Abstract
Background
Patients with pneumococcal meningitis often die or have severe neurological damage despite optimal antibiotic therapy. New or improved therapy is required. The delivery of new interventions will require an improved understanding of the disease pathogenesis. Our objective was to learn more about the pathophysiology of severe meningitis through the interpretation of differences in the proteomic profile of cerebrospinal fluid (CSF) from patients with meningitis.
Methods
Two-dimensional polyacrylamide gel electrophoresis of CSF from normal subjects (controls, n = 10) and patients with pneumococcal meningitis (n = 20) was analyzed. Spot differences were compared and identified between controls, nonsurvivors (n = 9), and survivors (n = 11).
Results
Protein concentration in CSF of patients with meningitis was 4-fold higher than in CSF of control subjects (7.0 mg/mL vs 0.23 mg/mL; P < .01). A mean of 2466 discrete protein spots was present in CSF of patients with meningitis. Thirty-four protein spots were differentially expressed in CSF of nonsurvivors, compared with survivors. None of these protein spots were observed in CSF of control subjects.
Conclusions
Proteomic screening of CSF yields potential biomarkers capable of differentiating control subjects from nonsurvivors and survivors of meningitis. Proteins involved in the inflammatory process and central metabolism were represented in the differentially expressed protein repertoire.
Streptococcus pneumoniae is the most common pathogen in bacterial meningitis beyond the neonatal period [1]. In underresourced countries such as Malawi, pneumococcal meningitis has a high fatality rate of 67% [2]. Survivors of the infection often develop long-term neurological sequelae, including hearing loss and other focal neurological deficits [2].
Endothelial cells separate blood from brain tissue compartments and form a protective blood-brain barrier (BBB). Pneumococci can use the human platelet-activating factor receptor to traverse the endothelial cells of the BBB. Later during the course of the disease, the integrity of the BBB is compromised by apoptosis of endothelial cells, allowing pneumococcal invasion of the cerebrospinal fluid (CSF) [3, 4]. Once in CSF, neuronal damage as a result of pneumococcal meningitis occurs by several mechanisms. The initial host inflammatory response is initiated by pneumococcal capsular polysaccharides and surface proteins, such as pneumolysin and PspA [5, 6]. This can result in cerebral ischemia, brain edema formation, hydrocephalus, or increased intracranial pressure [7]. In animal models, this has been shown to contribute to neuronal cell death via 3 distinct pathways: classic caspase-3–dependent cell death (ie, apoptosis), caspase-3–independent cell death (ie, pyknosis), and necrosis [8].
Proteomic methods allow quantitative analysis of expressed proteins present in CSF [9]. Proteins that are found in increased concentrations include both pneumococcal proteins and host proteins. Host proteins may result from increased BBB permeability (and, therefore, an excess of plasma proteins in CSF) or may originate locally in the central nervous system. We tested the hypothesis that, in addition to differences between CSF of controls and patients with meningitis, there would also be differences in the CSF proteome between nonsurvivors and survivors of meningitis.
Materials and Methods
Subjects
Patients were recruited to a double-blind, randomized, placebo-controlled trial of dexamethasone adjuvant therapy in adults with bacterial meningitis in Malawi [10]. As part of this study, a CSF specimen was collected from 465 patients, the majority of whom also had human immunodeficiency virus (HIV) infection [10]. Patients were not treated in an intensive care unit, as described elsewhere [10]. Patients were treated in hospital for a minimum of 10 days and were evaluated at 40 days and at 6 months. Clinically evident adverse events were recorded systematically throughout the trial period. At follow-up, patients had a standardized neurologic examination and a hearing assessment. Patients who did not return for follow-up appointments were visited at home. No other underlying diseases were specifically looked for other than HIV infection and malaria. However, some patients volunteered a past medical history.
Patient categories
Of those who survived, 103 patients survived to 1 month with no neurological impairment or recorded disability, 66 survivors had detectable hearing loss without complete deafness, and 47 patients had disability (paresis, intellectual impairment, blindness, complete deafness debility, or recurrent seizures). Two hundred forty-nine of these patients died, of whom 92 survived for ≥10 days but died before 40 days after presentation, and 157 patients died within 10 days after presentation to hospital. Patients in Malawi have basic medical care as described elsewhere [10]. The eventual cause of death was often not known. However, we have initiated a new study (2010–2012) to determine causes of death in a new prospective cohort. Initial group definitions included the following: survival with no neurological impairment or recorded disability (n = 103), deaf (n = 66), maimed (n = 47), protracted death (n = 92), and rapid death (n = 157). Recruitment included consent for diagnostic samples to be used in meningitis pathogenesis research at a later date.
Diagnosis
CSF specimens were examined as described elsewhere [10]. CSF was examined for cell and differential counts by microscopy. A Gram stain procedure was performed if the sample was turbid or had >10 white cells per cubic millimeter. Centrifuged CSF deposits were incubated on sheep blood agar in a candle-extinction jar at 37°C for 48 h, and isolates were identified by means of standard techniques. Blood was cultured at 37°C for a minimum of 48 h (BacT/Alert; bioMe´rieux). In the first 51 consecutive CSF specimens for which Gram stain and culture were negative, polymerase chain reaction for meningococcus and pneumococcus was performed [10].
Proven bacterial meningitis was defined as identification of an organism from a CSF specimen by means of microscopy, culture, or polymerase chain reaction or from blood culture in the context of a CSF specimen containing a white cell count of >100 cells/mm3 with >50% neutrophils. Probable bacterial meningitis was defined as a CSF specimen containing a cell count of >100 cells/mm3 with >50% neutrophils without identification of an organism [10]. Data regarding CSF and blood culture are provided in Table 1.
Table 1. Clinical Details of Patients.
| Characteristic | Control (n = 10) |
Nonsurvivors (n = 9) |
Survivors (n = 11) |
|---|---|---|---|
| Age, mean years ± SD | 27.8 ± 9.5 | 28.3 ± 4.8 | 35.0 ± 14.7 |
| Male sex | 3 (30) | 2 (22) | 5 (45) |
| Glasgow coma score, mean score ± SD | 9.4 ± 4.7 | 11.6 ± 2.8) | 8.0 ± 3.0) |
| Time to presentation, mean h (IQR) | 55.2 (12–96) | 50 (18–72) | 86.5 (10–168) |
| No of antimicrobials received previously | 2 | 0 | 0 |
| Positive CSF culture | … | 9 (100) | 11 (100) |
| Positive blood culture | … | 3 (33) | 4 (36) |
| Blood culture unavailable | … | 1 (11) | 0 (0) |
| Hemoglobin level, mean g/dL ± SD | … | 9.8 ± 1.4 | 10.2 ± 3.3 |
| Steroid treatment received | … | 0 (0) | 0 (0) |
| Steroid placebo received | … | 9 (100) | 11 (100) |
| HIV positive | … | 9 (100) | 11 (100) |
| HIV status not known | 10 (100) | … | … |
| Survival at day 10 | … | 0 (0) | 11 (100) |
NOTE. Data are no (%) of patients, unless otherwise indicated. CSF, cerebrospinal fluid; HIV, human immunodeficiency virus; IQR, interquartile range; SD, standard deviation.
CSF samples for 2-dimensional polyacrylamide gel electrophoresis (2D PAGE) analysis
The 2D PAGE analysis included samples from the rapid death group (nonsurvivors, n = 11) and the survival with no neurological impairment or recorded disability group (survivors, n = 9). Control CSF specimens were obtained from patients who presented with clinical features suggestive of meningitis but who were subsequently found to have a normal CSF profile (n = 10). Control patients were routinely discharged well and with no alternative diagnosis passed. Patients for whom any alternative diagnosis (eg, cryptococcal meningitis) was found were not included as controls. Information regarding patients is presented in Table 1.
The study was approved by the research ethics committees of the University Of Malawi College Of Medicine and the Liverpool School of Tropical Medicine. Patients or their legal guardians provided written informed consent or, if they were unable to read or write, independently witnessed verbal consent before recruitment.
Sample preparation
CSF samples were stored at −20°C within 1 h of sampling and at −80°C from 24 h until analysis. Archived CSF specimens were thawed at 4°C and desalted using a centrifugal filter (YM3, Amicon and Microcon; Millipore). The concentration of protein in each sample was determined using the Bradford assay. Samples with protein concentrations <1 mg/mL were concentrated by vacuum centrifugation to a volume sufficient for analysis with 2D PAGE.
2D PAGE comparison
The abundance of protein spots on the 2D PAGE gels was analyzed by scanning the spots with a PowerLook 111 scanner (UMAX). The intensity of each protein spot was measured using Progenesis software PG220 (Nonlinear Dynamics). Gels were grouped into “normal” (controls), nonsurvivors, and survivors. Spots were accepted as representative of the group if present in at least 50% of the gels in each group.
Protein identification
Protein spots demonstrating differential expression from the 2D PAGE comparison were manually excised from the stained gels and subjected to in-gel tryptic digestion. The resulting digest was sequenced by matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometry (MS) or liquid chromatography (LC)-MS/MS (Appendix, which appears only in the online version of the Journal). The protein digest spectra were analyzed using the Swiss Prot and mass spectral database (MSDB) nonredundant protein databases against Homo sapiens and Streptococcus pneumoniae with MASCOT software (Matrix Science; http://www.matrixscience.com).
Results
Sample information
Analysis was performed on 30 CSF samples which were representative of the spectrum of protein profiles among the CSF samples. This sample size was used for the analysis, because we found that spot saturation occurred in the analysis beyond this number of samples. Saturation occurred as a result of themes developing among all the gel spots and development of metathemes (ie, spot themes within the groups). These themes and metathemes included basic spot patterns as a result of endogenous proteins on the basis of spot intensities and spot volumes.
In addition, these samples had the largest range of protein concentration found in the collection of CSF specimens. Therefore, the proteins that were present in these 20 meningitis samples were most likely to represent the proteomic profile of the CSF collection. All patients with meningitis received a diagnosis of pneumococcal meningitis due to S. pneumoniae. All subjects were HIV positive, and all samples were collected before any treatment commenced. The median CD4 cell count at hospital admission for 101 consecutively admitted HIV-positive patients during recruitment to the trial was 102 cells/mm3 (interquartile range, 51–169 cells/mm3). We did not measure CD4 counts in all but 1 of these patients, because antiretrovirals were not routinely available until mid-2004 in Malawi, and therefore, there was no clinical imperative and no advantage to the patient to know their CD4 count. None of these patients were receiving antiretroviral therapy. All had World Health Organization stage III or IV disease (1990 criteria). As described elsewhere [10], normal CSF was obtained from patients who had negative results for meningitis or any other pathogen. The HIV status of the normal control patients was not known.
CSF protein concentrations
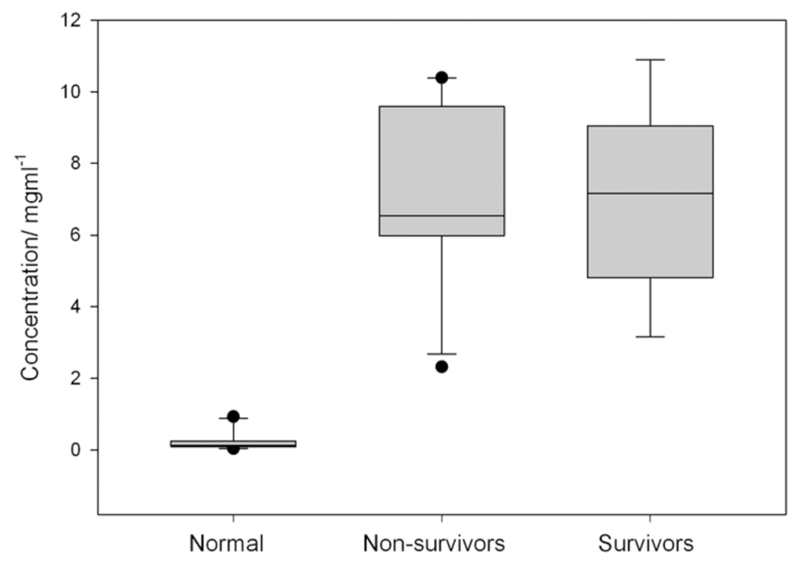
The concentration of protein in each sample was measured using the Bradford assay. The normal samples had a protein concentration range of 0.05–0.94 mg/mL (mean, 0.23 mg/mL). The nonsurvivors had a protein concentration of 2.33–10.40 mg/mL (mean, 6.94 mg/mL). The survivors had a protein concentration range of 3.16–10.89 mg/mL (mean, 7.07 mg/mL; Figure 1). The protein concentration in CSF of control patients was significantly less than that from patients with meningitis (P < .01). There was no significant difference between the protein concentration in nonsurvivors and survivors.
Figure 1.
The protein concentrations in cerebrospinal fluid did not vary significantly between survivors and nonsurvivors. However, cerebrospinal fluid from both nonsurvivors and survivors had a significantly greater concentration of protein, compared with cerebrospinal fluid from normal control subjects.
2D PAGE comparison
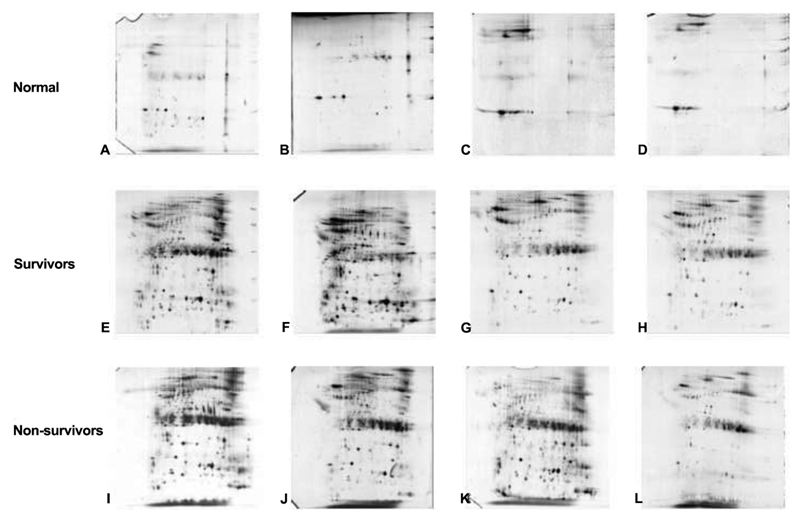
The gels were analyzed in the following 3 groups; normal (control), nonsurvivors, and survivors (Figure 2). CSF of normal patients was found to have an average of 638 protein spots/gel. The average number of protein spots identified for nonsurvivors was 2386, and the samples of survivors yielded a mean of 2546 protein spots per gel.
Figure 2.
Cerebrospinal fluid was separated using 2-dimensional polyacrylamide gel electrophoresis. The gels were grouped into the following 3 groups: normal (controls), survivors, and nonsurvivors. Two-dimensional polyacrylamide gel electrophoresis separates proteins on the basis of isoelectric point and molecular weight. The proteins are visualized using silver stain and analyzed for differences. It can be observed that cerebrospinal fluid of controls has less protein, compared with that of the survivors and nonsurvivors. It can also be observed that the determination of differences between survivors and nonsurvivors is complex.
Initially the 2D PAGE results of CSF from controls were compared with the pooled results of patients with meningitis. There were 20 protein spots from the CSF of controls that were common to the CSF of the pooled patients with meningitis, and therefore, >2400 proteins were considered to be present as a result of meningitis.
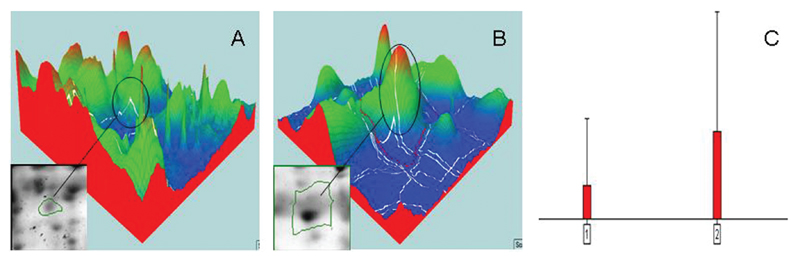
There were 469 common spots between CSF of nonsurvivors and survivors (present in a minimum of 50% of gels). All spots were analyzed as a “normalized volume,” which is a 3 dimensional computerized construct that nullifies variations in intensity, as illustrated in Figure 3 [11]. The normalized volume data for each spot on each 2D PAGE gel of nonsurvivors was compared with that of survivors. This comparison identified 34 proteins with a minimum 2-fold difference in normalized volume between nonsurvivors and survivors (Figure 4). None of the 20 protein spots that were common to CSF of both patients with meningitis and controls were in the list of spot differences found between nonsurvivors and survivors.
Figure 3.
To accurately compare the abundance of a protein spots across 2-dimensional gels, it is essential to compensate for variations from external sources, such as variation in staining time. Normalization is the process of removing such variation. In this analysis, a normalized volume was used, which is a computerized construct that creates a 3-dimensional visualization of the protein spot on the basis of the intensity of the spot. For example in panel A, a spot is represented as having a low normalized volume in a nonsurvivor gel; the same spot is demonstrated in panel B. This normalized volume can be compared between gels to determine whether there is a difference in intensity (C).
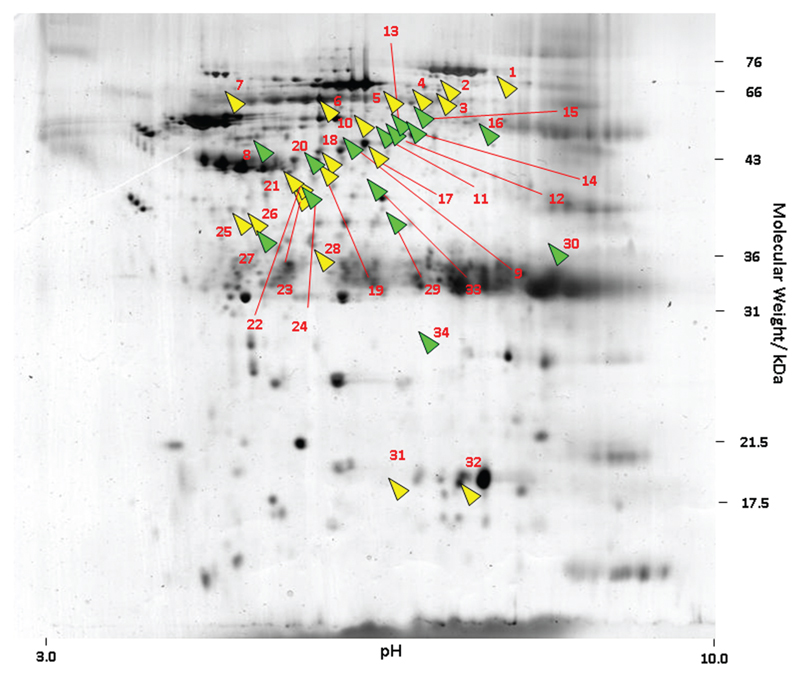
Figure 4.
Protein differences were identified in the nonsurvivor average gel, compared with the survivor average gel. Green indicates proteins found to have a minimum 2-fold up-regulation in nonsurvivors. Yellow indicates proteins found to have a minimum 2-fold down-regulation in nonsurvivors.
Protein identification
The protein spot differences identified using MALDI-TOF and LC-MS/MS are summarized in Table 2 (Data regarding the normalized volumes of the protein spots are provided in Table A1, which appears only in the online version of the Journal). The identified protein spots were functionally clustered into the following groups: membrane and skeletal proteins (n = 4), transporters (n = 2), glycoproteins (n = 2), G proteins of the Ras family (n = 1), metabolic enzymes (n = 4), cellular defense proteins (n = 3), globins (n = 1), kinases (n = 2), phosphatases (n = 3), chaperone (n = 1), protease (n = 1), translation proteins (n = 6), others (n = 3), and proteins of unknown function (n = 1).
Table 2. Functional Clustering of Identified Proteins.
| Functional category, protein | Reference | Up- or down-regulation in nonsurvivors | Fold expression in nonsurvivors |
|---|---|---|---|
| Cellular defense proteins | |||
| Complement C3 precursor | [15] | Down | 4.8 |
| Chitotriosidase | [14] | Up | 5.1 |
| Complement C1q tumor necrosis factor–related protein 9 | [16] | Up | 3.4 |
| Chaperones: T-complex protein 1 subunit zeta | [27] | Down | 2.8 |
| Metabolic enzymes | |||
| Phosphoglucomutase-like protein 5 | [23] | Down | 4.9 |
| Glyoxalase domain–containing protein 4 | [25] | Down | 3.6 |
| 26S protease regulatory subunit 7 | [28] | Up | 2.5 |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 9, mitochondrial | [29] | Up | 2.8 |
| Translation | |||
| Retinoic acid receptor RXR-gamma | [30] | Down | 2.5 |
| Cleavage stimulation factor (CSTFT) 64 kDa subunit, tau variant | [31] | Up | 3.3 |
| Heterogeneous nuclear ribonucleoproteins C1/C2 | [32] | Up | 2.5 |
| Zinc finger protein 179 | [33] | Up | 2.0 |
| Zinc finger protein 1 | [34] | Down | 2.0 |
| Eukaryotic translation initiation factor 2, subunit 2 beta | [35] | Down | 2.2 |
| Transporters | |||
| Serotransferrin | [36] | Down | 5.5 |
| Solute carrier family 25 (member 16) | [18] | Up | 4.0 |
| Glycoproteins | |||
| β-2-glycoprotein 1 precursor | [19] | Up | 3.6 |
| Zinc alpha 2-glycoprotein precursor | [37] | Down | 2.9 |
| G proteins of the Ras family: Ras-related protein Rab-37 (RAB37) | [24] | Down | 3.8 |
| Globins: haptoglobin | [38] | Up | 2.9 |
| Kinases | |||
| Brain-enriched guanylate kinase-associated protein (BEGAIN) | [20] | Down | 5.0 |
| Pyruvate kinase | [39] | Up | 2.1 |
| Proteases: tryptophan/serine protease | [40] | Up | 3.1 |
| Phosphatases | |||
| Lysosomal acid phosphatase | [41] | Up | 2.7 |
| Serine/threonine phosphatase 2-alpha 65K regulatory chain | [42] | Down | 2.7 |
| Serine/threonine/tyrosine-interacting–like protein 1 | [43] | Down | 2.2 |
| Membrane and skeletal proteins | |||
| Fibrinogen | [21] | Down | 2.5 |
| Fascin | [44] | Up | 3.0 |
| Mutant desmin | [45] | Up | 2.7 |
| Ankyrin repeat domain–containing protein 42 | [46] | Down | 2.5 |
| Others | |||
| Alpha 1 antitrypsin precursor | [47] | Down | 3.0 |
| Nuclear localized factor 1 | [48] | Down | 2.1 |
| Human serum albumin | [49] | Down | 2.0 |
| Unknown: cancer-associated gene 1 | [50] | Down | 2.5 |
Proteins associated with rapid death due to meningitis included chitotriosidase (CHIT 1, 5-fold up-regulation), serotransferrin and brain-enriched guanylate kinase-associated (BEGAIN, 5-fold down-regulation), complement C3 precursor and phosphoglucomutase-like protein 5 (4-fold down-regulation), cleavage stimulation factor (CSTFT; 64 kDa subunit tau variant), β-2-glycoprotein 1 precursor, solute carrier family 25 (member 16), fascin (singed-like protein), complement C1q tumor necrosis factor–related protein 9, tryptophan/serine protease (3-fold up-regulation), Ras-related protein Rab-37, and glyoxalase domain-containing protein 4 (3-fold down-regulation).
Discussion
In this analysis, 34 protein differences were observed between nonsurvivors and survivors of pneumococcal meningitis. These 34 proteins were subsequently identified with MS. Functional clustering revealed that most of the proteins identified were associated with inflammation, metabolism, or protein transcription and translation.
This is the first time that 2D PAGE has been applied to the analysis of CSF from adult patients who have received a diagnosis of pneumococcal meningitis. Previous work has focused on the analysis of inflammatory proteins, including cytokines, chemokines, and other inflammatory proteins, and has identified the proteins IL-1β, IL-6, IL-10, MIF, and TNF-α; CXC chemokines (ENA-78, GRO, IL-8, IP-10, and NAP-2); CC chemokines (MCP-1, MCP-2, and MIP-3α); and the growth factors HGF, IGFBP-1, and MCSF as being significant in the progression of pneumococcal disease toward fatal outcome due to meningitis. Similarly, work on CSF from patients with malaria has identified the analogous proteins IP-10, IL-1ra, IL-8, IP-10, PDGFbb, MIP-1β, Fas-L, sTNFR1, and sTNF-R2 in Ghanian children [12]. None of these proteins were identified in our list of protein differences. This may be because the differences between nonsurvivors and survivors were too subtle to be observed by 2D PAGE analysis. However, a more likely explanation is that the concentration of large proteins, which was very high, may have acted as a confounding factor in the analysis. As a result, many of these proteins may have been lost in the analysis. Therefore, an alternative approach may have been to fractionate the CSF and perform 2D PAGE of the individual fractions. However, the volume of CSF that can be collected from a lumbar puncture is low, and therefore, this would have been impractical to perform.
There was a great degree of variability in the proteomic profile of CSF samples visualized by 2D PAGE. The proteome consisted of host proteins that were either native to CSF or originated from breakdown of the BBB and pneumococcal proteins. In addition, it was possible that other pathogenic proteins were also present in CSF. HIV coinfection of patients may also have contributed to the protein profile observed in the 2D PAGE gels of CSF. HIV infection can increase CSF protein levels and cause pleocytosis (the appearance of leukocytes in the CSF). Pleocytosis can occur intermittently during HIV infection and most likely reflects an interruption of the BBB and trafficking of white blood cells from the blood into the CSF [13].
There were extensive protein spots detected that overlap between survivors and nonsurvivors, which highlights the complexity of the pathology of meningitis. This may have caused the coefficient of variation of the protein spots between samples to be high. It is probable that our data underestimated the numbers of small proteins that differ between the groups because of technical limitations of 2D PAGE as a result of the amount of large proteins present in meningitis-affected CSF. A more powerful separation technique may have allowed these low-abundance proteins to be observed. One such technique is multidimensional LC-MS/MS.
Three proteins implicated primarily in cellular defense were found, including CHIT-1, which is expressed by activated macrophages during inflammation. It has been suggested that CHIT-1 activity is a biochemical marker of macrophage activation. CHIT-1 can be considered to be an inflammatory protein because it is solely secreted by activated macrophages and may be used as an important predictor of neuronal disease severity [14]. Complement C3 is another defense protein which was down-regulated in nonsurvivors. Complement activity is essential for the opsonization of S. pneumoniae [15]. Its down-regulation in nonsurvivors suggests a link between reduced opsonization and subsequent death. The third defense protein identified was C1q tumor necrosis factor–related protein 9 (C1qTNF9). Its functions have not been fully characterized [16].
Several cytoskeletal associated proteins were also found among the spot differences. These proteins were most likely linked with the breakdown of the BBB or as a result of neuronal cell death. Fascin is involved in the assembly of actin filament bundles present in microspikes, membrane ruffles, and stress fibers. In the brain, fascin expression has been localized to neurons, glial cells, and endothelial cells. The second cytoskeletal protein identified was desmin, which is found in the endothelial cells of the meninges. The third cytoskeletal protein was fibrinogen (a coagulation factor), which was down-regulated in nonsurvivors [17].
Other proteins with an increased expression of >3-fold included solute carrier protein 25 member 16 (a mitochondrial carrier protein) [18]. Its up-regulation may be associated with the release of the pore-forming toxin pneumolysin from pneumococci, which is known to degrade the mitochondrial membrane as well as the cell membrane, leading to neuronal death. β-2-glycoprotein I (implicated in processes such as coagulation and atherosclerosis) was most likely up-regulated in nonsurvivors as a direct result of blood clotting during the damage to the BBB [19]. Finally, cleavage stimulation factor (64 kDa subunit tau variant) plays a significant role in mRNA polyadenylation and is directly involved in binding to pre-mRNAs. Up-regulation in nonsurvivors was most likely attributable to a rapid increase in the production of protein, possibly linked to the severity of the disease.
The proteins that had a reduced expression of at least 3-fold included the BEGAIN protein, a novel postsynaptic density component associated with the PSD-95/synapse associated protein 90 [20]. It is found in many neurons, in particular, those of the hippocampus. Its presence in CSF was most likely a result of cellular damage. However, the observation of reduced expression in nonsurvivors suggests that it may have an alternate unknown role. Serotransferrin is an iron-binding transport protein with antibacterial properties [21]. The breakdown of the BBB is the most likely source. However, the down-regulation of this protein suggests an association with iron, which is essential for the nutrient growth of organisms. The ability to acquire iron under low-iron conditions is related to the virulence of a variety of bacterial pathogens. Iron concentration in CSF is low, and S. pneumoniae acquire iron through pneumococcal uptake proteins, such as PiA and PiU. Therefore, it is possible that pneumococci were using transferrin as a source of iron in CSF, leading to its reduction in nonsurvivors [22]. Phosphoglucomutase-like protein 5 is a protein component of adherens-type cell-cell and cell-matrix junctions. It has not been previously identified in the brain; however, PGM-RP is part of this dystrophin–utrophin complex [23]. In the brain, utrophin is present in the choroid plexus epithelium and vascular endothelial cells, whereas the short C-terminal isoform of dystrophin (Dp71) is localized in the glial end-feet, surrounding blood vessels. Both proteins have also been localized in specific types of neurons in the brain and were most likely to be the source of this protein in CSF, either as a result of breakdown of the BBB or direct neuronal damage. Rab 37 is a protein that belongs to the small GTPase superfamily, which regulates cellular functions, including signal transduction, cytoskeletal organization, and membrane trafficking. These proteins are commonly found in endothelial cells that compose the BBB [24]. Therefore, it is most likely that this protein was leaked into the CSF when the BBB disintegrated. Glyoxalase domain-containing protein 4 (GLOD4) is a member of the glyoxalase system, a set of enzymes that play a role in detoxification of methylglyoxal and the other reactive aldehydes produced during metabolism [25]. Methylglyoxal has been shown to induce neurotoxicity through the impairment of the detoxification pathway and depletion of reduced glutathione. Widespread apoptotic cell death occurs as a result, because of the convergence of both mitochondrial and Fas-receptor pathways. Thus, down-regulation of this protein may have contributed to neuronal cell death in nonsurvivors.
Some of the proteins identified may be a result of the breakdown of the BBB and may be present in blood plasma. There were no differences in expression of pneumococcal proteins identified in the nonsurvivors, compared with survivors. Again, this may have been attributable to the high abundance of host proteins in the CSF. Some of the proteins expressed in this analysis appeared to correlate with gene expression data of Coimbra et al [26]. These proteins include Desmin and complement C3. The level of protein in CSF suggest that some of the other gene expression products identified by Coimbra and colleagues may yet be discovered in these CSF samples.
Proteomic approaches allow the analysis of a large number of proteins, compared with other methods. It also allows the identification of proteins that might otherwise be overlooked. This study has illustrated that several thousands of proteins are present in the CSF, and thus, targeted measurements of single markers are unlikely to build up a whole picture capable of explaining the pathogenesis of the disease. 2D PAGE has the ability to inspect multiple proteins at once and, hence, provide potential new targets that may have been overlooked previously. There is potential for the technique to be applied to various time points in the disease. We have shown that the separation of proteins with 2D PAGE and mass spectrometry allows an insight into the proteome of patients with severe meningitis. Such information could lead to the identification of new biomarkers for the disease and possibly novel drug targets. We have also shown that there are protein differences between patient CSF samples from survivors and nonsurvivors. These proteins may form the basis of clinical point assays to assess the presence and activity of inflammation in meningitis, with a particular emphasis on disease monitoring. However, the clinical relevance of the proteins would require testing before the data could have any significance.
Supplementary Material
Acknowledgments
We acknowledge the support of the NIHR BRC in microbial diseases (Dr Gavin Laing) and the NWDA for infrastructural support.
Financial support: The Wellcome Trust (grant number 061231 to S.B.G.) and LSTM Studentship (to U.R.G.).
Footnotes
Potential conflicts of interest: none reported.
References
- 1.Weisfelt M, de GJ, van der PT, van de BD. Pneumococcal meningitis in adults: new approaches to management and prevention. Lancet Neurol. 2006;5(4):332–342. doi: 10.1016/S1474-4422(06)70409-4. [DOI] [PubMed] [Google Scholar]
- 2.van de Beek D, de Gans J, Tunkel AR, Wijdicks EF. Community-acquired bacterial meningitis in adults. N Engl J Med. 2006;354(1):44–53. doi: 10.1056/NEJMra052116. [DOI] [PubMed] [Google Scholar]
- 3.Bermpohl D, Halle A, Freyer D, et al. Bacterial programmed cell death of cerebral endothelial cells involves dual death pathways. J Clin Invest. 2005;115(6):1607–1615. doi: 10.1172/JCI23223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber JR, Tuomanen EI. Cellular damage in bacterial meningitis: an interplay of bacterial and host driven toxicity. J Neuroimmunol. 2007;184(1–2):45–52. doi: 10.1016/j.jneuroim.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Gillespie SH, Balakrishnan I. Pathogenesis of pneumococcal infection. J Med Microbiol. 2000;49(12):1057–1067. doi: 10.1099/0022-1317-49-12-1057. [DOI] [PubMed] [Google Scholar]
- 6.Zysk G, Bethe G, Nau R, et al. Immune response to capsular polysaccharide and surface proteins of Streptococcus pneumoniae in patients with invasive pneumococcal disease. J Infect Dis. 2003;187(2):330–333. doi: 10.1086/367701. [DOI] [PubMed] [Google Scholar]
- 7.Koedel U, Scheld WM, Pfister HW. Pathogenesis and pathophysiology of pneumococcal meningitis 1. Lancet Infect Dis. 2002;2(12):721–736. doi: 10.1016/s1473-3099(02)00450-4. [DOI] [PubMed] [Google Scholar]
- 8.Marriott HM, Dockrell DH. Streptococcus pneumoniae: The role of apoptosis in host defense and pathogenesis. Int J Biochem Cell Biol. 2006;38(11):1848–1854. doi: 10.1016/j.biocel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Yuan X, Desiderio DM. Proteomics analysis of human cerebrospinal fluid. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815(1–2):179–189. doi: 10.1016/j.jchromb.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 10.Scarborough M, Gordon SB, Whitty CJ, et al. Corticosteroids for bacterial meningitis in adults in sub-Saharan Africa 1. N Engl J Med. 2007;357(24):2441–2450. doi: 10.1056/NEJMoa065711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishihara JC, Champion KM. Quantitative evaluation of proteins in one- and two-dimensional polyacrylamide gels using a fluorescent stain. Electrophoresis. 2002;23(14):2203–2215. doi: 10.1002/1522-2683(200207)23:14<2203::AID-ELPS2203>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 12.Armah HB, Wilson NO, Sarfo BY, et al. Cerebrospinal fluid and serum biomarkers of cerebral malaria mortality in Ghanaian children 3. Malar J. 2007;6:147. doi: 10.1186/1475-2875-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DM, Zarate S, Shao H, et al. Pleocytosis is associated with disruption of HIV compartmentalization between blood and cerebral spinal fluid viral populations 1. Virology. 2009;385(1):204–208. doi: 10.1016/j.virol.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isman FK, Kacira T, Kucur M, et al. Cerebrospinal fluid and serum chitotriosidase levels in patients with aneurysmal subarachnoid haemorrhage: preliminary results 2. Turk Neurosurg. 2007;17(4):235–242. [PubMed] [Google Scholar]
- 15.Yuste J, Sen A, Truedsson L, et al. Impaired opsonization with C3b and phagocytosis of Streptococcus pneumoniae in sera from subjects with defects in the classical complement pathway 1. Infect Immun. 2008;76(8):3761–3770. doi: 10.1128/IAI.00291-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaffler A, Scholmerich J, Salzberger B. Adipose tissue as an immunological organ: Toll-like receptors, C1q/TNFs and CTRPs 2. Trends Immunol. 2007;28(9):393–399. doi: 10.1016/j.it.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 17.Brueton MJ, Breeze GR, Stuart J. Fibrin-fibrinogen degradation products in cerebrospinal fluid 1. J Clin Pathol. 1976;29(4):341–344. doi: 10.1136/jcp.29.4.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusuhara H, Sugiyama Y. Active efflux across the blood-brain barrier: role of the solute carrier family. NeuroRx. 2005;2(1):73–85. doi: 10.1602/neurorx.2.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sodin-Semrl S, Rozman B. Beta2-glycoprotein I and its clinical significance: from gene sequence to protein levels 4. Autoimmun Rev. 2007;6(8):547–552. doi: 10.1016/j.autrev.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Yao I, Iida J, Nishimura W, Hata Y. Synaptic and nuclear localization of brain-enriched guanylate kinase-associated protein 2. J Neurosci. 2002;22(13):5354–5364. doi: 10.1523/JNEUROSCI.22-13-05354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenau A, Martins K, Amor S, et al. Evaluation of the ability of Streptococcus agalactiae strains isolated from genital and neonatal specimens to bind to human fibrinogen and correlation with characteristics of the fbsA and fbsB genes 1. Infect Immun. 2007;75(3):1310–1317. doi: 10.1128/IAI.00996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tai SS, Lee CJ, Winter RE. Hemin utilization is related to virulence of Streptococcus pneumoniae 1. Infect Immun. 1993;61(12):5401–5405. doi: 10.1128/iai.61.12.5401-5405.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin S, Almo SC, Satir BH. Functional diversity of the phosphoglucomutase superfamily: structural implications 1. Protein Eng. 1999;12(9):737–746. doi: 10.1093/protein/12.9.737. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Fan XG, Chen R, et al. Comparative proteome analysis of untreated and Helicobacter pylori-treated HepG2 16. World J Gastroenterol. 2005;11(22):3485–3489. doi: 10.3748/wjg.v11.i22.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thornalley PJ. Endogenous alpha-oxoaldehydes and formation of protein and nucleotide advanced glycation endproducts in tissue damage 6. Novartis Found Symp. 2007;285:229–243. doi: 10.1002/9780470511848.ch17. [DOI] [PubMed] [Google Scholar]
- 26.Coimbra RS, Voisin V, de Saizieu AB, et al. Gene expression in cortex and hippocampus during acute pneumococcal meningitis 1. BMC Biol. 2006;4:15. doi: 10.1186/1741-7007-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis VA, Hynes GM, Zheng D, Saibil H, Willison K. T-complex polypeptide-1 is a subunit of a heteromeric particle in the eukaryotic cytosol 1. Nature. 1992;358(6383):249–252. doi: 10.1038/358249a0. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, Chen CF, Baker PR, Chen P, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46(11):3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 29.Baens M, Chaffanet M, Cassiman JJ, Van den BH, Marynen P. Construction and evaluation of a hncDNA library of human 12p transcribed sequences derived from a somatic cell hybrid 4. Genomics. 1993;16(1):214–218. doi: 10.1006/geno.1993.1161. [DOI] [PubMed] [Google Scholar]
- 30.Royal W, III, Gartner S, Gajewski CD. Retinol measurements and retinoid receptor gene expression in patients with multiple sclerosis 1. Mult Scler. 2002;8(6):452–458. doi: 10.1191/1352458502ms858oa. [DOI] [PubMed] [Google Scholar]
- 31.Dass B, McDaniel L, Schultz RA, Attaya E, MacDonald CC. The gene CSTF2T, encoding the human variant CstF-64 polyadenylation protein tauCstF-64, lacks introns and may be associated with male sterility 1. Genomics. 2002;80(5):509–514. [PubMed] [Google Scholar]
- 32.Christian KJ, Lang MA, Raffalli-Mathieu F. Interaction of heterogeneous nuclear ribonucleoprotein C1/C2 with a novel cis-regulatory element within p53 mRNA as a response to cytostatic drug treatment 1. Mol Pharmacol. 2008;73(5):1558–1567. doi: 10.1124/mol.107.042507. [DOI] [PubMed] [Google Scholar]
- 33.Seki N, Hattori A, Muramatsu M, Saito T. cDNA cloning of a human brain finger protein, BFP/ZNF179, a member of the RING finger protein family 4. DNA Res. 1999;6(5):353–356. doi: 10.1093/dnares/6.5.353. [DOI] [PubMed] [Google Scholar]
- 34.Denisenko ON, O’Neill B, Ostrowski J, Van S I, Bomsztyk K. Zik1, a transcriptional repressor that interacts with the heterogeneous nuclear ribonucleoprotein particle K protein 8. J Biol Chem. 1996;271(44):27701–27706. doi: 10.1074/jbc.271.44.27701. [DOI] [PubMed] [Google Scholar]
- 35.Laurino JP, Thompson GM, Pacheco E, Castilho BA. The beta subunit of eukaryotic translation initiation factor 2 binds mRNA through the lysine repeats and a region comprising the C2-C2 motif 1. Mol Cell Biol. 1999;19(1):173–181. doi: 10.1128/mcb.19.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sass-Kuhn SP, Moqbel R, Mackay JA, Cromwell O, Kay AB. Human granulocyte/pollen-binding protein: recognition and identification as transferrin 1. J Clin Invest. 1984;73(1):202–210. doi: 10.1172/JCI111192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassan MI, Waheed A, Yadav S, Singh TP, Ahmad F. Zinc alpha 2-glycoprotein: a multidisciplinary protein 1. Mol Cancer Res. 2008;6(6):892–906. doi: 10.1158/1541-7786.MCR-07-2195. [DOI] [PubMed] [Google Scholar]
- 38.Jung SM, Lee K, Lee JW, et al. Both plasma retinol-binding protein and haptoglobin precursor allele 1 in CSF: candidate biomarkers for the progression of normal to mild cognitive impairment to Alzheimer’s disease 1. Neurosci Lett. 2008;436(2):153–157. doi: 10.1016/j.neulet.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Royds JA, Timperley WR, Taylor CB. Levels of enolase and other enzymes in the cerebrospinal fluid as indices of pathological change 26. J Neurol Neurosurg Psychiatry. 1981;44(12):1129–1135. doi: 10.1136/jnnp.44.12.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark HF, Gurney AL, Abaya E, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment 1. Genome Res. 2003;13(10):2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tappel A. Lysosomal enzymes and initiation of breast cancer 1. Med Hypotheses. 2005;64(2):288–289. doi: 10.1016/j.mehy.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Hemmings BA, ms-Pearson C, Maurer F, et al. Alpha- and beta-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure 3. Biochemistry. 1990;29(13):3166–3173. doi: 10.1021/bi00465a002. [DOI] [PubMed] [Google Scholar]
- 43.Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer 2. Cancer Metastasis Rev. 2008;27(2):253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- 44.Prasadarao NV, Wass CA, Stins MF, Shimada H, Kim KS. Outer membrane protein A-promoted actin condensation of brain microvascular endothelial cells is required for Escherichia coli invasion 1. Infect Immun. 1999;67(11):5775–5783. doi: 10.1128/iai.67.11.5775-5783.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niebroj-Dobosz I, Dziewulska D, Janik P. Auto-antibodies against proteins of spinal cord cells in cerebrospinal fluid of patients with amyotrophic lateral sclerosis (ALS) 1. Folia Neuropathol. 2006;44(3):191–196. [PubMed] [Google Scholar]
- 46.Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution 2. Protein Sci. 2004;13(7):1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greene C, Taggart C, Lowe G, Gallagher P, McElvaney N, O’Neill S. Local impairment of anti-neutrophil elastase capacity in community-acquired pneumonia 1. J Infect Dis. 2003;188(5):769–776. doi: 10.1086/377238. [DOI] [PubMed] [Google Scholar]
- 48.Warton K, Foster NC, Gold WA, Stanley KK. A novel gene family induced by acute inflammation in endothelial cells 1. Gene. 2004;342(1):85–95. doi: 10.1016/j.gene.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 49.Zunszain PA, Ghuman J, Komatsu T, Tsuchida E, Curry S. Crystal structural analysis of human serum albumin complexed with hemin and fatty acid 3. BMC Struct Biol. 2003;3:6. doi: 10.1186/1472-6807-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho B, Lim Y, Lee DY, et al. Identification and characterization of a novel cancer/testis antigen gene CAGE. Biochem Biophys Res Commun. 2002;292(3):715–726. doi: 10.1006/bbrc.2002.6701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.