Among hospitalized subjects exposed to antimicrobials, a high abundance of Lactobacillus spp. characterized the fecal microbiome of patients who did not acquire multidrug-resistant organisms during their hospitalization, consistent with a protective role for Lactobacillus spp.
Keywords: microbiome, acquisition, risk factors, hospital, multidrug-resistant organisms
Abstract
Background. The emergence and dissemination of multidrug-resistant organisms (MDROs) is a global threat. Characterizing the human microbiome among hospitalized patients and identifying unique microbial signatures among those patients who acquire MDROs may identify novel infection prevention strategies.
Methods. Adult patients admitted to 5 general medical-surgical floors at a 649-bed, tertiary care center in Boston, Massachusetts, were classified according to in-hospital antimicrobial exposure and MDRO colonization status. Within 48 hours of hospital admission (baseline) and at discharge (follow-up), rectal swab samples were obtained, and compared with samples from an external control group of healthy persons from the community. DNA was extracted from samples, next-generation sequencing performed, and microbial community structure and taxonomic features assessed, comparing those who acquired MDROs and those who had not, and the external controls.
Results. Hospitalized patients (n = 44) had reduced microbial diversity and a greater abundance of Escherichia spp. and Enterococcus spp. than healthy controls (n = 26). Among hospitalized patients, 25 had no MDROs at the time of the baseline sample and were also exposed to antimicrobials. Among this group, 7 (28%) acquired ≥1 MDRO; demographic and clinical characteristics were similar between MDRO-acquisition and MDRO-nonacquisition groups. Patients in the nonacquisition group had consistently higher Lactobacillus spp. abundance than those in the acquisition group (linear discriminant score, 3.97; P = .04).
Conclusions. The fecal microbiota of the hospitalized subjects had abnormal community composition, and Lactobacillus spp. was associated with lack of MDRO acquisition, consistent with a protective role.
The emergence and dissemination of multidrug-resistant organisms (MDROs) is a well-recognized health threat [1]. Compared with infections caused by antimicrobial-susceptible bacteria, those caused by MDROs are associated with 2–5 times higher mortality rates and contribute to a substantial economic burden [2]. Despite the ongoing implementation of infection control strategies, rates of MDROs continue to rise, indicating that novel approaches to curb acquisition and dissemination are needed [3, 4].
The human microbiome plays an important role in protecting the host from de novo colonization with exogenous pathogenic bacteria [5]. This colonization resistance is disrupted by exposure to antimicrobials, a leading risk factor for MDRO acquisition [6]. Advances in sequencing technologies and bioinformatic tools have allowed detailed analyses of the microbiome and have begun to uncover the contribution of specific microbial populations to the acquisition of pathogens such as Clostridium difficile and vancomycin-resistant enterococci (VRE) [7, 8]. Reintroduction of specific microbial populations has also been associated with protection against colonization with pathogenic bacteria [9]. Understanding the specific microbial community characteristics that may predispose to or prevent MDRO acquisition may lead to the development of novel infection control strategies.
In the current study, we hypothesized that hospitalized patients have reduced microbial diversity compared with healthy controls and that there are particular microbial signatures that can identify patients at high risk of MDRO acquisition. We tested these hypotheses by comparing the fecal microbiota of healthy and hospitalized subjects, with special reference to those who acquired MDROs during their hospital stay.
METHODS
Study Population
The study population consisted of adult patients hospitalized on 5 medical-surgical floors in a 649-bed, tertiary center in Boston, Massachusetts. The exclusion criteria eliminated (1) patients who were discharged before completing the study and in whom follow-up swab samples could therefore not be obtained, (2) patients in whom perianal/rectal cultures could not be obtained (colostomy or rectal bags), and (3) patients in whom the baseline rectal sample was not successfully sequenced. The hospital's institutional review board approved the protocol, and informed consent was obtained from all subjects before specimen and data collection.
Patient Screening and Data and Specimen Collection
Participants were identified using the hospital's electronic medical record system. If the patient agreed to participate, a first visit was performed within 48 hours of their admission, at which time clinical and epidemiological data and an initial rectal sample (baseline sample) were collected. A second visit, just before hospital discharge, was performed to collect data from their hospitalization and a second rectal sample (follow-up sample). Study data, including patient demographics, medical history, and history of antimicrobial exposure, were collected using the study hospital's electronic medical records and managed using REDCap (Research Electronic Data Capture) [10].
Baseline and follow-up specimens were processed for MDROs (methicillin-resistant Staphylococcus aureus, VRE, and multidrug-resistant gram-negative bacteria). Multidrug-resistant gram-negative bacteria were defined as gram-negative bacteria resistant to ≥3 of the following antimicrobials: ampicillin-sulbactam or piperacillin-tazobactam, ceftriaxone or ceftazidime (ceftazidime only for Pseudomonas spp.), ciprofloxacin, gentamicin, and meropenem. MDRO acquisition was defined as occurring in a patient in whom MDROs were not identified in the baseline specimen but were found in the follow-up specimen.
Subjects were classified into 4 mutually exclusive groups, according to in-hospital antimicrobial exposure and MDRO status (Table 1). Group 1 included patients not exposed to antimicrobials during their hospitalization and not colonized with MDROs either at baseline or follow-up; group 2, patients exposed to antimicrobials in whom MDRO were recovered at follow-up but not at baseline (MDRO acquisition); group 3, patients exposed to antimicrobials who were colonized with MDROs at baseline regardless of the MDRO colonization status at follow-up; and group 4, patients exposed to antimicrobials who did not have MDROs either at baseline or at follow-up (MDRO nonacquisition). To compare the fecal microbiome of our study subjects with that of persons in the community, an external control group (group 5) was also included. This group consisted of healthy persons, >50 years old, living in the United States. Data for these patients was obtained from the Human Microbiome Project database. The publically available V4 region sequences of the bacterial 16S ribosomal RNA (rRNA) gene were obtained from the MG-RAST server under accession number qiime:850 [12].
Table 1.
Definition of Study Groups, According to In-Hospital Antimicrobial Exposure, Multidrug-resistant Organism (MDRO) Colonization at Baseline and Follow-up, and MDRO Acquisition
| Study Group | In-Hospital Antimicrobial Exposure | MDROs Recovered From Swab Sample |
MDRO Acquisition | |
|---|---|---|---|---|
| Baseline | Follow-up | |||
| Group 1 | No | No | No | No |
| Group 2 | Yes | No | Yes | Yes |
| Group 3 | Yes | Yes | Yes or no | No |
| Group 4 | Yes | No | No | No |
| Group 5a | Not hospitalized, healthy controls | … | … | … |
Abbreviation: MDRO, multidrug-resistant organism.
a Group 5 included 16S DNA sequences obtained from fecal samples of healthy American individuals, >50 years old, inhabitants of metropolitan areas (St Louis, Missouri) from the published Human Microbiome Project study [11].
Microbiological Methods
Rectal samples were obtained using a sterile double-tipped swab (Starswab II; Starplex Scientific) inserted 0.5–1 cm into the anus, and they were processed within 1–2 hours after collection. MDRO identification was performed on both baseline and follow-up swab samples, using the first tip of the swab. The second tip was placed in a vial containing 20% glycerol and stored at −80°C for the subsequent microbial community profiling of baseline swab samples. Identification of MDROs and antimicrobial susceptibility testing was performed as described elsewhere, in accordance with Clinical and Laboratory Standards Institute methods [11, 13].
Amplicon Generation and High-Throughput Sequencing
DNA was extracted from baseline samples by enzymatic digestion and bead-beating steps, followed by the use of the QIAamp Stool DNA Mini Kit (Qiagen), according to the manufacturer's protocol. After determining the concentration of the extracted DNA by Nanodrop, 16S rRNA gene amplicons were generated by polymerase chain reaction (PCR) with a bar-coded primer set targeted to the V4 variable region, as described elsewhere [14]. PCR reactions were carried out in triplicate and then pooled; reactions yielding no amplicons or those for which negative controls were positive were repeated. The DNA concentration of each successful PCR was determined with a Quanti-iT assay (Invitrogen), according to the manufacturer's recommendations. Amplicons were then pooled in equimolar concentrations, purified using Agencourt Ampure XP beads (Beckman-Coulter), and sequenced on an Illumina MiSeq Sequencer at the Tufts University Core Facility, using a paired-end- 250–base pair protocol, with reads merged as described elsewhere [14].
Statistical Methods and Bioinformatics
Patient-Level Analysis
Clinical and demographic characteristics of the study population were described by reporting means with standard deviations (SDs) or medians with interquartile ranges for continuous variables and absolute frequencies with percentages for categorical variables. Relevant characteristics were compared between groups using analysis of variance or Kruskal–Wallis rank test for continuous and Fisher exact test for categorical variables. A significance level of P < .05 was used for all statistical tests. All statistical analyses were performed using Stata software, version 13.0 (StataCorp).
Fecal Microbiome Assessment
The community characteristics of the fecal microbiota were determined by comparing alpha and beta diversity metrics between the groups of hospitalized patients and the external control group. To detect signatures in the fecal microbiome potentially associated with protection and/or risk of MDRO acquisition, features were compared specifically between the group of patients who acquired MDROs (group 2) and the group that did not (group 4).
Initial bioinformatics analysis was done using the QIIME (Quantitative Insights Into Microbial Ecology) software package, version 1.7 [15]. Sequences were demultiplexed and then clustered into operational taxonomic units (OTUs) based on ≥99% identity, using the UCLUST algorithm [16]. The most frequent sequence within each OTU was aligned using PyNAST [17] with the core reference database Greengenes for assignment of taxonomy and construction of an OTU table (closed reference OTU picking method) [18]. Differences in community composition between study groups were analyzed using both abundance-based distance metrics (weighted UniFrac) and incidence-based measures (unweighted UniFrac) [19, 20]. Differences in UniFrac distances between groups were tested using the analysis of similarity (ANOSIM) method. The relative abundances of bacterial taxa at any phylogenetic level between the groups were investigated using linear discriminant analysis effect size analysis [21]. An LDA score of 3.0 was set as a cutoff, and an α level of .05 was used for nonparametric testing between groups.
RESULTS
Study Population and Antimicrobial Exposure
From 27 May 2013 to 24 January 2014, a total of 201 patients were approached; of these, 81 patients signed the informed consent form, of whom 49 (60.5%) completed the study with collection of both baseline and follow-up samples. A total of 32 patients (39.5%) were discharged before follow-up swab samples could be obtained and were therefore excluded from the study. Five patients were excluded owing to unsuccessful sequencing of the baseline sample, leaving a final study population of 44 patients.
There were no significant differences in demographic and clinical characteristics among groups 1–4, with the following exceptions: residency in a healthcare-associated facility in the previous 12 months, with 100% of subjects in group 3 (P = .04), and reason for admission (infectious disease), with 0% in group 1 (P < .01). The total intervals from admission to baseline swab samples and from baseline to follow-up swab samples were not significantly different among groups (P = .49 and P = .09, respectively) (Table 2).
Table 2.
Clinical and Demographic Characteristics of the Study Population
| Patient Characteristic | Patients, No. (%)a |
P Value | |||
|---|---|---|---|---|---|
| Group 1 (No Antimicrobial Exposure) (n = 8) | Group 2 (MDRO Acquisition) (n = 7) | Group 3 (Baseline MDRO Colonization) (n = 11) | Group 4 (No MDRO Acquisition) (n = 18) | ||
| Age, mean (SD), y | 68.4 (8.9) | 61.9 (17.9) | 72.4 (14. 7) | 64.6 (12.6) | .36 |
| Male sex | 6 (75.0) | 6 (85.7) | 9 (81.8) | 13 (72.2) | .92 |
| Race/ethnicity | |||||
| White | 6 (75.0) | 5 (71.4) | 9 (81.8) | 10 (55.6) | .87 |
| Hispanic/Latino | 1 (12.5) | 2 (28.6) | 2 (18.2) | 4 (22.2) | |
| African/American | 1 (12.5) | 0 | 0 | 3 (16.7) | |
| Asian | 0 | 0 | 0 | 1 (5.6) | |
| Residents in an HCAF <12 m before enrollment | 4 (50.0) | 4 (57.2) | 11 (100) | 12 (66.7) | .04 |
| BMI, mean (SD), kg/m2 | 27.0 (8.5) | 27.1 (8.6) | 26.1 (7.3) | 27.4 (4.0) | .91 |
| Diabetes mellitus | 3 (37.5) | 2 (28.6) | 6 (54.6) | 10 (55.6) | .62 |
| Charlson comorbidity index, mean (SD) | 6.25 (2.8) | 3.71 (2.9) | 6.91 (2.8) | 4.67 (2.8) | .07 |
| Current use of probiotic use | 0 | 0 | 2 (18.2) | 0 | .50 |
| Long-term proton pump inhibitors | 4 (50.0) | 2 (28.6) | 6 (54.6) | 7 (38.9) | .73 |
| Long-term histamine H2 receptor antagonists | 0 | 1 (14.3) | 0 | 2 (11.1) | .55 |
| History of gastrointestinal disease | 4 (50.0) | 3 (42.9) | 7 (63.6) | 7 (38.9) | .64 |
| Reason for hospitalization | |||||
| Infectious disease | 0 | 3 (42.9) | 9 (81.8) | 11 (61.1) | <.01 |
| Noninfectious disease | 8 (100) | 4 (57.1) | 2 (18.2) | 7 (38.9) | |
| Interval between admission and baseline sample, mean (SD), d | 1.25 (0.5) | 1.57 (0.5) | 1.27 (0.5) | 1.5 (0.6) | .49 |
| Interval between baseline and follow-up samples, mean (SD), d | 1.88 (0.8) | 2.14 (0.4) | 2.27 (0.9) | 3.11 (1.8) | .09 |
Abbreviations: BMI, body mass index; HCAF, healthcare-associated facility; MDRO, multidrug-resistant organism; SD, standard deviation.
a Data represent No. (%) of patients unless otherwise specified.
All 4 groups were exposed to antimicrobials in the 12 months before admission (P = .12). The number of antimicrobial courses in the prior 12 months differed significantly between groups (P = .01), with greater exposure among those colonized at baseline (group 3) (Supplementary Table 1). Thirty-six patients (81.8%) were exposed to antimicrobials during their hospitalization (groups 2–4) and were receiving antimicrobials at the time of both baseline and follow-up swab samples. Thirty-one patients (86.1%) received combinations of ≥2 antimicrobials. The mean (SD) number of antimicrobials received at the time of the baseline sample was 3.0 (1.4), without significant differences between groups (P = .46). The antimicrobials most frequently used during hospitalization were intravenous vancomycin (61.1% of patients), β-lactams (63.9%), and agents with antianaerobic activity (69.4%). There were no significant differences in the types of antimicrobials received during hospitalization between patients who acquired MDROs (group 2) and those who did not (group 4), except for cefazolin (P < .01) (Supplementary Tables 2 and 3).
MDRO Colonization at Baseline and MDRO Hospital Acquisition
Eleven patients were colonized with ≥1 MDRO at baseline (group 3), among whom 7 (63.6%) had a history of MDRO colonization in the previous year. The MDROs isolated at baseline were Escherichia coli in 4 patients; Klebsiella pneumoniae, VRE, and methicillin-resistant S. aureus in 3 patients each; and Proteus mirabilis, Citrobacter freundii, and Pseudomonas aeruginosa in 1 patient each. MDRO acquisition (group 2) was detected in 7 patients (1 patient acquired 2 different MDROs). None were colonized with MDROs the year before admission. The acquired MDROs were VRE in 5 patients and E. coli, K. pneumoniae, and P. aeruginosa in 1 patient each.
16S rRNA Gene Sequencing Output
A total of 44 baseline perianal/rectal specimens were successfully sequenced. After quality filtering, the 44 specimens provided 3 638 570 high-quality 16S rRNA gene sequences, with a median of 84 752 (27 661) sequences per sample. All samples were analyzed at a sequencing depth of 7300 sequences per sample, close to the saturation point for new species discovery. A total of 5057 unique OTUs were identified from sequenced specimens.
Microbial Community Assessment of Baseline Samples
Alpha Diversity
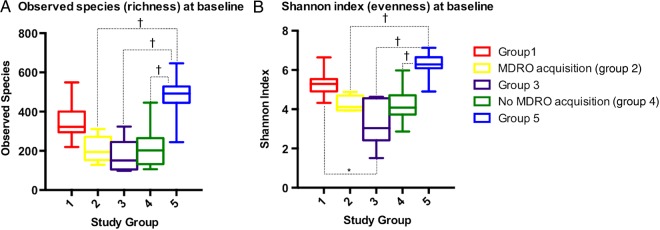
At baseline, the median Shannon diversity index differed significantly between study groups, as did the median number of observed species (overall P < .01 for each metric) (Figure 1). Other alpha diversity indicators calculated showed similar results (Supplementary Table 4). Healthy controls (group 5) presented the highest alpha diversity. Among groups 1–4, patients not exposed to antimicrobials (group 1) presented the highest diversity. In contrast, the patients exposed to antimicrobials and with baseline MDRO colonization (group 3) had the lowest diversity (Figure 1).
Figure 1.
Alpha diversity analyses of the baseline samples show the comparison between groups at a sequencing depth of 7300 sequences per sample. Differences between groups were estimated using the Dunn multiple comparisons test as the posttest after a Kruskal–Wallis test for overall differences. Only statistically significant differences between groups are shown. *P < .05; †P < .001. Abbreviation: MDRO, multidrug-resistant organism.
Beta Diversity
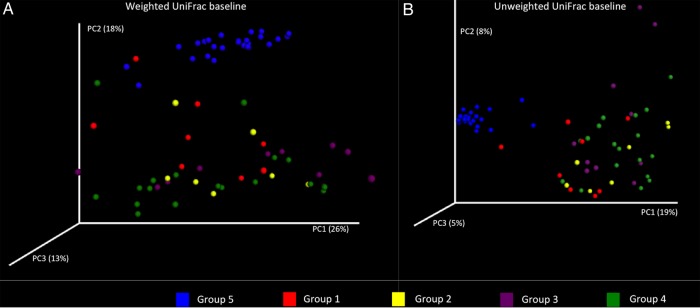
The analysis of the community structure showed that samples of hospitalized subjects (groups 1–4) clustered separately from those obtained from healthy individuals (group 5). According to the ANOSIM analysis of weighted and unweighted UniFrac distances, the differences between groups were statistically significant (R = 0.58 and P = .001 for weighted and R = 0.70 and P = .001 for unweighted UniFrac distances) (Figure 2). In addition, the intragroup distances among healthy controls were significantly lower than the distances within each group of hospitalized subjects, indicating that samples from healthy individuals were more similar to one another than were the samples from hospitalized patients (Supplementary Figure 1).
Figure 2.
Principal coordinates (PCs) analysis of beta diversity metrics between study groups at baseline, showing weighted (A) and unweighted (B) UniFrac distances. Samples from healthy controls (group 5; blue dots) clustered apart from those obtained from hospitalized individuals (groups 1–4).
Fecal Microbiome Composition
Differentially abundant features at any phylogenetic level were analyzed, first comparing hospitalized patients and healthy individuals and then comparing hospitalized patients exposed to antimicrobials who acquired MDROs (group 2) or those who did not (group 4). Among hospitalized patients, the most abundant phyla were Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. Compared with healthy controls, hospitalized subjects had higher relative abundance of Actinobacteria and lower relative abundance of Firmicutes and Clostridia. At the genus level, microbiota belonging to the genera Faecalibacterium, Bacteroides, Ruminococcus, Blautia, and Parabacteroides were more abundant among healthy controls. Conversely, well-recognized human pathogens, such as Escherichia spp. and Enterococcus spp., were more abundant among hospitalized subjects compared with healthy controls. The mean relative abundance of Escherichia spp. was 3.3% (SD, 12.7%) among hospitalized patients and <0.01% among healthy controls (P < .001). Similarly, for Enterococcus spp., the mean relative abundance was 4.5% (SD, 16.7%) among hospitalized patients and <0.01% among healthy controls (P < .001) (Supplementary Figure 2).
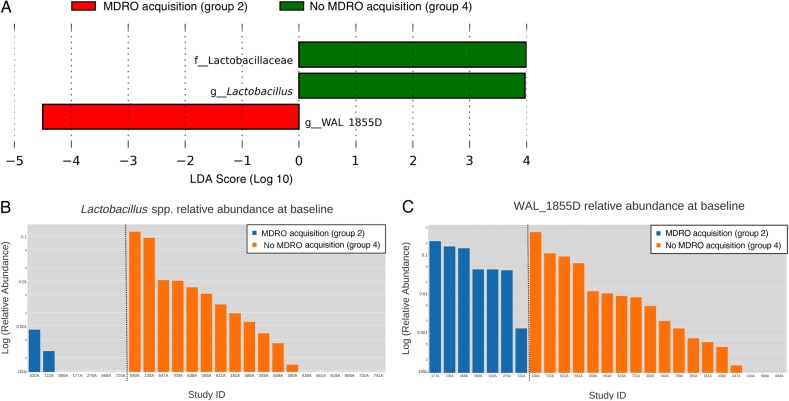
Analysis of features that were differentially abundant between patients who acquired MDROs and those who did not (groups 2 and 4, respectively) showed that patients who remained uncolonized with MDROs during their hospitalization had a statistically significant and biologically consistent higher Lactobacillus spp. abundance compared with the group that acquired MDROs. Because prior studies have shown that the relative abundance of Lactobacillus spp. in healthy human subjects is 0.01%, with a 10-fold increase if exposed to antimicrobials [22, 23], we used a relative abundance of 0.1% to categorize patients into high or low Lactobacillus abundance groups. According to this classification, the risk ratio for remaining uncolonized with MDROs among patients with high Lactobacillus abundance (≥0.1%) was 1.78 (95% confidence interval, 1.09–2.91; P = .02). Finally, a higher abundance of taxon WAL 1855_D was observed among patients who acquired MDROs (Figure 3).
Figure 3.
Features that were differentially abundant between patients who acquired multidrug-resistant organisms (MDROs) during their hospital stay and those who did not. A, Linear discriminant analysis (LDA) effect size output and LDA score. B, C, Bar charts representing the logarithm of the relative abundance of Lactobacillus spp. and WAL 1855D among patients who acquired MDROs (group 2) and those who did not (group 4). Abbreviations: f_, family; g_, genus.
DISCUSSION
In this study, we compared the fecal microbiome composition and structure between (1) hospitalized patients and healthy controls and (2) hospitalized patients who acquired MDROs and those who did not. When hospitalized patients were compared with healthy controls, relevant differences were identified. First, the fecal microbiome of hospitalized patients was significantly reduced in bacterial diversity, a finding possibly due to extensive prior antimicrobial exposure [6, 24]. The impact of reduced diversity in hospitalized patients has yet to be determined. Data from prior studies have shown that reduced diversity is associated with C. difficile infection, VRE bacteremia, and poor clinical outcomes [25–27]. Second, community composition differed remarkably between healthy controls and hospitalized patients. Although higher relative abundances of anaerobes, such as Blautia spp. [28], were more commonly found among healthy controls, the fecal microbiome of hospitalized patients had comparatively greater abundance of Escherichia spp. and Enterococcus spp. This finding probably explains the predominance of nosocomial infections caused by these organisms.
Comparison of the fecal microbiome of patients exposed to antimicrobials who acquired MDROs during hospitalization and those who did not also identified potentially important differences in microbiome abundances. In particular, high relative abundance of Lactobacillus spp. was a signature of the fecal microbiome of patients who remained uncolonized with MDROs. The difference in Lactobacillus spp. abundance was found in samples obtained during the first 48 hours of hospitalization, preceding detection of MDROs by several days. Important confounders, such as duration of follow-up, and antimicrobial exposure before MDRO acquisition were similar between those who acquired MDROs and those who did not. These findings suggest that Lactobacillus spp. may play a role in protecting the human host against MDRO acquisition. Numerous other commensal bacteria that confer protection against intestinal pathogens have been identified. Examples include Bacterioides thetaiotaomicron, which secretes a factor that represses toxin production by enterohemorrhagic E. coli [29], Barnesiella intestihominis, which reduces the risk of VRE colonization [9], and Clostridium scindens, which protects against C. difficile [30].
Studies have also identified beneficial effects specific to the presence of Lactobacillus spp. In 1 study of patients with C. difficile infection, coadministration of Lactobacillus spp. [31] with oral vancomycin prevented recurrences more frequently than treatment with oral vancomycin alone [32]. Another study showed that a high Lactobacillus spp. abundance was a microbial signature associated with colonization resistance against C. difficile infection [33]. Several controlled clinical trials have shown a benefit of using Lactobacillus spp. as a probiotic in the treatment of infectious diarrhea in children and in preventing antimicrobial-associated diarrhea [31]. In contrast, a recent study did not find a beneficial effect of Lactobacillus rhamnosus in preventing MDRO acquisition; however, this study was limited by a small sample size and the use of a single Lactobacillus species at a small dose [34]. The association between the presence of Lactobacillus spp. and lack of MDRO acquisition we observed does not prove a causal role. The properties of Lactobacillus spp., including growth inhibition of several pathogens, direct bactericidal effects, and competitive exclusion of pathogenic bacteria, provide biological plausibility for a causal role [35]. This should be further investigated in animal models and in larger human studies, including longer follow-up periods, assessment of resistance determinants, and characterization of the Lactobacillus species associated with a beneficial effect. The association between the uncommon bacterial taxon, WAL 1855D and MDRO acquisition will require further investigation.
Our study has several limitations. First, our small cohort is subject to unrecognized bias. Second, MDROs were detected using rectal swab samples, which have a low sensitivity if patients are not receiving antimicrobials [11, 36]. To avoid potential misclassification, only patients exposed to antimicrobials were considered for estimating associations between the fecal microbiome and MDRO acquisition. Third, as a cross-sectional analysis, our study did not account for the temporal variation of the human microbiome [22]. However, the goal of this study was to identify potential organisms associated with MDRO acquisition or its absence; therefore, only baseline swab samples, collected before the outcome of interest, were examined. Finally, in-hospital antimicrobial exposure was similar between patients who acquired MDROs and those who did not, with the exception of cefazolin exposure, which was more frequent in the group that acquired MDROs. Because patients received many other antimicrobials concurrently, the difference in cefazolin exposure is unlikely to explain differences in MDRO acquisition or microbiome composition.
Although current strategies to control the emergence of MDROs have efficacy, including infection control efforts and antimicrobial stewardship programs, the continuous emergence of new resistant determinants [37] and the increasing MDRO prevalence across the globe [38] highlight the need for new therapeutic alternatives. Preventing MDRO acquisition by manipulating the human microbiome is a promising approach [39, 40]. This study provides data to support a possible role of Lactobacillus spp. in the prevention of MDRO acquisition.
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. Anne Kane, MD, from the Phoenix Laboratory at the Tufts Medical Center, Boston, who performed the 16S ribosomal RNA gene library preparation and provided technical documentation.
Financial support. This work was supported in part by the National Institutes of Health (grant R01 DK090989); the Diane Belfer Program for Human Microbial Ecology (M. J. B.); Becas Chile, Comision Nacional de Investigacion Cientifica y Tecnologica, Gobierno de Chile (R. A.); and the National Institute of Allergy and Infectious Diseases (grant K24 AI119158 to E. M. C. D.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Laxminarayan R, Duse A, Wattal C et al. Antibiotic resistance—the need for global solutions. Lancet Infect Dis 2013; 13:1057–98. [DOI] [PubMed] [Google Scholar]
- 2.Howell WL; World Economic Forum Risk Response Network. Global risks 2013. Cologny, Switzerland: World Economic Forum, 2013. [Google Scholar]
- 3.Spellberg B, Bartlett J, Gilbert D. The future of antibiotics and resistance. N Engl J Med 2013; 368:299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tosh PK, McDonald LC. Infection control in the multidrug-resistant era: tending the human microbiome. Clin Infect Dis 2012; 54:707–13. [DOI] [PubMed] [Google Scholar]
- 5.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 2013; 13:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci 2011; 108:4554–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britton RA, Young VB. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology 2014; 146:1547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taur Y, Pamer EG. The intestinal microbiota and susceptibility to infection in immunocompromised patients. Curr Opin Infect Dis 2013; 26:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubeda C, Bucci V, Caballero S et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 2013; 81:965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder GM, D'Agata EMC. Diagnostic accuracy of surveillance cultures to detect gastrointestinal colonization with multidrug-resistant gram-negative bacteria. Am J Infect Control 2012; 40:474–6. [DOI] [PubMed] [Google Scholar]
- 12.Yatsunenko T, Rey FE, Manary MJ et al. Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing. Supplement M100-S21, 2011. [PubMed]
- 14.Caporaso JG, Lauber CL, Walters WA et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012; 6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460–1. [DOI] [PubMed] [Google Scholar]
- 17.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010; 26:266–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald D, Price MN, Goodrich J et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012; 6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005; 71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaScience 2013; 2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segata N, Izard J, Waldron L et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011; 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huttenhower C, Gevers D, Knight R et al. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Sullivan O, Coakley M, Lakshminarayanan B et al. Alterations in intestinal microbiota of elderly Irish subjects post-antibiotic therapy. J Antimicrob Chemother 2013; 68:214–21. [DOI] [PubMed] [Google Scholar]
- 24.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008; 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang JY, Antonopoulos DA, Kalra A et al. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 2008; 197:435–8. [DOI] [PubMed] [Google Scholar]
- 26.Taur Y, Xavier JB, Lipuma L et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis 2012; 55:905–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taur Y, Jenq RR, Perales M-A et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood 2014; 124:1174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenq RR, Taur Y, Devlin SM et al. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant 2015; 21:1373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Sablet T, Chassard C, Bernalier-Donadille A, Vareille M, Gobert AP, Martin C. Human microbiota-secreted factors inhibit Shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun 2009; 77:783–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buffie CG, Bucci V, Stein RR et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2014; 517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patel R, DuPont HL. New approaches for bacteriotherapy: prebiotics, new-generation probiotics, and synbiotics. Clin Infect Dis 2015; 60:S108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wullt M, Hagslätt M-LJ, Odenholt I. Lactobacillus plantarum 299v for the treatment of recurrent Clostridium difficile-associated diarrhoea: a double-blind, placebo-controlled trial. Scand J Infect Dis 2003; 35:365–7. [DOI] [PubMed] [Google Scholar]
- 33.Abujamel T, Cadnum JL, Jury LA et al. Defining the vulnerable period for re-establishment of Clostridium difficile colonization after treatment of C. difficile infection with oral vancomycin or metronidazole. PLoS One 2013; 8:e76269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon JH, Bommarito KM, Reske KA et al. Randomized controlled trial to determine the impact of probiotic administration on colonization with multidrug-resistant organisms in critically ill patients. Infect Control Hosp Epidemiol 2015; 36:1451–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lievin-Le Moal V, Servin AL. Anti-infective activities of Lactobacillus strains in the human intestinal microbiota: from probiotics to gastrointestinal anti-infectious biotherapeutic agents. Clin Microbiol Rev 2014; 27:167–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agata EMD, Gautam S, Green WK, Tang Y-W. High rate of false-negative results of the rectal swab culture method in detection of gastrointestinal colonization with vancomycin-resistant enterococci. Clin Infect Dis 2002; 34:167–72. [DOI] [PubMed] [Google Scholar]
- 37.Rossi F, Diaz L, Wollam A et al. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med 2014; 370:1524–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Price LS, Poirel L, Bonomo RA et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 2013; 13:785–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halpin AL, McDonald LC. The dawning of microbiome remediation for addressing antibiotic resistance. Clin Infect Dis 2016; 62:1487–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crum-Cianflone NF, Sullivan E, Ballon-Landa G. Fecal microbiota transplantation and successful resolution of multidrug-resistant-organism colonization. J Clin Microbiol 2015; 53:1986–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.