Summary
The acclimation of plants to changes in light intensity requires rapid responses at several different levels. These include biochemical and biophysical responses as well as alterations in the steady‐state level of different transcripts and proteins. Recent studies utilizing promoter::reporter constructs suggested that transcriptional responses to changes in light intensity could occur within seconds, rates for which changes in mRNA expression are not routinely measured or functionally studied. To identify and characterize rapid changes in the steady‐state level of different transcripts in response to light stress we performed RNA sequencing analysis of Arabidopsis thaliana plants subjected to light stress. Here we report that mRNA accumulation of 731 transcripts occurs as early as 20–60 sec following light stress application, and that at least five of these early response transcripts play an important biological role in the acclimation of plants to light stress. More than 20% of transcripts accumulating in plants within 20–60 sec of initiation of light stress are H2O2‐ and ABA‐response transcripts, and the accumulation of several of these transcripts is inhibited by transcriptional inhibitors. In accordance with the association of rapid response transcripts with H2O2 and ABA signaling, a mutant impaired in ABA sensing (abi‐1) was found to be more tolerant to light stress, and the response of several of the rapid response transcripts was altered in mutants impaired in reactive oxygen metabolism. Our findings reveal that transcriptome reprogramming in plants could occur within seconds of initiation of abiotic stress and that this response could invoke known as well as unknown proteins and pathways.
Keywords: Arabidopsis thaliana, transcription, RNA‐Seq, light stress, ultra‐fast
Significance Statement
Transcriptome reprogramming in plants could occur within seconds of abiotic stress initiation and could invoke known as well as unknown proteins and pathways. Here we use light stress as a case study to show that ultra‐fast transcriptional responses can reveal important transcripts for abiotic stress acclimation and/or for pathogen resistance.
Introduction
Playing a principal role in sustaining life on Earth, plants convert solar radiation into bio‐available energy. Unable to avoid abiotic stress by means of relocation, plants have evolved sophisticated acclimation mechanisms to cope with changes in their environment. These include sensing, signal transduction and stress protection proteins and pathways (Nakashima and Yamaguchi‐Shinozaki, 2006; Bailey‐Serres and Voesenek, 2008; Cavanagh et al., 2008; Munns and Tester, 2008; Chinnusamy and Zhu, 2009; Mittler and Blumwald, 2010). Although changes in environmental conditions could occur within seconds in nature, studies attempting to dissect the responses of plants to abiotic stress have traditionally focused on events occurring 10–30 min or hours after application of abiotic stress (Mittler et al., 2012). Recent studies have nonetheless indicated that the response of plants to abiotic stress could occur much faster than previously measured, and that changes in environmental conditions such as temperature or light intensity could cause immediate alterations in the level or structure of different proteins, metabolites and RNA molecules, as well as changes in the redox status of different molecules (Mittler, 2002; Miller et al., 2009; Mittler et al., 2012; Suzuki et al., 2013a; Gilroy et al., 2014; Dietz, 2015). Even a gradual change in environmental conditions could trigger a rapid response once a particular physiological or biochemical sensing threshold is passed (Mittler et al., 2012). Alterations in metabolites and RNA species could result from stress‐induced differential enzymatic co‐efficiencies, RNA transcription and processing, or metabolite and RNA stability. These could in turn reprogram the cell metabolome and transcriptome and trigger specific sensors for abiotic stress response that would in turn activate multiple signal transduction pathways and result in the mounting of a full‐scale acclimation response (Mittler et al., 2012). Although much is known about the different signaling and downstream pathways that mediate the acclimation of plants to stress, virtually nothing is known about the rapid changes in the metabolome and transcriptome of plants that occur within seconds to minutes of initiation of abiotic stress (Mittler et al., 2012; Suzuki et al., 2013a,b; Gilroy et al., 2014; Miller et al., 2009).
We recently reported on the detection of ultra‐fast changes in the metabolome of plants subjected to high‐light stress, with changes in many metabolites occurring as early as 15 sec after the application of light stress (Suzuki et al., 2013a). Using a Zat12::luciferase reporter system, we also reported on the existence in plants of a rapid local and systemic signal termed the reactive oxygen species (ROS) wave that is activated by different abiotic stresses and propagates at rates of up to 8.4 cm min−1 (Miller et al., 2009; Mittler et al., 2011). Those reports, as well as a recent report on the existence of a rapid local and systemic ‘calcium wave’ (Choi et al., 2014), and the possible integration of the two (Gilroy et al., 2014), suggest that transcription, RNA stability and/or RNA processing responses in plants could occur at a much faster rate than is typically studied. Moreover, if such rapid responses occur, and have an important biological function, then the lack of data for early time points in many of the abiotic stress‐response transcriptome studies deposited in different gene and data banks (e.g. Hruz et al., 2008; https://genevestigator.com/gv/) could hamper our attempts to develop crops with enhanced tolerance to abiotic stresses because many of the important early response genes will be missed. In support of the possibility that the steady‐state transcript level of many genes could be enhanced within seconds of initiation of abiotic stress are measurements of transcription rates in eukaryotic cells nearing or exceeding 50 kb min−1 (Maiuri et al., 2011), and the discovery that many genes in eukaryotic cells contain stalled RNA polymerases at their promoters and could mount a rapid transcriptional response following changes in environmental conditions (Nechaev and Adelman, 2008; Levine, 2011; Kwak and Lis, 2013).
Here we uncover the ultra‐fast transcriptome response of plants triggered by light stress. This response includes the ordered and clustered mRNA accumulation of 731 transcripts that occurs as early as 20–60 sec after application of light stress. We further determined that five of the transcripts involved in this ultra‐fast response play an important biological role in the acclimation of plants to light stress, that many of the ultra‐fast light stress‐response transcripts are H2O2‐ or ABA‐response transcripts, and that the accumulation of several of the ultra‐fast response transcripts is inhibited by transcriptional inhibitors. We also report that a mutant impaired in ABA sensing (abi‐1) is more tolerant to light stress, and that the response of several of the rapid response transcripts was altered in mutants impaired in ROS metabolism/signaling. Our findings reveal that transcriptome reprogramming in plants occurs at a much faster rate than is typically studied and that this response could involve known as well as unknown transcripts and pathways. Because several of the genes identified as ultra‐fast light response genes appear to be important for acclimation to light stress, our studies highlight the need to study the ultra‐fast transcriptional response of plants to other abiotic or biotic stresses that may include important, but as‐yet unidentified, genes for acclimation to abiotic stress and/or plant resistance to pathogens and pests.
Results
RNA sequencing (RNA‐Seq) analysis of the ultra‐fast response of Arabidopsis to light stress
Arabidopsis thaliana plants subjected to light stress for 0, 20, 30, 60 and 90 sec were used for transcriptome (RNA‐Seq; 0, 20, 60 sec) and quantitative (q)PCR analyses and H2O2 measurements (0, 20, 30, 60 or 90 sec) in three biological replicates (Figure 1 and Figure S1a in Supporting Information). Each biological replicate contained three technical replicates of 15–20 plants each, grown in pots for 18–21 days, exposed to a light intensity of 1000 μmol m−2 sec−1 at 21°C, and immediately dipped in liquid nitrogen. The steady‐state level of 731 transcripts was found to be significantly enhanced in response to light stress within 20–60 sec. These were divided into three clusters based on their expression pattern (Figure 1a, Tables S1–S3), indicating a complex response pattern that involved differential timing. Out of the 731 transcripts shown in Figure 1, 49 were found not to be annotated on the ATH1 Affymetrix chips (Table S4). Interestingly, only 34 transcripts out of the 731 shown in Figure 1(a) increased in expression from a very low basal level [below 2.0 fragments per kilobase of transcript per million fragments mapped (FPKM); Table S5]. The steady‐state level of 419 and 668 transcripts significantly declined at 20 and 60 sec of light stress, respectively (Tables S6 and S7). As shown in Figure 1(b), qPCR analysis confirmed the expression of selected transcripts from Cluster I. It should be noted that few transcripts included in Cluster I in some experiments could be found in Clusters II or III in other experiments. These differences, detected by qPCR, could result from small variations in the physiological pre‐conditioning of plants used for the different experiments or from the stochastic nature of the transcriptional response in different cells belonging to the same tissue (Stegle et al., 2015). Interestingly, enhanced cellular levels of H2O2 were not detected during early stages of light stress (Figure 1b). This finding could be linked to the high content of antioxidants in plants, generating a buffer against rapid changes in ROS (Mittler, 2002; Halliwell, 2006). In accordance with this hypothesis, the level of ascorbic acid rapidly decreased in response to light stress in Arabidopsis in a process that was dependent on the presence of ascorbate peroxidase 1 (APX1; Davletova et al., 2005a), an ascorbate‐dependent H2O2‐scavenging enzyme (Figure 1c). This finding demonstrated that APX1 could be directly involved in scavenging of H2O2 produced during the initial response to light stress, and that during this process the stored levels of ascorbic acid are utilized as part of the Asada–Foyer–Halliwell pathway (Figure 1c; Halliwell, 2006).
Figure 1.

Ultra‐fast alterations in transcript steady‐state level in response to light stress in Arabidopsis thaliana detected by RNA sequencing.
(a) Three different clusters of transcript alterations distinguished by their pattern of response to light stress.
(b) Accumulation of H2O2 and selected transcripts determined by quantitative PCR during the early stages of light stress acclimation.
(c) Rapid changes in the level of ascorbic acid in response to light stress in wild‐type (WT) plants and knockout mutants lacking the ascorbate‐dependent H2O2‐scavenging enzyme ascorbate peroxidase 1 (apx1).
The majority of transcripts belonging to all ultra‐fast response clusters were annotated as stress, abiotic or biotic response transcripts based on their Gene Ontology (GO) annotation (Figure 2a), and genes encoding the bulk of these transcripts were scattered on all Arabidopsis chromosomes, with only a few clustering at certain locations (Figure S1b). A breakdown of the overlap between responses to different biotic/abiotic stresses and the different transcripts found in each of the ultra‐fast response cluster indicated that many of the transcripts with an enhanced steady‐state level in response to light stress are also responsive to abiotic stresses such as drought, cold, heat and salinity, demonstrating their possible involvement in tolerance of abiotic stress in plants (Figure 2b; ATH1 chip data were obtained from the supplementary material of Huang et al., 2008; Matsui et al., 2008; Larkindale and Vierling, 2008; Kleine et al., 2007; Ding et al., 2014; Tosti et al., 2006; Consales et al., 2012; Truman et al., 2006). A comparison, shown in a Venn diagram in Figure 2(c), between the transcriptomes of the ultra‐fast response to light stress (this work), the response of Arabidopsis to H2O2 (Davletova et al., 2005b; data obtained from the supplementary material therein) and the response of Arabidopsis to a 3‐h light treatment (Kleine et al., 2007; data obtained from the supplementary material therein), revealed that 502 transcripts found to be induced between 20 and 60 sec after light stress application were not found to be induced by the longer light stress or H2O2 treatments. Out of these 502 transcripts, 245 transcripts (Table S8) were also not found to be included in the response of plants to the stresses shown in Figure 2(b). Of course, 49 of these were not represented in the ATH1 chips (Table S4). These transcripts could therefore represent a unique group of transcripts that are relatively more specific to the ultra‐fast response (Table S8).
Figure 2.
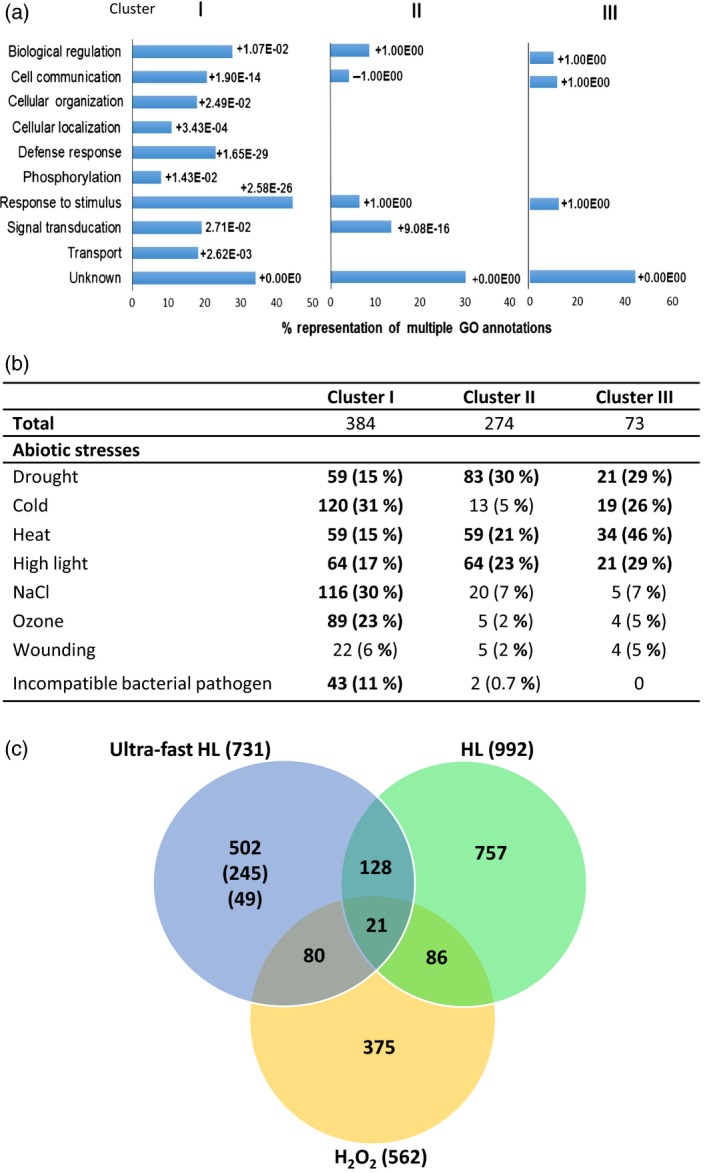
Meta‐analysis of ultra‐fast light stress‐response transcripts in Arabidopsis.
(a) Gene Ontology annotation of the different transcripts found in the three ultra‐fast response clusters shown in Figure 1(a).
(b) Distribution of abiotic and biotic stress‐response transcripts between the different ultra‐fast response clusters. Transcript representation higher than 10% is highlighted in bold.
(c) Venn diagram showing the overlap between ultra‐fast response transcripts to light stress, transcripts accumulating in Arabidopsis following a 3‐h light stress treatment and transcripts accumulating in Arabidopsis following a 1‐h treatment with H2O2. Out of the 502 transcripts shown to be unique to ultra‐fast high light (HL), 245 do not overlap with any of the abiotic stresses shown in (b). Out of those 245 transcripts, 49 do not appear on the ATH1 Affymetrix chips.
Functional characterization of ultra‐fast response transcripts in Arabidopsis
To determine whether some of the ultra‐fast response transcripts play a biological role in the acclimation of plants to light stress, we obtained from the SALK collection knockout mutants for 70 genes encoding transcripts with altered expression at 20 sec of light stress (O'Malley and Ecker, 2010) and screened them for tolerance to light stress. Seven knockouts showed enhanced cell death in response to light stress and a second independent knockout was obtained for these and screened again. As shown in Figure 3(a), five genes were found to be important for acclimation to light stress using two independent knockouts. They encoded two proteins of unknown function (At5g10695 and At3g10020), a Golgi‐associated protein (At5g51430), a RAV transcription factor (At1g25560) and a glycosyl hydrolase (At3g13750), and were required to prevent light‐induced cell death in Arabidopsis leaves. It should also be mentioned that a sixth protein identified by our analysis, Zat12 (Figure 1b), was previously shown to be required for acclimation to light stress (Iida et al., 2000). Interestingly, as shown in Figure 3(b), none of the five transcripts functionally characterized in Figure 3(a) was induced in response to the 3‐h light stress treatment (Kleine et al., 2007). Also, none of the five transcripts shown in Figure 3(a) was found to be enhanced in a time‐course experiment subjecting Arabidopsis to light stress as reported in Davletova et al. (2005a; Figures S2 and S3). The five ultra‐fast transcripts assayed in Figure 3(a) were therefore primarily induced during early stages of the response of Arabidopsis to light stress (with two of them also induced by H2O2; Figure 3b). Our finding that these transcripts were primarily induced during the rapid response of plants to light stress, but not during later stages of this response (Figures 3b, S2 and S3; Davletova et al., 2005a; Kleine et al., 2007), highlight their potential biological role in protecting plants from light stress (Figure 3a), thus demonstrating that the rapid response is important for the tolerance of plants to light stress.
Figure 3.
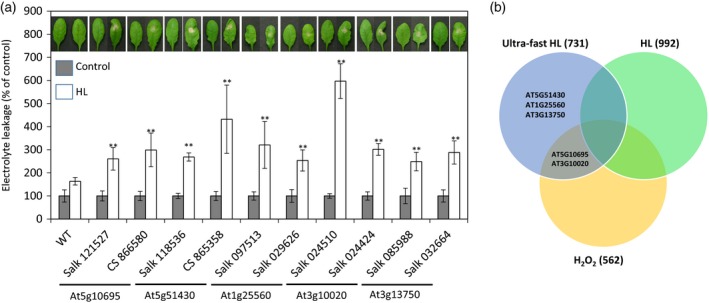
Functional analysis of selected ultra‐fast response transcripts in Arabidopsis.
(a) Light stress‐induced cell death in knockout mutants for five different genes encoding ultra‐fast response transcripts. Two independent alleles for each gene were subjected to light stress and cell death was photographed and measured by electrolyte leakage. **P < 0.01. HL, high light.
(b) Venn diagram showing the overlap between the five transcripts tested in (a), the ultra‐fast response to light stress, the response of Arabidopsis to a 3‐h light stress treatment and the response of Arabidopsis to a 1‐h treatment with H2O2.
Transcriptional regulation of ultra‐fast response transcripts
To determine if some of the ultra‐fast transcripts identified by our analysis (Figure 1) were regulated, at least partially, at the transcriptional level, we used qPCR to test whether transcriptional inhibitors such as α‐amanitin and actinomycin D would suppress their accumulation. We selected four different transcripts with a defined response pattern confirmed by qPCR for this analysis (Figure 4). As can be seen in Figure 4(a), changes in transcript accumulation, visualized by RNA‐Seq read alignment maps, could be seen for the four selected transcripts across the entire length of their corresponding genes with some genes changing in expression two‐ to three‐fold within 20 sec of light stress. Pre‐treatment with α‐amanitin or actinomycin D suppressed the accumulation of all four selected transcripts (Figure 4b), demonstrating that at least some of the transcripts accumulating at 20 sec of light stress could require active transcription for their regulation.
Figure 4.
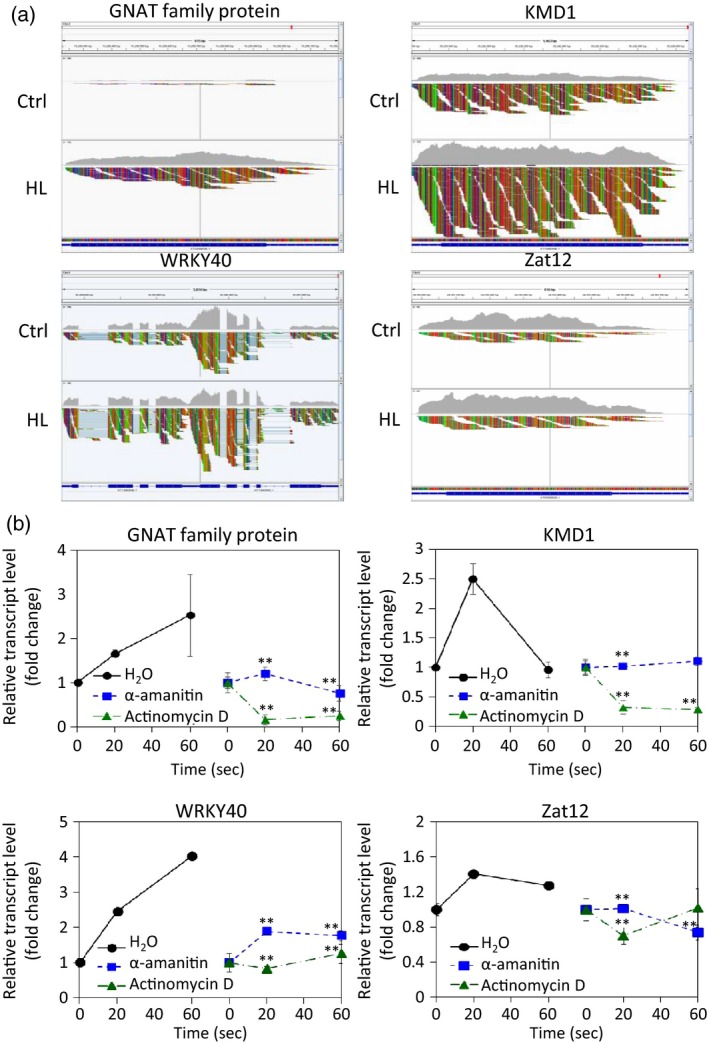
Suppression of light stress‐induced ultra‐fast accumulation of transcripts by transcriptional inhibitors.
(a) Standardized RNA sequencing read maps for four selected genes encoding ultra‐fast response transcripts at 20 sec of light stress exposure. HL, high light.
(b) Transcript accumulation for the four genes shown in (a) measured by quantitative PCR in plants treated or untreated with two different transcriptional inhibitors (α‐amanitin or actinomycin D) prior to light stress treatment. **P < 0.01.
To further determine if the steady‐state transcript level of some of the ultra‐fast response transcripts is enhanced transcriptionally within seconds after light stress, we constructed different reporter genes in which the WRKY40 and the Zat12 promoters were fused to an unstable variant of the luciferase gene (Lucu). As can be seen in Figure 5(a), subjecting three independent homozygous lines for each of these reporter genes to light stress for 20 sec resulted in enhanced luciferase activity that occurred within 40–60 sec in the Zat12::Lucu and 110–140 sec in the WRKY40::Lucu constructs. In addition, we used qPCR to measure the accumulation of the luciferase transcripts in Zat12::luc (Miller et al., 2009; regular stable luciferase fused to the same Zat12 promoter fragment) plants 20 and 60 sec following application of light stress. As shown in Figure 5(b), luciferase transcripts driven by the Zat12 promoter could be detected as early as 20 sec following light stress. The findings shown in Figures 4(b) and 5 suggest that at least some of the transcripts with enhanced steady‐state levels at 20 and 60 sec following application of light stress in Arabidopsis are driven by the activation of their promoters.
Figure 5.

Analysis of the ultra‐fast response to light stress using promoter::reporter constructs.
(a) Luciferase activity measurements of the ultra‐fast light stress response using an unstable luciferase gene fused to the promoters of the Zat12 (measured from 0 to 100 sec) or WRKY40 (measured from 0 to 200 sec) genes. Three independent transgenic lines per construct are shown.
(b) Accumulation of the luciferase transcript, determined by quantitative PCR, during the early stages of light stress acclimation in Zat12::luc plants. **P < 0.01.
Because the steady‐state level of many of the transcripts shown in Figure 1 could also be enhanced in response to light stress due to changes in their RNA stability (i.e. by increased stability during the early stages of light stress), we determined the content of 11 different RNA‐destabilizing sequences (Ohme‐Takagi et al., 1993; Narsai et al., 2007) in these transcripts and compared it with the content of the same RNA‐destabilizing sequences in transcripts induced by light stress at 3 h (Kleine et al., 2007). As shown in Figure S4, the content of the destabilizing sequences was very similar in these two transcript groups. However, further studies are needed to determine the role of RNA stability in the ultra‐fast response of Arabidopsis to light stress.
Involvement of ABA and ROS in the ultra‐fast response of Arabidopsis to light stress
Meta‐analysis of the transcriptomic response at 20 and 60 sec of light stress revealed that 12–22% of the transcripts with an enhanced steady‐state level in all clusters are ABA‐response transcripts (Figure 6a; ATH1 chip data was obtained from the supplementary material of Nemhauser et al., 2006; Blanco et al., 2009; Davletova et al., 2005b; Scarpeci et al., 2008; Gadjev et al., 2006). Twelve to 21% of transcripts from Cluster I were also brassinolide‐, jasmonate‐ and H2O2‐response transcripts. These findings suggest that Cluster I is distinct from Clusters II and III in its content of hormone‐response transcripts (Figure 6a). To further test the dependence of the ultra‐fast response on ABA signaling we studied the response of selected mutants impaired in ABA and retrograde signaling to light stress using the assay described in Figure 3(a). For this analysis we used aba‐1, deficient in ABA biosynthesis, abi‐1, deficient in ABA sensing via protein phosphatase 2C that is involved in NADPH oxidase activation in response to ABA, and abi‐4 and gun‐1, deficient in retrograde signaling from the chloroplast to the nuclei (Koussevitzky et al., 2007; Suzuki et al., 2013a). As shown in Figure 6(b), of the different mutants tested only the abi‐1 mutant displayed enhanced tolerance to light stress, potentially due to an enhanced content of stress‐response transcripts or deficiency in ABA‐induced NADPH oxidase activation. Because protein phosphatase 2C (ABI‐1) activity could be directly suppressed by enhanced H2O2 levels, or by enhanced ABA levels (through PYR/PYL; Mittler and Blumwald, 2015), the finding that the abi‐1 mutant is impaired in the response to light stress could suggest that this protein is involved, via H2O2 and/or ABA, in early light stress responses in Arabidopsis. Interestingly, as shown in Figure 6(c), a considerable overlap was found between the transcripts upregulated in untreated abi‐1 plants (Hoth et al., 2002; data obtained from the supplementary material therein) and the transcripts enhanced in plants subjected to 3‐h light stress (Kleine et al., 2007) or the ultra‐fast high light (HL) treatment described in this work. This finding could suggest that in Arabidopsis deficiency in ABI‐1 mimics some aspects of the light stress response and that the abi‐1 mutant is more tolerant to light stress (Figure 6b) because it already activates some of these responses.
Figure 6.
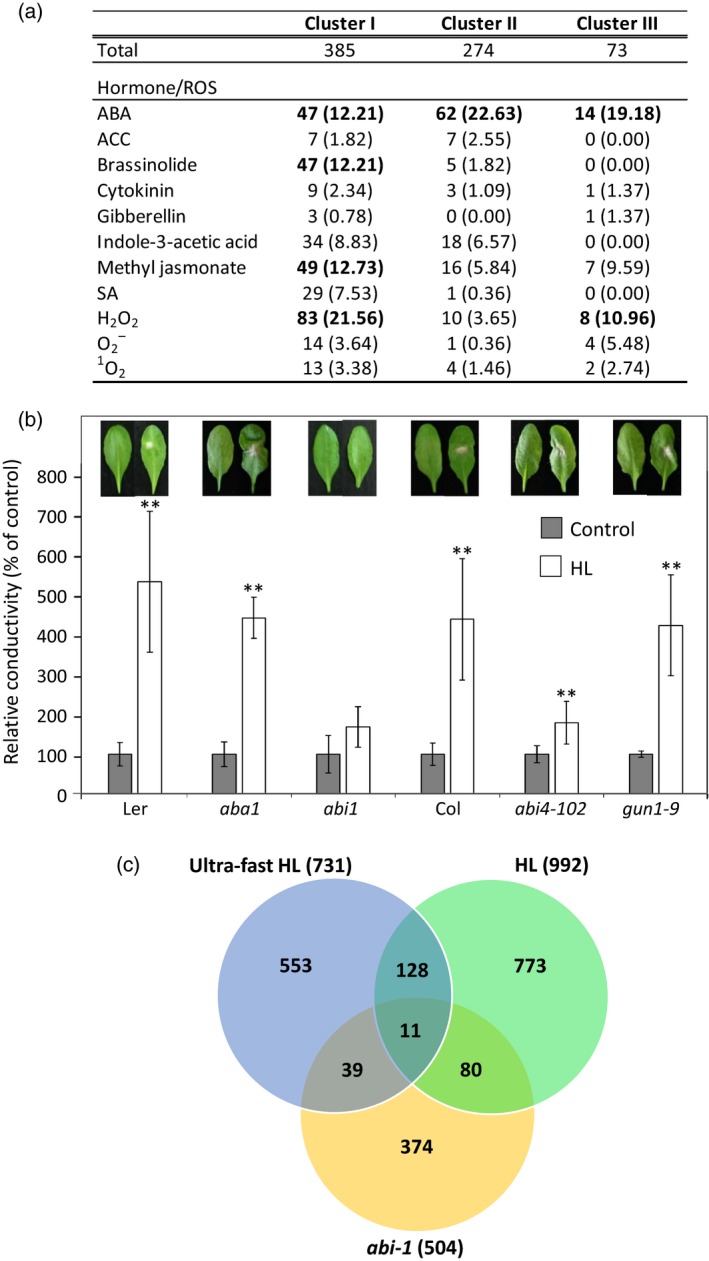
Distribution of hormone‐ and reactive oxygen species (ROS)‐response transcripts between the different ultra‐fast response clusters and tolerance of different mutants impaired in ABA and retrograde signaling to light stress.
(a) Meta‐analysis of the different ultra‐fast clusters showing that Cluster I is distinct from Clusters II and III in its content of hormone‐response transcripts. Transcript representation higher than 10% is highlighted in bold. ACC, ethylene; SA, salicylic acid.
(b) Light stress‐induced cell death in different mutants impaired in ABA and retrograde signaling. **P < 0.01. HL, high light.
(c) Venn diagram showing the overlap between transcripts accumulating in the abi‐1 mutant in the absence of stress, ultra‐fast response transcripts to light stress and transcripts accumulating in Arabidopsis following a 3‐h treatment of light stress.
To further test the dependence of some ultra‐fast response transcripts on H2O2/ROS we analyzed the expression of selected ultra‐fast response transcripts in mutants impaired in ROS scavenging/signaling such as apx1, cat2 and rbohD. apx1 and cat2 are impaired in H2O2 scavenging whereas rbohD is impaired ROS signaling (Torres et al., 2002; Vanderauwera et al., 2011). As can be seen in Figure 7, some of the transcripts tested were found to be dependent on ROS scavenging (WRKY40), some were independent of ROS signaling/scavenging (WRKY18 and KMD1) and some were only partially dependent on ROS scavenging (Zat12). Interestingly, the response of all transcripts tested was unaltered in the rbohD mutant, suggesting that RBOHD may not be involved in the regulation of these transcripts and that the possible function of ABI1 in this response could be different from its function in classical ABA sensing. Further studies are of course required to unravel the role of ROS and ABA in the ultra‐fast response of plants to light stress.
Figure 7.

Expression of four selected ultra‐fast transcripts in two genetic backgrounds impaired in reactive oxygen species (ROS) scavenging (apx1 and cat2) and in a genetic background impaired in ROS signaling (rbohD).
(a) WRKY40.
(b) WRKY18.
(c) Zat12.
(d) KMD1.
*P < 0.05; **P < 0.01.
Discussion
Several lines of evidence suggest that the ultra‐fast transcriptome response reported here is biologically‐important. It was composed of 731 transcripts that demonstrated an ordered and clustered response (Figure 1, Tables S1–S3 and S5–S7); it included several ultra‐fast response‐specific transcripts that were required for acclimation to light stress (Figure 3; Iida et al., 2000); the different clusters included in the response were composed of transcripts with differential responsiveness to different abiotic/biotic stresses, ROS or hormones (Figures 2 and 6); expression of some of the transcripts involved in the response could be suppressed by transcriptional inhibitors (Figure 4); and it included transcripts that responded differentially when studied in a genetic background that was altered in ROS scavenging responses (Figure 7). Taken together, these observations indicate that very rapid responses at the mRNA level in plants could play an important biological role in the acclimation of plants to stress.
It is possible that many of the ultra‐fast changes observed in the steady‐state transcript level in response to light stress in our study (Figure 1a, b, Tables S1–S3 and S5–S7) were caused by post‐transcriptional alterations in RNA stability. Furthermore, many of the ultra‐fast response transcripts contained RNA‐destabilizing sequences, albeit not at a proportion higher than late (3 h) light stress‐response transcripts (Figure S4). Nevertheless, the findings that transcriptional inhibitors suppressed the expression of some ultra‐fast response transcripts (Figure 4), and that the ultra‐fast response could be observed via promoter::reporter constructs (Figure 5), combined with several reports of stalled RNA polymerases found on the promoters of eukaryotic genes (Nechaev and Adelman, 2008; Levine, 2011; Kwak and Lis, 2013), strongly suggest that some of the ultra‐fast response transcripts reported here are regulated at the transcriptional level and could contain stalled but active RNA polymerases at their promoters. This possibility, combined with reported transcription rates of over 50 kb min−1 in eukaryotic cells (Maiuri et al., 2011), could explain the rapid accumulation of some of the transcripts observed in our experiments (Figures 1, 2, 3, 4, Tables S1–S3), especially considering the relatively short length of Arabidopsis genes (average of 2000 bp; Arabidopsis Genome Initiative, 2000). Differential RNA stability and stalled RNA polymerases could therefore provide a mechanistic explanation for the ultra‐fast transcriptome reprograming observed in response to light stress in Arabidopsis.
In contrast to the rapid accumulation of different transcripts, the ultra‐fast transcriptome response of Arabidopsis to light stress included a rapid decline in the level of many transcripts (Figure S1a, Tables S6 and S7). Genome‐wide analyses of decline of mRNA in Arabidopsis have revealed three different groups of transcripts that are prone to rapid mRNA decline: transcripts encoded by intronless genes, transcripts possessing destabilizing sequences in their 3′ end and transcripts that are targets of microRNA regulation (Narsai et al., 2007). In addition, uncapping‐mediated mRNA degradation was found to be associated with abiotic stress responses (Zhang et al., 2013). Nevertheless, the majority of studies focusing on mRNA decline in plants did not use short time points such as 20 and 60 sec and further analysis is needed to decipher the mechanisms that regulate mRNA decline under the conditions described in this study.
Taking into account the ‘harsh’ biological assay we used to define a function in light stress acclimation (i.e. cell death; Figure 3), it is likely that many other transcripts identified by our transcriptome analysis could play a role in light stress acclimation, albeit less significant than prevention of cell death. The relationship between ROS and the expression of many of the genes identified by our analysis is unclear at present. High levels of ROS did not accumulate in cells during the first 0–90 sec of light stress (Figure 1b), but the expression of some ultra‐fast response genes was dependent on ROS signaling (Figure 7). This finding suggests that the high antioxidant capacity of plant cells is able to handle the initial rise in ROS produced during early stages of light stress (Figure 1c), but that several genes that are highly sensitive to ROS levels still react (Figures 1b and 7). In addition to these, proteins inhibited by H2O2, such as ABI‐1, could also be affected by low levels of ROS (Figure 6b, c). The rapid APX1‐dependent depletion of ascorbic acid levels in response to light stress (Figure 1c) supports the hypothesis that plants have a high buffering capacity for rapid changes in ROS levels. Key players in the regulation of rapid transcriptional responses could therefore be localized and/or low‐level ROS signals, calcium signatures or ABI‐1 and/or conjugated ABA (Miller et al., 2009; Mittler et al., 2011, 2012; Suzuki et al., 2013a; Choi et al., 2014; Gilroy et al., 2014; Mittler and Blumwald, 2015).
Abscisic acid was shown to play a key role in the response of plants to abiotic stresses, and ABA and ROS were shown to coordinate responses to the ROS wave in systemic tissues (Suzuki et al., 2013a; Mittler and Blumwald, 2015). Our findings that many of the ultra‐fast response transcripts are also ABA‐response transcripts (Figure 6a) and that the abi‐1 mutant has an impaired response to light stress (Figure 6b) is therefore in line with the potential role of ABA in this response. Because, in addition to its function in the ABA pathway, the ABI‐1 protein could also be directly inhibited by H2O2 (Mittler and Blumwald, 2015), an alternative explanation for our findings could also be that during the ultra‐fast response to light stress the little H2O2 that is accumulated is directly inhibiting this protein. In this case the deficiency in ABI‐1 protein in the abi‐1 mutant will mimic the response of these plants to light stress and pre‐condition them to resist light (Figure 6b, c). Further studies are required to address these possibilities.
Virtually all time‐course omics studies of plant or animal responses to abiotic stress lack early time points (seconds to minutes; e.g. Hruz et al., 2008). Our findings demonstrate that during these early stages of the stress response the cell undergoes a reprogramming of its transcriptome that could affect the activation of signal transduction pathways and the establishment of successful acclimation. The large number of transcripts that respond to light stress within 20–60 sec in Arabidopsis, uncovered by our analysis (Figure 1, Tables S1–S3 and S5–S7), combined with the important role that some of these early response transcripts play in light stress tolerance (Figure 3; Iida et al., 2000), highlight the importance of these early stages for plant acclimation. Further characterization of these transcripts, as well as additional studies of the ultra‐fast response of plants to other abiotic and biotic stresses, could lead to the development of new and novel approaches to enhance the tolerance of plants and crops to different environmental stresses using pathways and compounds that were not previously known, or considered to be involved in abiotic stress.
Experimental procedures
Plant material and growth conditions
Arabidopsis thaliana Col‐0 (cv. Columbia‐0), WS‐0 (cv. Wassilewskija‐0), Ler‐0 (cv. Landsberg erecta), rbohD, apx1, aba1, abi1, antisense Cat2 (Torres et al., 2002; Davletova et al., 2005a; Koussevitzky et al., 2007; Vanderauwera et al., 2011; Suzuki et al., 2013a,b) and confirmed knockout lines for 70 genes encoding high‐light‐response transcripts (O'Malley and Ecker, 2010; Table S1) were grown in peat pellets (Jiffy‐7, Jiffy, http://www.jiffygroup.com/en/) or soil mixture (MetroMix 200, SUN GRO, http://www.sungro.com/) in 9 x 9 x 6 cm3 pots covered with a fiberglass screen net at 23°C under constant low light (50 μmol m−2 sec−1). Knockout lines were obtained from ABRC (http://abrc.osu.edu/) and bulked together with wild‐type seeds under carefully controlled growth conditions as previously described (Suzuki et al., 2011; Luhua et al., 2013). Zat12::luc plants were obtained as described in Miller et al. (2009). A similar strategy was also used to fuse the promoters of Zat12 (Miller et al., 2009) and WRKY40 (1.2 kb), to the unstable form of Luciferase [Promega, http://www.promega.com/; pGL4.11(luc2p), designated here as LucU] (Davletova et al., 2005b; Miller et al., 2009). Luciferase activity was imaged as described by Miller et al. (2009). One leaf of 16‐ to 20‐day‐old plants was sprayed with 1 mm luciferin (GOLD Biotechnology, https://www.goldbio.com/), exposed to light stress (1500 μmol m−2 sec−1) using a goose neck bulb, and imaged using a NightOWL LB983 NC100 (Berthold, https://www.berthold.com/) imager. Images were captured every 10 sec for 300 sec. Bioluminescence in photon counts sec−1 was measured using indigo v.2.0.3.0 (Berthold). Treated samples were compared to their respective controls and graphed.
Light stress treatment
For RNA‐Seq, qPCR analyses and measurements of H2O2 and ascorbic acid, 15–20 plants grown in pots for 18–21 days as described above were exposed to a light intensity of 1000 μmol m−2 sec−1 at 21°C for periods of 0, 20 and 60 sec, 0, 10, 60 and 300 sec, or 0, 20, 30, 60 and 90 sec in a growth chamber (E‐30‐HB, Percival Scientific, http://www.percival-scientific.com/). Samples were collected by immediately dipping the pots in liquid nitrogen. Frozen tissues were then cut onto aluminum foil, ground and transferred into 1.5‐ml tubes (about 100–150 mg per tube). Samples were kept frozen during the entire collecting process and stored at −80°C. For treatment with transcriptional inhibitors, plants grown in pots were sprayed with water, 10 μm α‐amanitin or 75 μg ml−1 actinomycin D and incubated for 90 min at room temperature (21°C) prior to exposure to light stress for periods of 0, 20 and 60 sec. To test the tolerance of plants to light stress, one fully expanded rosette leaf of 21‐ to 25‐day‐old plants grown on peat pellets was exposed to 2000 μmol m−2 sec−1 high light for 1 h using a gooseneck light source (ACE I; Schott, http://www.amscope.com/). Leaves were then photographed and analyzed for electrolyte leakage as described below.
RNA‐Seq, qRT‐PCR, meta‐analyses and ascorbic acid measurements
For RNA‐Seq analysis, three independent biological replicates, each composed of leaves pooled from at least 30 different plants in three technical repeats, were used for each experimental condition. Total RNA was isolated and purified as described in Suzuki et al. (2013a) and RNA‐Seq analysis was conducted using an Illumina HiSeq2000 at the University of Wisconsin‐Madison Biotechnology Gene Expression Center. Gene Ontology annotations of the transcripts identified by our RNA‐seq analyses were performed using PANTHER (http://www.pantherdb.org/pathway/) or obtained from TAIR (https://www.arabidopsis.org/tools/bulk/go/index.jsp). Quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed as previously described (Miller et al., 2009; Suzuki et al., 2013a) using a StepOnePlusTM Real‐Time PCR System (Applied Biosystems, http://www.appliedbiosystems.com/). The qPCR data were analyzed with steponeplus software v.2.0.1 (Applied Biosystems). Threshold cycle values for Zat12, WRKY18, WRKY40, APX1, RbohD, GNAT5 and KMD1 were calculated with the CT of EF1‐a as an internal control. Primer pairs used for amplifications are shown in Table S9. The overlap between transcripts enhanced in leaves in response to short‐term high‐light exposure and transcripts enhanced in response to ABA, ethylene (ACC), brassinolide (BL), cytokinin (CK), gibberellin (GA), auxin (IAA), methyl jasmonate (MJ), salicylic acid (SA), H2O2, or 1O2 (Davletova et al., 2005b; Gadjev et al., 2006; Nemhauser et al., 2006; Scarpeci et al., 2008; Blanco et al., 2009), or in response to different abiotic stresses (Tosti et al., 2006; Truman et al., 2006; Kleine et al., 2007; Huang et al., 2008; Larkindale and Vierling, 2008; Matsui et al., 2008; Consales et al., 2012; Ding et al., 2014), was determined as previously described (Miller et al., 2009; Suzuki et al., 2013a). Levels of ascorbic acid were measured by gas chromatography–mass spectrometry (GC‐MS) analysis and expressed as relative to an internal control (ribitol) as previously described (Rizhsky et al., 2004; Suzuki et al., 2013a).
H2O2 measurement
The accumulation of H2O2 in tissues exposed to the short‐term light stress was measured using Amplex Red (Molecular Probes, Invitrogen, http://www.invitrogen.com/). Five hundred microliters of 50‐mm sodium phosphate buffer (pH 7.4) containing 50 μm Amplex Red and 0.05 U ml−1 horseradish peroxidase was added to ground tissues and samples were centrifuged at 12 000 g for 12 min at 4°C. Following the centrifugation, 450 μl of supernatant was transferred into fresh tubes and incubated for 30 min at room temperature in the dark. Absorbance at 560 nm was then measured and the concentration of H2O2 in each sample was determined from a standard curve consisting of 0, 0.5, 1, 3, 6 and 9 μm of H2O2. Following the measurement of absorbance, tissue samples were completely dried using a speed vacuum concentrator at 30°C for 30 min and H2O2 accumulation per mg dry weight was calculated.
Electrolyte leakage assay
Electrolyte leakage was measured as described by Sung and Guy (2003) with minor modifications. Briefly, one fully expanded leaf exposed to high light as described above was excised and immersed in 10 ml of distilled, deionized water in a 50‐ml Falcon tube. Samples were shaken for 1 h at room temperature and the conductivity of the water was measured using a conductivity meter (Sung and Guy, 2003). Samples were then heated to 95°C for 20 min using a heat block, shaken for 1 h at room temperature and the conductivity of the water was measured again. The percentage of electrolyte leakage was calculated as the percentage of the conductivity before heating over that after heating.
Statistical analysis
We performed next generation RNA‐Seq for differential expression profiling and characterization of transcript processing events. Three biological replicates were obtained as described above. Paired‐end Illumina sequencing generated on average 21 million read pairs per sample, with each sequence read having a length of 101 nucleotides. We utilized the services of frequently used, publicly available RNA‐Seq analysis software, namely bowtie (Langmead et al., 2009), tophat (Trapnell et al., 2009) and cufflinks (Trapnell et al., 2010), for alignment of paired‐end reads onto the reference genome, parsing the alignment to infer the exon–exon splice junctions, and performing the differential expression analysis of annotated genes. Transcripts expressing differentially in two (or more) conditions were identified by examining the difference in their abundance under the two conditions. The abundance of a transcript is measured in terms of FPKM, normalized for the transcript length and total number of cDNA fragments for a sample replicate. The difference in expression was obtained as the log of fold change in abundance between the two conditions. A test of statistical significance for differential expression of each transcript was performed based on a negative binomial model estimated from the data (Trapnell et al., 2010). The fold change of genes with multiple isoforms was assessed by summing up the FPKMs for all isoforms of a gene and then measuring the difference between the two conditions (Trapnell et al., 2010). Other statistical analyses were performed by one‐tailed Student's t‐test as previously described in Suzuki et al. (2013a) . Results are presented as mean ± SD (*P < 0.05; **P < 0.01).
We extracted three distinctive patterns representing elevation in steady‐state transcript level at 20 or 60 sec or both using a Fortran code on a Unix platform: Cluster I, elevation of steady‐state transcript level at 20 sec followed by decline at 60 sec; Cluster II, elevation of steady‐state transcript level at 60 sec with no significant change at 20 sec; and Cluster III, elevation of steady‐state transcript level at 20 sec and 60 sec. Elevation or decline in steady‐state transcript level at a particular time point indicated a statistically significant increase or decrease, respectively, in steady‐state transcript level relative to the previous time point. Steady‐state transcript level refers to transcript abundance which is measured in terms of FPKM, normalized for the transcript length and total number of cDNA fragments for a replicate sample.
Author contributions
NS, RM, GM, VS, RA designed and supervised the research. NS, ES, RA, LS, AB and AD performed the research and analyzed the data. RM and NS wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Supporting information
Figure S1. (a) Histogram showing the fold change distribution of all transcripts that had a significant change in their steady‐state transcript level (increase or decrease) in response to light stress.
Figure S1. (b) Distribution of genes encoding transcripts up‐regulated at 20 sec of light stress on all Arabidopsis chromosomes.
Figure S2. Changes in the steady‐state transcript level of the five genes tested with mutants in Figure 3 in response to light stress.
Figure S3. Heat map showing changes in the steady‐state transcript level of the 682 transcripts that appear in the ATH1 Affymetrix chips (out of 731 from Clusters I, II and II; Figure 1) in response to light stress.
Figure S4. Content of RNA‐destabilizing sequences in ultra‐fast response transcripts and transcripts responding to a 3 h light stress treatment.
Table S1. Transcripts with a significant enhancement in steady state transcript level at 20‐sec high‐light exposure (Cluster I).
Table S2. Transcripts with a significant enhancement in steady state transcript level at 60‐sec high‐light exposure (Cluster II).
Table S3. Transcripts with a significant enhancement in steady state transcript level at 20‐ and 60‐sec high‐light exposure (Cluster III).
Table S4. Transcripts in Clusters I, II and III, not found in Affymetrix ATH1 chips.
Table S5. Transcripts in Clusters I, II and III divided into those enhanced from less than a 2.0 FPKM value and from more than a 2.0 FPKM value.
Table S6. Transcripts with a significantly declined steady‐state transcript level at 20‐sec high‐light exposure.
Table S7. Transcripts with a significantly declined steady‐state transcript level at 60‐sec high‐light exposure.
Table S8. Transcripts in Clusters I, II and III not found to be enhanced by a longer (3 h) light stress treatment or any of the other abiotic stresses tested (Figure 2b).
Table S9. Primer pairs used for quantitative real‐time PCR.
Acknowledgements
This work was supported by funding from the National Science Foundation (IOS‐1353886, IOS‐0639964, IOS‐0743954), the University of North Texas, College of Arts and Sciences, Sophia University, Faculty of Science and Technology, Israel Science Foundation (grant no. 938/11) and Marie Curie Actions‐International Career Integration Grant (grant no. 293999). The funders had no role in the design, data collection, analysis, decision to publish or preparation of the manuscript.
Accession numbers: Arabidopsis Genome Initiative locus identifiers for genes mentioned in this article are as follows: Zat12 (At5g59820), WRKY18 (At4g31800), WRKY40 (At1g80840), APX1 (At1g07890), RbohD (At5g47910), Cat2 (At4g35090), GNAT family protein (At2g39030), KMD1 (At1g80440), EF1‐a (At5g60390), proteins of unknown function (At5g10695 and At3g10020), a Golgi‐associated protein (At5g51430), a RAV transcription factor (At1g25560) and a glycosyl hydrolase (At3g13750). RNA‐Seq data for this work are deposited under the GEO accession number GSE60865.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana . Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bailey‐Serres, J. and Voesenek, L. (2008) Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339. [DOI] [PubMed] [Google Scholar]
- Blanco, F. , Salinas, P. , Cecchini, N.M. et al. (2009) Early genomic responses to salicylic acid in Arabidopsis . Plant Mol. Biol. 70, 79–102. [DOI] [PubMed] [Google Scholar]
- Cavanagh, C. , Morell, M. , Mackay, I. and Powell, W. (2008) From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11, 215–221. [DOI] [PubMed] [Google Scholar]
- Chinnusamy, V. and Zhu, J. (2009) Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W.G. , Toyota, M. , Kim, S.H. , Hilleary, R. and Gilroy, S. (2014) Salt stress‐induced Ca2+ waves are associated with rapid, long‐distance root‐to‐shoot signaling in plants. Proc. Natl Acad. Sci. USA, 111, 6497–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consales, F. , Schweizer, F. , Erb, M. , Gouhier‐Darimont, C. , Bodenhausen, N. , Bruessow, F. , Sobhy, I. and Reymond, P. (2012) Insect oral secretions suppress wound‐induced responses in Arabidopsis . J. Exp. Bot. 63, 727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova, S. , Rizhsky, L. , Liang, H. , Shengqiang, Z. , Oliver, D.J. , Coutu, J. Shulaev, V. , Schlauch, K. and Mittler, R. (2005a) Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis . Plant Cell, 17, 268–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova, S. , Schlauch, K. , Coutu, J. and Mittler, R. (2005b) The zinc‐finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis . Plant Physiol. 139, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz, K.J. (2015) Efficient high light acclimation involves rapid processes at multiple mechanistic levels. J. Exp. Bot. 66, 2401–2414. [DOI] [PubMed] [Google Scholar]
- Ding, F. , Cui, P. , Wang, Z. , Zhang, S. , Ali, S. and Xiong, L. (2014) Genome‐wide analysis of alternative splicing of pre‐mRNA under salt stress in Arabidopsis . BMC Genomics, 15, 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev, I. , Vanderauwera, S. , Gechev, T.S. , Laloi, C. , Minkov, I.N. , Shulaev, V. Apel, K. , Inzé, D. , Mittler, R. and Van Breusegem, F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis . Plant Physiol. 141, 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy, S. , Suzuki, N. , Miller, G. , Choi, W. , Toyota, M. , Devireddy, A.R. and Mittler, R. (2014) A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 19, 623–630. [DOI] [PubMed] [Google Scholar]
- Halliwell, B. (2006) Reactive species and antioxidants. redox biology is a fundamental theme of aerobic life. Plant Physiol. 141, 312–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoth, S. , Morgante, M. , Sanchez, J.P. , Hanafey, M.K. , Tingey, S.V. and Chua, N.H. (2002) Genome‐wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1‐1 mutant. J. Cell Sci. 115, 4891–4900. [DOI] [PubMed] [Google Scholar]
- Hruz, T. , Laule, O. , Szabo, G. , Wessendorp, F. , Bleuler, S. , Oertle, L. , Widmayer, P. , Gruissem, W. and Zimmermann, P. (2008) Genevestigator v3: a reference expression database for the meta‐analysis of transcriptomes. Adv. Bioinform, 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D. , Wu, W. , Abrams, S.R. and Cutler, A.J. (2008) The relationship of drought‐related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, A. , Kazuoka, T. , Torikai, S. , Kikuchi, H. and Oeda, K. (2000) A zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis . Plant J. 24, 191–203. [DOI] [PubMed] [Google Scholar]
- Kleine, T. , Kindgren, P. , Benedict, C. , Hendrickson, L. and Strand, A. (2007) Genome‐wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky, S. , Nott, A. , Mockler, T.C. , Hong, F. , Sachetto‐Martins, G. , Surpin, M. , Lim, J. , Mittler, R. and Chory, J. (2007) Multiple signals from damaged chloroplasts converge on a common pathway to regulate nuclear gene expression. Science, 316, 715–719. [PubMed] [Google Scholar]
- Kwak, H. and Lis, J.T. (2013) Control of transcriptional elongation. Annu. Rev. Genet. 47, 483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. and Salzberg, S.L. (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale, J. and Vierling, E. (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol. 146, 748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M. (2011) Paused RNA polymerase II as a developmental checkpoint. Cell, 145, 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhua, S. , Hegie, A. , Suzuki, N. et al. (2013) Linking genes of unknown function with abiotic stress responses by high‐throughput phenotype screening. Physiol. Plant. 148, 322–333. [DOI] [PubMed] [Google Scholar]
- Maiuri, P. , Knezevich, A. , De Marco, A. et al. (2011) Fast transcription rates of RNA polymerase II in human cells. EMBO Rep. 12, 1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, A. , Ishida, J. , Morosawa, T. et al. (2008) Arabidopsis transcriptome analysis under drought, cold, high‐salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. [DOI] [PubMed] [Google Scholar]
- Miller, G. , Schlauch, K. , Tam, R. , Cortes, D. , Torres, M.A. , Shulaev, V. Dangl, J.L. and Mittler, R. (2009) The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2, ra45. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Mittler, R. and Blumwald, E. (2010) Genetic engineering for modern agriculture: challenges and perspectives. Annu. Rev. Plant Biol. 61, 443–462. [DOI] [PubMed] [Google Scholar]
- Mittler, R. and Blumwald, E. (2015) The roles of ROS and ABA in systemic acquired acclimation. Plant Cell, 27, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. , Vanderauwera, S. , Suzuki, N. , Miller, G. , Tognetti, V.B. , Vandepoele, K. Gollery, M. , Shulaev, V. and Van Breusegem, F. (2011) ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. [DOI] [PubMed] [Google Scholar]
- Mittler, R. , Finka, A. and Goloubinoff, P. (2012) How do plants feel the heat? Trends Biochem. Sci. 37, 118–125. [DOI] [PubMed] [Google Scholar]
- Munns, R. and Tester, M. (2008) Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Nakashima, K. and Yamaguchi‐Shinozaki, K. (2006) Regulons involved in osmotic stress‐responsive and cold stress‐responsive gene expression in plants. Physiol. Plant. 126, 62–71. [Google Scholar]
- Narsai, R. , Howell, K.A. , Millar, A.H. , O'Toole, N. , Small, I. and Whelan, J. (2007) Genome‐wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana . Plant Cell, 19, 3418–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev, S. and Adelman, K. (2008) Promoter‐proximal pol II: when stalling speeds things up. Cell Cycle, 7, 1539–1544. [DOI] [PubMed] [Google Scholar]
- Nemhauser, J.L. , Hong, F. and Chory, J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell, 126, 467–475. [DOI] [PubMed] [Google Scholar]
- Ohme‐Takagi, M. , Taylor, C.B. , Newman, T.C. and Green, P.J. (1993) The effect of sequences with high AU content on mRNA stability in tobacco. Proc. Natl Acad. Sci. USA 90, 11811–11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley, R.C. and Ecker, J.R. (2010) Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61, 928–940. [DOI] [PubMed] [Google Scholar]
- Rizhsky, L. , Liang, H. , Shuman, J. , Shulaev, V. , Davletova, S. and Mittler, R. (2004) When defense pathways collide: the response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 134, 1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpeci, T.E. , Zanor, M.I. , Carrillo, N. , Mueller‐Roeber, B. and Valle, E.M. (2008) Generation of superoxide anion in chloroplasts of Arabidopsis thaliana during active photosynthesis: a focus on rapidly induced genes. Plant Mol. Biol. 66, 361–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle, O. , Teichmann, S.A. and Marioni, J.C. (2015) Computational and analytical challenges in single‐cell transcriptomics. Nat. Rev. Genet. 16, 133–145. [DOI] [PubMed] [Google Scholar]
- Sung, D.Y. and Guy, C.L. (2003) Physiological and molecular assessment of altered expression of Hsc70‐1 in Arabidopsis, evidence for pleiotropic consequences. Plant Physiol. 132, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. , Sejima, H. , Tam, R. , Schlauch, K. and Mittler, R. (2011) Identification of the MBF1 heat‐response regulon of Arabidopsis thaliana . Plant J. 66, 844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. , Miller, G. , Salazar, C. et al. (2013a) Temporal‐spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell, 25, 3553–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. , Miller, G. , Sejima, H. , Harper, J. and Mittler, R. (2013b) Enhanced seed production under prolonged heat stress conditions in Arabidopsis thaliana plants deficient in cytosolic ascorbate peroxidase 2. J. Exp. Bot. 64, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, M.A. , Dangl, J.L. and Jones, J.D. (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl Acad. Sci. USA, 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosti, N. , Pasqualini, S. , Borgogni, A. , Ederli, L. , Falistocco, E. , Crispi, S. and Paolocci, F. (2006) Gene expression profiles of O3‐treated Arabidopsis plants. Plant, Cell Environ. 29, 1686–1702. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Pachter, L. and Salzberg, S.L. (2009) TopHat: discovering splice junctions with RNA‐seq. Bioinformatics (Oxford, England), 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. et al. (2010) Transcript assembly and quantification by RNA‐seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman, W. , Zabala, M.T. and Grant, M. (2006) Type III effectors orchestrate a complex interplay between transcriptional networks to modify basal defence responses during pathogenesis and resistance. Plant J. 46, 14–33. [DOI] [PubMed] [Google Scholar]
- Vanderauwera, S. , Suzuki, N. , Miller, G. et al. (2011) Extranuclear protection of chromosomal DNA from oxidative stress. Proc. Natl Acad. Sci. USA 108, 1711–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Mao, Z. and Chong, K. (2013) A global profiling of uncapped mRNAs under cold stress reveals specific decay patterns and endonucleolytic cleavages in Brachypodium distachyon . Genome Biol. 14(8), R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (a) Histogram showing the fold change distribution of all transcripts that had a significant change in their steady‐state transcript level (increase or decrease) in response to light stress.
Figure S1. (b) Distribution of genes encoding transcripts up‐regulated at 20 sec of light stress on all Arabidopsis chromosomes.
Figure S2. Changes in the steady‐state transcript level of the five genes tested with mutants in Figure 3 in response to light stress.
Figure S3. Heat map showing changes in the steady‐state transcript level of the 682 transcripts that appear in the ATH1 Affymetrix chips (out of 731 from Clusters I, II and II; Figure 1) in response to light stress.
Figure S4. Content of RNA‐destabilizing sequences in ultra‐fast response transcripts and transcripts responding to a 3 h light stress treatment.
Table S1. Transcripts with a significant enhancement in steady state transcript level at 20‐sec high‐light exposure (Cluster I).
Table S2. Transcripts with a significant enhancement in steady state transcript level at 60‐sec high‐light exposure (Cluster II).
Table S3. Transcripts with a significant enhancement in steady state transcript level at 20‐ and 60‐sec high‐light exposure (Cluster III).
Table S4. Transcripts in Clusters I, II and III, not found in Affymetrix ATH1 chips.
Table S5. Transcripts in Clusters I, II and III divided into those enhanced from less than a 2.0 FPKM value and from more than a 2.0 FPKM value.
Table S6. Transcripts with a significantly declined steady‐state transcript level at 20‐sec high‐light exposure.
Table S7. Transcripts with a significantly declined steady‐state transcript level at 60‐sec high‐light exposure.
Table S8. Transcripts in Clusters I, II and III not found to be enhanced by a longer (3 h) light stress treatment or any of the other abiotic stresses tested (Figure 2b).
Table S9. Primer pairs used for quantitative real‐time PCR.


