
(b) Quantitative RT‐PCR analysis of FRO2 and IRT1 transcript levels in the roots of intact and decapitated Col‐0 seedlings that were grown under standard conditions (+Fe) and treated or not with P. simiae WCS417 VOCs. Seedlings were treated with VOCs in a two‐compartment plate assay when 12 days old, and samples were collected 2 days later. Shoot decapitation was performed immediately prior to the VOC treatment.
(c) Quantitative RT‐PCR analysis of MYB72,FRO2 and IRT1 transcript levels in the roots of control and WCS417 VOC‐treated Col‐0 seedlings that were grown with sufficient iron (50 μm Fe(III)EDTA) under the indicated conditions. Decapitation and darkness treatments were started just prior to application of VOCs. Root samples were harvested 2 days after VOC application.
(d) Representative images of segments of the late maturation zone of GUS‐stained roots of pMYB72:GFP‐GUS seedlings exposed to WCS417 VOCs under the indicated conditions (+Fe). Carbon dioxide (CO 2) was trapped by adding a 1 m KOH solution in the three‐compartment plate assay, just prior to VOC treatment. Chemical inhibition of photosynthesis was achieved by transferring 11‐day‐old seedlings to norflurazon‐supplemented plates 24 h before VOC treatment. Images were taken 2 days after VOC application. Scale bars = 50 μm.
Values in (a–c) are means ± SD of three replicates. Gene expression was normalized to that of ACTIN7. Asterisks indicate statistically significant differences compared with mock‐treated roots (Student's t test; P < 0.05).
Spatial expression of MYB72 and strategy I in response to bacterial VOCs and iron deficiency
Given the intimate relationship between MYB72 and iron‐uptake processes (Zamioudis et al., 2014), and the fact that MYB72 is co‐expressed with the strategy I iron‐uptake genes FRO2 and IRT1 (Figure 3), we compared the spatial expression of the strategy I response in the Arabidopsis root in response to WCS417 VOCs or iron deprivation. To this end, we utilized the pMYB72:GFP‐GUS reporter and GUS staining to monitor the expression patterns of MYB72 in the primary root of control and VOC‐treated seedlings that were grown in the light under conditions of sufficient iron availability to recapitulate systemic regulation, and in the primary root of decapitated light‐treated seedlings and intact darkness‐treated seedlings that were subjected to iron deficiency to recapitulate local regulation. In addition, we tested intact seedlings subjected to iron deficiency under a normal photoperiod to address any interaction between local and systemic regulation of MYB72 and related iron deficiency‐inducible mechanisms. Similar to WCS417‐colonized roots (Figure 1d), WCS417 VOCs activated the MYB72 promoter in the late maturation zone of roots of seedlings grown under a normal photoperiod (Figure 5a). By contrast, iron limitation in roots of decapitated light‐treated seedlings and intact darkness‐treated seedlings specifically activated the MYB72 promoter in the elongation and early maturation zones (Figure 5a). Iron‐deprived seedlings growing under light conditions expressed the GUS reporter in the whole primary root, with GUS expression in the apical zone that was stronger than that in decapitated or darkness‐treated seedlings (Figure 5a). The latter result suggests that putative photosynthesis‐related systemic signals prime the apical root for enhanced expression of MYB72 and related iron‐deficiency responses upon iron limitation in the rhizosphere. Similar locally and systemically regulated MYB72 expression patterns were observed in the lateral roots of 3‐week‐old seedlings, although MYB72 expression in the basal primary root was less prominent (Figure 5b–e).
Figure 5.
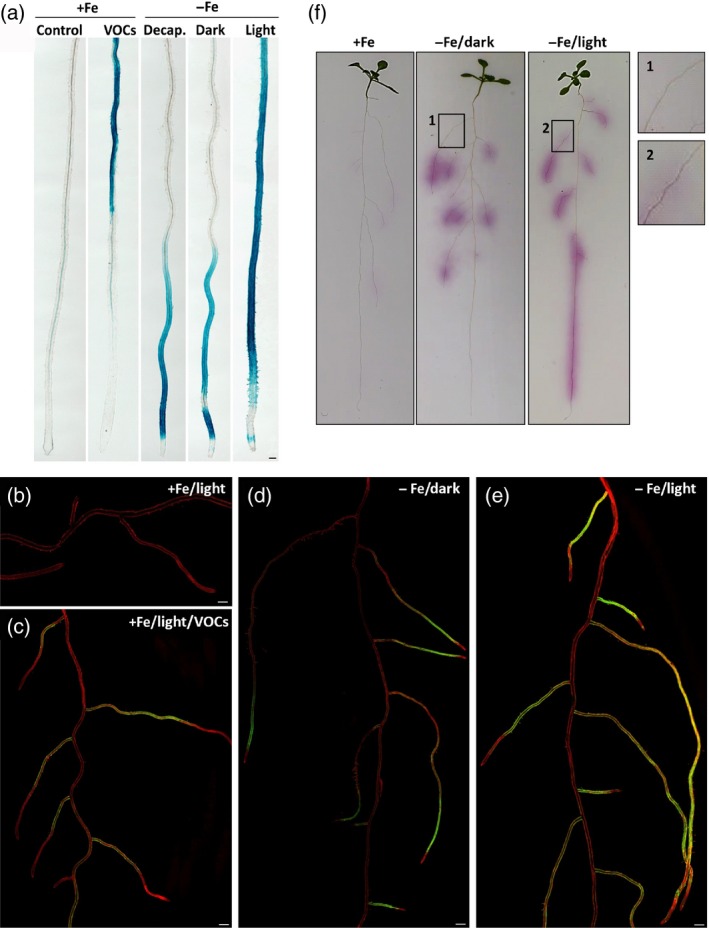
Local and systemic activation of iron‐uptake responses in Arabidopsis roots.
(a) Representative images of GUS‐stained primary roots of 2‐week‐old pMYB72:GFP‐GUS seedlings subjected to P. simiae WCS417 VOCs under standard growth conditions (+Fe) in the light, or to iron deficiency (−Fe, 300 μm Ferrozine) with decapitation (decap.), in complete darkness (dark) or under a normal photoperiod (light). GUS staining was performed 3 days after treatment. Scale bar = 50 μm.
(b–e) Representative confocal images of roots of 3‐week‐old pMYB72:GFP‐GUS seedlings growing under (b) standard conditions (+Fe, light), (c) upon exposure to WCS417 VOCs (+Fe, light, VOCs), (d) upon exposure to iron‐limited conditions and complete darkness (−Fe, dark), and (e) upon exposure to iron‐limited conditions under a normal photoperiod (−Fe, light). Images were obtained 3 days after treatment using the tile scan function of the confocal laser scanning microcope. Consecutive pictures were stitched using a 15% overlay at each border. Scale bars = 500 μm.
(f) In situ localization of ferric chelate reductase activity in the roots of Col‐0 seedlings under sufficient iron or iron‐limited conditions in darkness or light, 3 days after treatment. The magnified images on the right show induction of ferric chelate reductase activity specifically in the late maturation zone of iron‐deprived light‐grown seedlings.
To further confirm that the observed MYB72 expression patterns in Figure 5(b–e) reflect the strategy I response, we analyzed the in situ activity of ferric chelate reductase in the roots of similarly treated seedlings. As shown in Figure 5(f), local induction (−Fe/Dark) and systemic induction (−Fe/Light) of ferric chelate reductase activity under conditions of iron deficiency showed a similar spatial expression pattern for ferric chelate reductase as for pMYB72:GFP‐GUS. Collectively, these data indicate that MYB72 expression and strategy I iron‐uptake mechanisms are activated in the apical root zone by a root‐autonomous iron‐sensing system that registers iron limitation in the root vicinity. For activation of these iron‐acquisition mechanisms in the late maturation zone, photosynthesis‐related systemic signals are also required, which enhance the root autonomous responses to iron deficiency in the apical root zone. Furthermore, our results indicate that bacterial VOCs only stimulate the photosynthesis‐dependent iron‐deficiency response in the late maturation zone of the root, irrespectively of the iron levels in the root vicinity.
FIT and NO regulate bacterial VOC‐induced MYB72
Under iron limitation, expression of the iron‐uptake genes FRO2 and IRT1 is under the control of the bHLH transcription factor FIT, which regulates their expression upon heterodimerization with members of the Ib sub‐group of the bHLH transcription factor gene family (Yuan et al., 2008; Wang et al., 2013). To investigate whether the same transcriptional network is required for WCS417 VOC‐induced expression of MYB72, we initially measured the expression levels of FIT and of bHLH38 and bHLH39, which encode two of the FIT‐interacting partners. As shown in Figure 6(a), the transcript levels of FIT, bHLH38 and bHLH39 were significantly up‐regulated in response to root treatment with WCS417 VOCs. To further investigate whether FIT is essential for VOC‐mediated induction of MYB72, we measured the expression of MYB72 in the loss‐of‐function mutant fit1‐2. WCS417 VOC‐induced expression of MYB72 was severely compromised in fit1‐2, indicating that FIT is an important regulator of bacterial VOC‐stimulated MYB72 expression (Figure 6b). In order to determine whether FIT regulates MYB72 expression upon heterodimerization with the bHLH38/39 transcription factors, we assessed MYB72 expression in transgenic lines over‐expressing FIT (oxFIT), bHLH38 (oxbHLH38), bHLH39 (oxbHLH39) or both FIT and bHLH38 (oxFIT/bHLH38). Over‐expression of either FIT, bHLH38 or bHLH39 alone did not strongly affect basal MYB72 transcription. However, simultaneous over‐expression of FIT and bHLH38 resulted in constitutive expression of MYB72, indicating that, like FRO2 and IRT1, MYB72 is synergistically regulated by FIT and its bHLH interaction partner(s) (Figure 6c).
Figure 6.
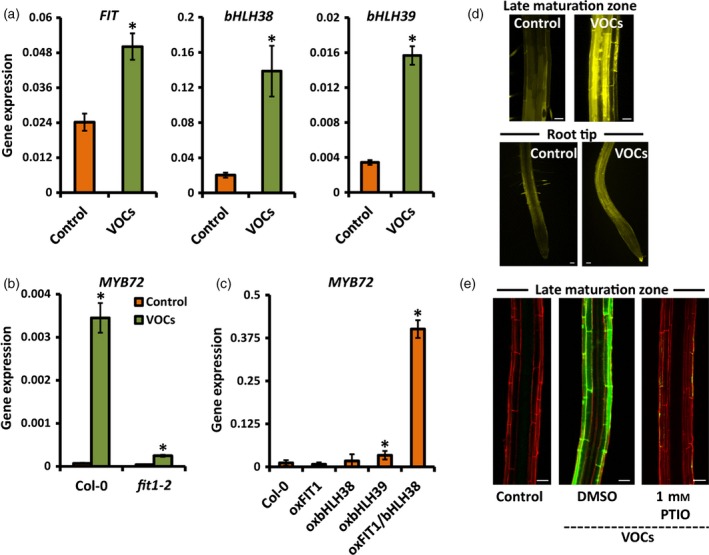
Bacterial VOC‐mediated induction of iron‐uptake responses in Arabidopsis roots depends on FIT and NO.
(a) Quantitative RT‐PCR analysis of FIT,bHLH38 and bHLH39 transcript levels in the roots of mock‐treated (control) and P. simiae WCS417 VOC‐treated Arabidopsis Col‐0 seedlings that were grown with sufficient iron (50 μm Fe(III)EDTA).
(b) MYB72 expression levels in the roots of mock‐treated (control) and WCS417 VOC‐treated wild‐type Col‐0 and mutant fit1‐2 seedlings that were grown with sufficient iron.
(c) MYB72 transcript levels in the roots of Col‐0 and transgenic lines over‐expressing FIT (oxFIT), bHLH38 (oxbHLH38), bHLH39 (oxbHLH39), or FIT and bHLH38 (oxFIT/bHLH38) under standard conditions (sufficient Fe, no VOCs).
(d) NO accumulation in the late maturation zone and root tip of mock‐treated (control) and VOC‐treated seedlings visualized by DAF‐FMDA fluorescence. Scale bars = 50 μm.
(e) Representative confocal images of pMYB72:GFP‐GUS roots that were either mock‐treated (control) or treated with WCS417 VOCs in the absence or presence of the NO scavenger PTIO. Scale bars = 50 μm.
Seedlings were treated with VOCs in a two‐compartment plate assay when 12 days old. Samples for quantitative RT‐PCR analysis and microscopy were collected 2 days after VOC treatment. Values in (a–c) are means ± SD of three replicates. Gene expression was normalized to that of ACTIN7. Asterisks indicate statistically significant differences compared to mock‐treated roots (Student's t test; P < 0.05).
NO has been shown to have critical functions in the physiological responses of roots expressing iron‐uptake responses upon iron limitation (Chen et al., 2010; Meiser et al., 2011). By using 4‐amino‐5‐methylamino‐2′,7′‐difluorofluorescein diacetate (DAF‐FMDA) staining and confocal microscopy, we found that NO accumulated in the late maturation zone and root tip of WCS417 VOC‐treated seedlings (Figure 6d). To determine whether NO is required for VOC‐mediated induction of MYB72, pMYB72:GFP‐GUS seedlings were transferred to medium supplemented with the NO scavenger 2‐phenyl‐4,4,5,5‐tetramethylimidazoline‐1‐oxyl 3‐oxide (PTIO). As shown in Figure 6(e), treatment with the NO scavenger completely abolished MYB72 expression in VOC‐treated seedlings, suggesting that NO‐mediated signaling is involved in bacterial VOC‐induced expression of MYB72 and related iron‐deficiency responses.
Discussion
Function of the root microbiome in improving iron nutrition
Soil‐borne microbes have critical roles in improving plant mineral nutrition. Such functions are well‐documented for plant symbioses with arbuscular mycorrhizal fungi and Rhizobium bacteria (Zamioudis and Pieterse, 2012). Non‐symbiotic plant growth‐promoting bacteria also have important roles in improving plant nutrition, either by enhancing the bioavailability of insoluble minerals or by improving the root system architecture of host plants, thus increasing the root's exploratory capacity for water and minerals (Lopez‐Bucio et al., 2007; Richardson and Simpson, 2011; Zamioudis et al., 2013). The root‐specific transcription factor MYB72 has been shown to play an important role in both rhizobacteria‐mediated ISR and iron acquisition (Van der Ent et al., 2008; Segarra et al., 2009; Palmer et al., 2013; Zamioudis et al., 2014). In this study, we demonstrated that airborne signals from ISR‐inducing rhizobacteria are important elicitors of MYB72 expression and that of the co‐expressed iron‐uptake genes FRO2 and IRT1 (Figure 2), and that a photosynthesis‐related systemic signal from the shoot is required for activation of their expression in the late maturation zone of the root (Figures 4 and 5). Moreover, we show that bacterial VOC‐induced expression of these iron‐acquisition responses functions independently of the iron availability in the root vicinity (Figure S4), thereby enhancing the iron‐uptake machinery. Such manipulation of iron homeostatic mechanisms by bacterial VOCs was previously demonstrated to contribute to plant fitness under conditions of iron limitation (Zhang et al., 2009).
Selected bacterial strains of the natural Arabidopsis root microbiota belonging to the genus Pseudomonas and the class Actinobacteria were particularly potent in activating expression of the iron‐deficiency marker MYB72 (Table S2), suggesting that they collectively participate in stimulation of iron nutrition in their host plant. In support of this, microbial communities derived from compost soil were shown to improve iron content in Arabidopsis when introduced to germ‐free soil (Carvalhais et al., 2013b). However, the availability of iron in the root vicinity as affected by soil microbial communities did not in itself explain the positive impact of root‐colonizing microbes on plant iron nutrition (Carvalhais et al., 2013b). Hence, in addition to directly enhancing iron mineralization and solubilization in the soil, stimulation of the iron‐acquisition machinery in the plant is an important function of specific rhizosphere microbiota.
Local and systemic induction of iron‐uptake responses
Plants and animals are equipped with iron‐sensing systems that register the cellular iron status. The existence of homeostatic mechanisms is essential for cellular viability because they ensure adequate iron supply while preventing cellular toxicity from iron overload. Although significant progress has been made during recent years in terms of elucidating transcriptional and physiological responses to iron deficiency, the sensing system(s) that register iron limitation in dicots are largely unknown (Vigani et al., 2013). Dedicated split‐root experiments in Arabidopsis and other plant species revealed that iron uptake is subjected to local and systemic regulation (Grusak and Pezeshgi, 1996; Schmidt et al., 1996; Vert et al., 2003; Giehl et al., 2009). In this study, we found that WCS417 VOC‐mediated induction of the iron‐deficiency response is expressed in the late maturation zone of Arabidopsis roots, and requires photosynthesis‐dependent signals from the shoot. Moreover, the bacterial VOC‐induced iron‐deficiency response functions independently of the iron status in the root vicinity. This in contrast to the iron‐deficiency response that is triggered by low‐iron conditions in the rhizosphere, which is not dependent on photosynthesis‐related signals and is predominantly expressed in the apical root zone. Previously, VOCs of plant‐beneficial rhizobacteria have been shown to promote plant growth and enhance the photosynthetic capacity of Arabidopsis (Ryu et al., 2003; Zamioudis et al., 2013). In the case of the plant growth‐promoting rhizobacterium B. subtilis GB03, stimulation of iron‐uptake mechanisms was linked to this process (Zhang et al., 2009). Because photosynthesis is an iron‐demanding process, it is plausible that the enhanced photosynthetic capacity mediated by bacterial VOCs directly or indirectly increases iron demands in the shoot, resulting in up‐regulation of strategy I and MYB72 expression in the roots. Future studies are required to reveal the exact mechanisms by which bacterial VOCs activate strategy I for iron uptake and up‐regulate MYB72 gene expression during the onset of ISR.
The FIT1 transcriptional network integrates local and systemic iron‐deficiency signals
The observation that expression of FER1, a molecular marker for cellular iron status, rapidly decreases in the Arabidopsis root in response to bacterial VOCs suggests that the VOC‐induced iron sensory signal(s) in the shoot systemically stimulate sequestration of iron from the root, resulting in activation of FIT‐dependent iron‐uptake mechanisms in the late maturation zone of the root. Because roots do not photosynthesize, other physiological processes are likely to deplete iron in the apical root zone, where the putative local sensory system is located. Young cells in the apical root zone are anticipated to be metabolically more active than older cells in the mature root, and thus more sensitive to local iron deprivation. In the fit mutant, expression of MYB72 (Figure 6c) and the iron‐uptake genes FRO2 and IRT1 (Colangelo and Guerinot, 2004) were shown to be severely compromised in response to WCS417 VOCs or under conditions of iron limitation, respectively, indicating that both local and systemic induction of iron‐uptake mechanisms are controlled in the roots by the central regulator FIT.
Role of NO in the VOC‐induced iron‐uptake response
Despite the fact that the shoot‐dependent iron‐uptake system does not stimulate the iron‐uptake response in the apical root, shoot‐borne signals appear to prime the root‐autonomous iron‐deficiency response for enhanced expression when iron is limited in the rhizosphere (Figure 5a). NO has a well‐established role in orchestrating adaptive responses to iron limitation (Chen et al., 2010; Garcia et al., 2010; Meiser et al., 2011). We observed that NO accumulated not only in the late maturation zone of roots of bacterial VOC‐exposed seedlings, but also in the apical root where the local iron‐sensing system operates (Figure 6d), suggesting that NO primes iron‐uptake mechanisms in the apical root under conditions of iron limitation. In further support of this, treatment with the NO donor S‐nitrosoglutathione was shown to specifically enhance FRO2 gene expression and root ferric chelate reductase activity only under conditions of iron limitation (Chen et al., 2010). NO generated during photosynthesis by chloroplast‐localized nitrite reductases may function itself as long‐distance priming molecule (Jasid et al., 2006). Alternatively, NO may be generated locally in the apical root upon perception of primary systemic shoot‐borne molecules.
Dual role of MYB72 in ISR and iron‐uptake responses
In this study, we show that the root‐specific MYB72 transcription factor gene is co‐regulated with the iron‐deficiency genes FRO2 and IRT1 in a FIT‐dependent manner. Originally we identified MYB72 as an essential component of ISR that is triggered by beneficial root‐colonizing Pseudomonas spp. and Trichoderma spp. strains and protects foliar tissues against a broad spectrum of plant pathogens (Van der Ent et al., 2008; Segarra et al., 2009). Interestingly, a link between iron homeostasis and enhanced disease resistance has also been reported for Arabidopsis plants growing under iron‐limited conditions (Kieu et al., 2012), or expressing induced resistance triggered by β‐aminobutyric acid (Koen et al., 2014). However, how this mechanistically relates to the MYB72‐dependent iron‐deficiency responses associated with rhizobacteria‐mediated ISR is currently unknown. How may the iron‐deficiency and ISR signaling networks be inter‐connected? Recently, we demonstrated that MYB72 regulates the expression of genes that are involved in the production and secretion of iron‐mobilizing phenolic metabolites (Zamioudis et al., 2014). Amongst the MYB72 target genes is BGLU42, which is activated in the same root cells as MYB72 and encodes a glucoside hydrolase that is involved in excretion of iron‐mobilizing phenolic compounds into the rhizosphere, but is also required for onset of rhizobacteria‐mediated ISR (Zamioudis et al., 2014). We therefore hypothesized that rhizobacteria‐mediated BGLU42 activity may also result in generation of a so far unidentified mobile ISR signal. Constitutive over‐expression of BGLU42 in Arabidopsis confers broad‐spectrum disease resistance (Zamioudis et al., 2014), supportive of this possibility. Alternatively, nutrient deficiencies have been shown to alter root exudation and the production of semiochemicals, which in turn leads to changes in the communication between plant roots and root‐associated bacteria (Yang and Crowley, 2000; Carvalhais et al., 2013a). Hence rhizobacteria‐mediated activation of MYB72 and BGLU42 and their downstream effects on the production and excretion of iron‐mobilizing phenolics may be required for establishment of a mutualistic plant–microbe relationship that in turn results in activation of ISR by the beneficial microbe. Either way, our study highlights the existence of an ingenious communication network between plants and mutualistic rhizobacteria, and provides insight into the molecular mechanisms of iron uptake and systemic immunity as stimulated by members of the root microbiota.
Experimental Procedures
Plant material and growth conditions
Arabidopsis thaliana accession Col‐0 was used as the wild‐type. The fit1‐2 mutant in the Col‐0 background (Colangelo and Guerinot, 2004) was obtained from the Nottingham Arabidopsis Stock Centre ( http://arabidopsis.info/). The following over‐expressor lines were used: oxFIT, oxbHLH38, oxbHLH39 and oxFIT/bHLH38 (Yuan et al., 2008). For generation of the pMYB72:GFP‐GUS reporter line, the 1.7 kb genomic region upstream of the start codon of MYB72 was amplified from genomic Col‐0 DNA using primers pMYB72‐FW and pMYB72‐RV (Table S3). The fragment was cloned into the pDONR221‐pGEMT‐Easy vector (Promega, http://nld.promega.com/) using the BP reaction, and recombined into the destination vector pBGWFS7.0 (Gateway Vectors, https://gateway.psb.ugent.be/search/index/transcriptional_reporters/any) using the LR reaction (Invitrogen, https://www.thermofisher.com/nl/en/home/brands/invitrogen.html) according to the manufacturer's instructions. The recombinant plasmid was transformed into Agrobacterium tumefaciens strain GV3101::pMP90, after which A. tumefaciens‐mediated plant transformation was performed using the floral‐dip method (Clough and Bent, 1998).
For in vitro growth of Arabidopsis seedlings, seeds were surface‐sterilized and sown on agar‐solidified 1× Murashige and Skoog (MS) medium supplemented with 1% sucrose. After 2 days of stratification at 4°C, the Petri dishes were positioned vertically and transferred to a growth chamber (22°C, 10 h light/14 h dark, light intensity 100 μmol m−2 sec−1). Uniform 5‐day‐old seedlings were transferred to new plates containing 1× modified Hoagland medium (Hoagland and Arnon, 1938) containing KNO3 (3 mm), MgSO4 (0.5 mm), CaCl2 (1.5 mm), K2SO4 (1.5 mm), NaH2PO4 (1.5 mm), H3BO3 (25 μm), MnSO4 (1 μm), ZnSO4 (0.5 μm), (NH4)6Mo7O24 (0.05 μm), CuSO4 (0.3 μm) and MES (2.5 mm). The concentration of Fe(III)EDTA was adjusted to 50 μm unless otherwise specified. The pH of the medium was adjusted to 5.8 using KOH. Transferred seedlings were allowed to continue growing for another 6–7 days on vertically positioned plates in the growth chamber before treatment (see below). Experiments involving direct application of bacteria to the roots were performed in square plates. For experiments involving bacterial VOCs, two‐compartment circular plates with a center partition were used. These plates physically separate seedlings and microbes, but allow gas exchange between compartments (Zamioudis et al., 2013). For the CO2‐depletion experiments, three‐compartment plates were used (see below).
Cultivation of microbes and treatments
The model rhizobacterial strains Pseudomonas simiae WCS417, Pseudomonas capeferrum WCS358, Pseudomonas defensor WCS374 (Van Wees et al., 1997; Berendsen et al., 2015) and the WCS417 siderophore mutants S680 and M634 (Duijff et al., 1993) were cultured at 28°C on King's medium B (King et al., 1954) supplemented with 50 μg ml−1 rifampicin. After 24 h of growth, cells were collected in 10 mm MgSO4, washed twice by centrifugation for 5 min at 5000 g, and finally resuspended in 10 mm MgSO4. The bacterial titer was adjusted to an OD600 of 0.01 (approximately 107 cfu ml−1). Ten microliters of bacterial suspension were then applied to the roots of each 12‐day‐old seedling, immediately below the hypocotyl. Bacterial VOCs were applied to 12‐day‐old seedlings in two‐compartment plates by transferring 50 μl of bacterial suspension with an OD600 of 0.1 into the plant‐free compartment containing 1× MS agar‐solidified medium.
Elicitor, chemical and iron‐deficiency treatments
For studies involving treatments of Arabidopsis roots with defense elicitors, flagellin (flg22) (GenScript, http://www.genscript.com/) or chitosan (Sigma, https://www.sigmaaldrich.com) were applied to the liquid medium (1× MS medium, 0.5% sucrose) for 2‐week‐old hydroponically grown seedlings to final concentrations of 250 nm or 0.001% v/v, respectively. Root samples were collected at defined time points after treatment. Iron deficiency was induced by transferring standard‐grown, 12‐day‐old seedlings to plates containing 1× modified Hoagland agar‐solidified medium from which iron was omitted. The complete elimination of iron was achieved by supplementing the medium with the Fe(II) chelator Ferrozine (Sigma) at a final concentration of 300 μm. Decapitation of the shoot was performed by cutting at the root–shoot junction immediately after transfer of the seedlings to iron‐deprived medium, or just prior to treatment of the seedlings with bacterial VOCs. Darkness treatment was applied by placing the plates containing the seedlings in complete darkness, immediately after the start of the VOC treatment. CO2 depletion was achieved by adding a KOH‐based trap (1 m KOH) in one section of a three‐compartment plate just prior to VOC treatment. Chemical inhibition of photosynthesis was achieved 24 h before VOC treatment by transferring 11‐day‐old seedlings to new plates in which the Hoagland medium was supplemented with 5 μm norflurazon (Sigma). Chemical inhibition of NO signaling was achieved 24 h before VOC treatment by transferring 11‐day‐old seedlings to Hoagland plates supplemented with the NO scavenger PTIO (Sigma) at a final concentration of 1 mm.
Chlorophyll measurements
Chlorophyll measurements were performed essentially as described by Hiscox and Israelstam (1979). In brief, leaf tissue from five pooled Arabidopsis seedlings was cut into small pieces and placed in a vial containing 3 ml dimethylsulfoxide per 100 mg fresh weight. Nine replicates of five pooled seedlings each were incubated for 45 min at 65°C. After cooling to room temperature, chlorophyll (a + b) extracts were transferred to a cuvette, and spectrophotometer readings were performed using a DU‐64 spectrophotometer (Beckman, https://www.beckmancoulter.com) at a wavelength of 652 nm. Chlorophyll concentrations were calculated as described by Hiscox and Israelstam (1979).
Fluorescence microscopy
Confocal laser scanning microscopy was performed using a Zeiss LSM 700 microscope ( http://www.zeiss.com). GFP was excited using a 488 nm argon laser, and fluorescence was detected at 500–550 nm. As a counter‐stain, roots were stained in 10 μg ml−1 propidium iodide solution for 2 min. For detection of endogenous NO, seedlings were incubated in a solution containing 5 μm DAF‐FMDA in buffered solution (10 mm Tris/HCl, pH 7.4) for 1 h at 25°C in the dark. Subsequently, seedlings were washed three times for 15 min each using fresh buffer (10 mm Tris/HCl, pH 7.4). The fluorescence emitted by DAF‐FMDA was detected by excitation at 495 nm and emission at 515 nm (Fernández‐Marcos et al., 2011).
GUS histohemical staining
Histochemical detection of GUS activity was performed using a GUS staining solution comprising 50 mm sodium phosphate, pH 7, 10 mm EDTA, 0.5 mm K4[Fe(CN)6], 0.5 mm K3[Fe(CN)6], 0.5 mm X‐Gluc and 0.01% Silwet L‐77 (Van Meeuwen, http://vanmeeuwen.com) at 37°C for 3 h. Stained roots were cleared in a mixture of chloral hydrate/glycerol/water (8:1:2), and observed using Nomarski optics.
Root ferric chelate reductase activity
Ferric chelate reductase activity was visualized by transferring seedlings onto agar‐solidified modified Hoagland medium containing 0.5 mm CaSO4, 0.5 mm Ferrozine and 0.5 mm Fe(III)EDTA for 20 min (Schmidt et al., 2000). Quantification of ferric chelate reductase activity was performed by transferring root tissues into liquid medium containing 0.1 mm Fe(III)EDTA and 0.3 mm Ferrozine (Yi and Guerinot, 1996). After 20 min incubation, the absorbance of the Fe(II)–Ferrozine complex was recorded at 562 nm. Reduction rates were calculated using an extinction coefficient of 28.6 mm −1 cm−1. Roots were briefly rinsed in 10 mm MgSO4 solution before starting the assays.
Microarray analysis
Two‐week‐old Arabidopsis Col‐0 seedlings growing in modified Hoagland agar medium were inoculated with WCS417 by applying 10 μl of a bacterial suspension (107 cfu ml−1) to the primary root of each seedling, immediately below the hypocotyl. Three biological replicates of mock‐ and WCS417‐treated roots were harvested 48 h later for microarray analysis, which was performed as described previously (Zamioudis et al., 2014). For gene annotations according to biological categories, the AmiGO Term Enrichment software was used (Carbon et al., 2009).
Quantitative RT‐PCR analysis
Total RNA was extracted using an RNeasy kit (Qiagen, https://www.qiagen.com) according to the manufacturer's instructions, and treated with TURBO™ DNase (Ambion, https://www.thermofisher.com). Then, cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Cycle thresholds were determined in duplicate per transcript in three biological replicates per sample using the ABI PRISM 7700 sequence detection system (Applied Biosystems, http://www.appliedbiosystems.com) and SYBR Green I (Thermo Fisher, https://www.thermofisher.com) as the reporter dye. Gene expression data were normalized using ACTIN7. The gene‐specific primers used are listed in Table S3.
Supporting information
Figure S1. Ferric chelate reductase activity in Arabidopsis Col‐0 roots colonized by Pseudomonas spp. strains.
Figure S2. Confocal images of pMYB72:GFP‐GUS roots colonized by Pseudomonas spp. strains.
Figure S3. Bacterial determinants involved in induction of the iron‐deficiency response in Arabidopsis roots.
Figure S4. Bacterial VOCs activate the strategy I iron‐deficiency response independently of the iron status in the root vicinity.
Table S1. Microarray data for selected up‐ and down‐regulated genes in Arabidopsis Col‐0 roots in response to colonization by Pseudomonas simiae WCS417 bacteria.
Table S2. The extent by which volatiles from selected members of the Arabidopsis natural root microbiome activate MYB72 expression in Arabidopsis roots.
Table S3. Arabidopsis Genome Initiative numbers and primers used in this study for cloning and studying expression of Arabidopsis genes.
Acknowledgments
This work was supported by Advanced Grants 269072 and 323094 from the European Research Council.
References
- Bauer, P. , Ling, H.Q. and Guerinot, M.L. (2007) FIT, the FER‐like iron deficiency induced transcription factor in Arabidopsis. Plant Physiol. Biochem. 45, 260–261. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Pieterse, C.M.J. and Bakker, P.A.H.M. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Van Verk, M.C. , Stringlis, I.A. , Zamioudis, C. , Tommassen, J. , Pieterse, C.M.J. and Bakker, P.A.H.M. (2015) Unearthing the genomes of plant‐beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genom. 16, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat, J.‐F. , Dubos, C. and Gaymard, F. (2015) Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20, 33–40. [DOI] [PubMed] [Google Scholar]
- Buckhout, T. , Yang, T. and Schmidt, W. (2009) Early iron‐deficiency‐induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genom. 10, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli, D. , Rott, M. , Schlaeppi, K. et al (2012) Revealing structure and assembly cues for Arabidopsis root‐inhabiting bacterial microbiota. Nature, 488, 91–95. [DOI] [PubMed] [Google Scholar]
- Bulgarelli, D. , Schlaeppi, K. , Spaepen, S. , Van Themaat, E.V.L. and Schulze‐Lefert, P. (2013) Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. [DOI] [PubMed] [Google Scholar]
- Carbon, S. , Ireland, A. , Mungall, C.J. , Shu, S. , Marshall, B. and Lewis, S. (2009) AmiGO: online access to ontology and annotation data. Bioinformatics, 25, 288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais, L.C. , Dennis, P.G. , Fan, B. , Fedoseyenko, D. , Kierul, K. , Becker, A. , von Wiren, N. and Borriss, R. (2013a) Linking plant nutritional status to plant–microbe interactions. PLoS ONE, 8, e68555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais, L.C. , Muzzi, F. , Tan, C.‐H. , Choo, J.H. and Schenk, P.M. (2013b) Plant growth in Arabidopsis is assisted by compost soil‐derived microbial communities. Fron. Plant Sci. 4, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.W. , Yang, J.L. , Qin, C. , Jin, C.W. , Mo, J.H. , Ye, T. and Zheng, S.J. (2010) Nitric oxide acts downstream of auxin to trigger root ferric‐chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol. 154, 810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Colangelo, E.P. and Guerinot, M.L. (2004) The essential basic helix‐loop‐helix protein FIT1 is required for the iron deficiency response. Plant Cell, 16, 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, E. and Guerinot, M. (2002) Iron stress in plants. Genome Biol. 3, 1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mortel, J.E.V. , Schat, H. , Moerland, P.D. , Van Themaat, E.V.L. , Van der Ent, S. , Blankestijn, H. , Ghandilyan, A. , Tsiatsiani, S. and Aarts, M.G.M. (2008) Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd‐hyperaccumulator Thlaspi caerulescens . Plan, Cell Environ. 31, 301–324. [DOI] [PubMed] [Google Scholar]
- De Vleesschauwer, D. and Höfte, M. (2009) Rhizobacteria‐induced systemic resistance In Plant Innate Immunity (Van Loon L.C., ed.). London: Academic Press Ltd/Elsevier Science Ltd, pp. 223–281. [Google Scholar]
- Dinneny, J.R. , Long, T.A. , Wang, J.Y. , Jung, J.W. , Mace, D. , Pointer, S. , Barron, C. , Brady, S.M. , Schiefelbein, J. and Benfey, P.N. (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science, 320, 942–945. [DOI] [PubMed] [Google Scholar]
- Duijff, B.J. , Meijer, J.W. , Bakker, P.A.H.M. and Schippers, B. (1993) Siderophore‐mediated competition for iron and induced resistance in the suppression of fusarium wilt of carnation by fluorescent Pseudomonas spp. Neth. J. Plant Pathol. 99, 277–289. [Google Scholar]
- Fernández‐Marcos, M. , Sanz, L. , Lewis, D.R. , Muday, G.K. and Lorenzo, O. (2011) Nitric oxide causes root apical meristem defects and growth inhibition while reducing PIN‐FORMED 1 (PIN1)‐dependent acropetal auxin transport. Proc. Natl Acad. Sci. USA, 108, 18506–18511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, M.J. , Lucena, C. , Romera, F.J. , Alcantara, E. and Perez‐Vicente, R. (2010) Ethylene and nitric oxide involvement in the up‐regulation of key genes related to iron acquisition and homeostasis in Arabidopsis . J. Exp. Bot. 61, 3885–3899. [DOI] [PubMed] [Google Scholar]
- Gaymard, F. , Boucherez, J. and Briat, J.F. (1996) Characterization of a ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem. J. 318, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl, R.F.H. , Meda, A.R. and Von Wiren, N. (2009) Moving up, down, and everywhere: signaling of micronutrients in plants. Curr. Opin. Plant Biol. 12, 320–327. [DOI] [PubMed] [Google Scholar]
- Grusak, M.A. and Pezeshgi, S. (1996) Shoot‐to‐root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 110, 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscox, J.D. and Israelstam, G.F. (1979) A method for the extraction of chlorophyll from leaf tissue without maceration. Can. J. Bot. 57, 1332–1334. [Google Scholar]
- Hoagland, D.R. and Arnon, D.I. (1938) The water culture method for growing plants without soil. Calif. Agric. Exp. Station Bull. 347, 36–39. [Google Scholar]
- Jakoby, M. , Wang, H.‐Y. , Reidt, W. , Weisshaar, B. and Bauer, P. (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana . FEBS Lett. 577, 528–534. [DOI] [PubMed] [Google Scholar]
- Jasid, S. , Simontacchi, M. , Bartoli, C.G. and Puntarulo, S. (2006) Chloroplasts as a nitric oxide cellular source. Effect of reactive nitrogen species on chloroplastic lipids and proteins. Plant Physiol. 142, 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieu, N.P. , Aznar, A. , Segond, D. , Rigault, M. , Simond‐Cote, E. , Kunz, C. , Soulie, M.C. , Expert, D. and Dellagi, A. (2012) Iron deficiency affects plant defence responses and confers resistance to Dickeya dadantii and Botrytis cinerea . Mol. Plant Pathol. 13, 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, E.O. , Ward, M.K. and Raney, D.E. (1954) Two simple media for the demonstration of phycocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Koen, E. , Trapet, P. , Brulé, D. et al (2014) β‐Aminobutyric acid (BABA)‐induced resistance in Arabidopsis thaliana: link with iron homeostasis. Mol. Plant Microbe Interact. 27, 1226–1240. [DOI] [PubMed] [Google Scholar]
- Lingam, S. , Mohrbacher, J. , Brumbarova, T. , Potuschak, T. , Fink‐Straube, C. , Blondet, E. , Genschik, P. and Bauer, P. (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3‐LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell, 23, 1815–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Bucio, J. , Campos‐Cuevas, J.C. , Hernandez‐Calderon, E. , Velasquez‐Becerra, C. , Farias‐Rodriguez, R. , Macias‐Rodriguez, L.I. and Valencia‐Cantero, E. (2007) Bacillus megatenum rhizobacteria promote growth and alter root‐system architecture through an auxin‐and ethylene‐independent signaling mechanism in Arabidopsis thaliana . Mol. Plant Microbe Interact. 20, 207–217. [DOI] [PubMed] [Google Scholar]
- Lundberg, D.S. , Lebeis, S.L. , Paredes, S.H. et al (2012) Defining the core Arabidopsis thaliana root microbiome. Nature, 488, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser, J. , Lingam, S. and Bauer, P. (2011) Posttranslational regulation of the iron deficiency basic helix‐loop‐helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiol. 157, 2154–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes, R. , Kruijt, M. , De Bruijn, I. et al (2011) Deciphering the rhizosphere microbiome for disease‐suppressive bacteria. Science, 332, 1097–1100. [DOI] [PubMed] [Google Scholar]
- Millet, Y.A. , Danna, C.H. , Clay, N.K. , Songnuan, W. , Simon, M.D. , Werck‐Reichhart, D. and Ausubel, F.M. (2010) Innate immune responses activated in Arabidopsis roots by microbe‐associated molecular patterns. Plant Cell, 22, 973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, C.M. , Hindt, M.N. , Schmidt, H. , Clemens, S. and Guerinot, M.L. (2013) MYB10 and MYB72 are required for growth under iron‐limiting conditions. PLoS Genet. 9, e1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C.M. and Bakker, P.A.H.M. (2014) Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. [DOI] [PubMed] [Google Scholar]
- Richardson, A.E. and Simpson, R.J. (2011) Soil microorganisms mediating phosphorus availability. Plant Physiol. 156, 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, N.J. , Procter, C.M. , Connolly, E.L. and Guerinot, M.L. (1999) A ferric‐chelate reductase for iron uptake from soils. Nature, 397, 694–697. [DOI] [PubMed] [Google Scholar]
- Ryu, C.‐M. , Farag, M.A. , Hu, C.H. , Reddy, M.S. , Wei, H.X. , Pare, P.W. and Kloepper, J.W. (2003) Bacterial volatiles promote growth in Arabidopsis. Proc. Natl Acad. Sci. USA, 100, 4927–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, C.‐M. , Farag, M.A. , Hu, C.H. , Reddy, M.S. , Kloepper, J.W. and Paré, P.W. (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, W. , Boomgaarden, B. and Ahrens, V. (1996) Reduction of root iron in Plantago lanceolata during recovery from Fe deficiency. Physiol. Plant. 98, 587–593. [Google Scholar]
- Schmidt, W. , Tittel, J. and Schikora, A. (2000) Role of hormones in the induction of iron deficiency responses in Arabidopsis roots. Plant Physiol. 122, 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segarra, G. , Van der Ent, S. , Trillas, I. and Pieterse, C.M.J. (2009) MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 11, 90–96. [DOI] [PubMed] [Google Scholar]
- Sivitz, A. , Grinvalds, C. , Barberon, M. , Curie, C. and Vert, G. (2011) Proteasome‐mediated turnover of the transcriptional activator FIT is required for plant iron‐deficiency responses. Plant J. 66, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Sivitz, A.B. , Hermand, V. , Curie, C. and Vert, G. (2012) Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT‐independent pathway. PLoS ONE, 7, e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent, S. , Verhagen, B.W.M. , Van Doorn, R. et al (2008) MYB72 is required in early signaling steps of rhizobacteria‐induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon, L.C. , Bakker, P.A.H.M. , Van der Heijdt, W.H.W. , Wendehenne, D. and Pugin, A. (2008) Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol. Plant Microbe Interact. 21, 1609–1621. [DOI] [PubMed] [Google Scholar]
- Van Wees, S.C.M. , Pieterse, C.M.J. , Trijssenaar, A. , Van't Westende, Y.A.M. , Hartog, F. and Van Loon, L.C. (1997) Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol. Plant–Microbe Interact. 10, 716–724. [DOI] [PubMed] [Google Scholar]
- Vert, G. , Grotz, N. , Dédaldéchamp, F. , Gaymard, F. , Guerinot, M.L. , Briat, J.‐F. and Curie, C. (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell, 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert, G.A. , Briat, J.‐F. and Curie, C. (2003) Dual regulation of the Arabidopsis high‐affinity root iron uptake system by local and long‐distance signals. Plant Physiol. 132, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigani, G. , Zocchi, G. , Bashir, K. , Philippar, K. and Briat, J.‐F. (2013) Signals from chloroplasts and mitochondria for iron homeostasis regulation. Trends Plant Sci. 18, 305–311. [DOI] [PubMed] [Google Scholar]
- Walker, E.L. and Connolly, E.L. (2008) Time to pump iron: iron‐deficiency‐signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11, 530–535. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Cui, Y. , Liu, Y. , Fan, H. , Du, J. , Huang, Z. , Yuan, Y. , Wu, H. and Ling, H.‐Q. (2013) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana . Mol. Plant, 6, 503–513. [DOI] [PubMed] [Google Scholar]
- Yang, C.‐H. and Crowley, D.E. (2000) Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 66, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, Y. and Guerinot, M.L. (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10, 835–844. [DOI] [PubMed] [Google Scholar]
- Yuan, Y.X. , Zhang, J. , Wang, D.W. and Ling, H.Q. (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res., 15, 613–621. [DOI] [PubMed] [Google Scholar]
- Yuan, Y. , Wu, H. , Wang, N. , Li, J. , Zhao, W. , Du, J. , Wang, D. and Ling, H.‐Q. (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis . Cell Res. 18, 385–397. [DOI] [PubMed] [Google Scholar]
- Zamioudis, C. (2012) Signaling in Arabidopsis roots in response to beneficial rhizobacteria. PhD Thesis, Utrecht University. [Google Scholar]
- Zamioudis, C. and Pieterse, C.M.J. (2012) Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150. [DOI] [PubMed] [Google Scholar]
- Zamioudis, C. , Mastranesti, P. , Dhonukshe, P. , Blilou, I. and Pieterse, C.M.J. (2013) Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 162, 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis, C. , Hanson, J. and Pieterse, C.M.J. (2014) β‐Glucosidase BGLU42 is a MYB72‐dependent key regulator of rhizobacteria‐induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204, 368–379. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Sun, Y. , Xie, X. , Kim, M.‐S. , Dowd, S.E. and Paré, P.W. (2009) A soil bacterium regulates plant acquisition of iron via deficiency‐inducible mechanisms. Plant J. 58, 568–577. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Ferric chelate reductase activity in Arabidopsis Col‐0 roots colonized by Pseudomonas spp. strains.
Figure S2. Confocal images of pMYB72:GFP‐GUS roots colonized by Pseudomonas spp. strains.
Figure S3. Bacterial determinants involved in induction of the iron‐deficiency response in Arabidopsis roots.
Figure S4. Bacterial VOCs activate the strategy I iron‐deficiency response independently of the iron status in the root vicinity.
Table S1. Microarray data for selected up‐ and down‐regulated genes in Arabidopsis Col‐0 roots in response to colonization by Pseudomonas simiae WCS417 bacteria.
Table S2. The extent by which volatiles from selected members of the Arabidopsis natural root microbiome activate MYB72 expression in Arabidopsis roots.
Table S3. Arabidopsis Genome Initiative numbers and primers used in this study for cloning and studying expression of Arabidopsis genes.


