Abstract
In the dividing eukaryotic cell the spindle assembly checkpoint (SAC) ensures each daughter cell inherits an identical set of chromosomes. The SAC coordinates the correct attachment of sister chromatid kinetochores to the mitotic spindle with activation of the anaphase-promoting complex/cyclosome (APC/C), the E3 ubiquitin ligase that initiates chromosome separation. In response to unattached kinetochores, the SAC generates the mitotic checkpoint complex (MCC), a multimeric assembly that inhibits the APC/C, delaying chromosome segregation. Here, using cryo-electron microscopy we determined the near-atomic resolution structure of an APC/C-MCC complex (APC/CMCC). We reveal how degron-like sequences of the MCC subunit BubR1 block degron recognition sites on Cdc20, the APC/C coactivator subunit (Cdc20APC/C) responsible for substrate interactions. BubR1 also obstructs binding of UbcH10 (APC/C’s initiating E2) to repress APC/C ubiquitination activity. Conformational variability of the complex allows for UbcH10 association, and we show from a structure of APC/CMCC in complex with UbcH10 how the Cdc20 subunit intrinsic to the MCC (Cdc20MCC) is ubiquitinated, a process that results in APC/C reactivation when the SAC is silenced.
The fidelity of chromosome separation at each cell division cycle ensures the inheritance of the correct complement of genetic material in successive generations of cells. The APC/C initiates sister chromatid separation by controlling the proteasomal degradation of securin and cyclin B 1,2. Their degradation allows separase to remove sister chromatid cohesin. Critical to the maintenance of chromosome integrity of dividing cells is the SAC 3,4. The SAC responds to unattached kinetochores by generating the MCC which functions to suppress APC/C-catalysed ubiquitination of securin and cyclin B.
Although the components of the SAC machinery are known 5,6, and some details of the molecular events that sense the absence of kinetochore attachment (and possibly intra-kinetochore tension) to signal MCC assembly have been characterized, important questions remain 4. Intrinsic to this process is the conversion of O-Mad2 to C-Mad2 catalysed by a C-Mad2-Mad1 complex at unattached kinetochores 7–9. Soluble C-Mad2 engages the N-terminus of Cdc20 (refs 10,11), the mitotic activating subunit of the APC/C, which then binds the BubR1-Bub3 dimer to form the MCC 12. Mad2 and BubR1 interact cooperatively with Cdc20 (refs 9,13–18), and synergistically inhibit the APC/C in mitosis 14,19.
In an important advance, it was proposed 2, and shown 20, that the tetrameric MCC inhibits the APC/C already in complex with Cdc20 (the regulatory subunit that recognizes D box, KEN box and ABBA motif degrons of APC/C substrates and that promotes the catalytically active conformation of the APC/C [APC/CCdc20]) 21,22. In addition to inhibiting the APC/C, the MCC contributes to APC/C reactivation after SAC silencing through proteasome-catalysed Cdc20 degradation 23,24. SAC-mediated Cdc20 proteolysis is APC/C, Mad2 and BubR1/Mad3-dependent 17,23–27, suggesting that Cdc20 ubiquitination occurs in the context of the APC/CMCC, an idea supported by findings that release from mitotic arrest, concomitant with Cdc20 destruction, requires the small APC/C subunit Apc15 (refs 28–30).
To obtain insights into reciprocal APC/C and MCC regulation, we reconstituted recombinant APC/CMCC and APC/CMCC with UbcH10 (APC/C’s initiating E2) complexes for structural and biochemical analysis. From a cryo-EM reconstruction of the APC/CMCC, we identify conformational variability of the complex that explains its capacity to repress substrate ubiquitination, but also allows for UbcH10 to catalyse intramolecular Cdc20MCC ubiquitination.
Reconstitution and overall features of APC/CMCC
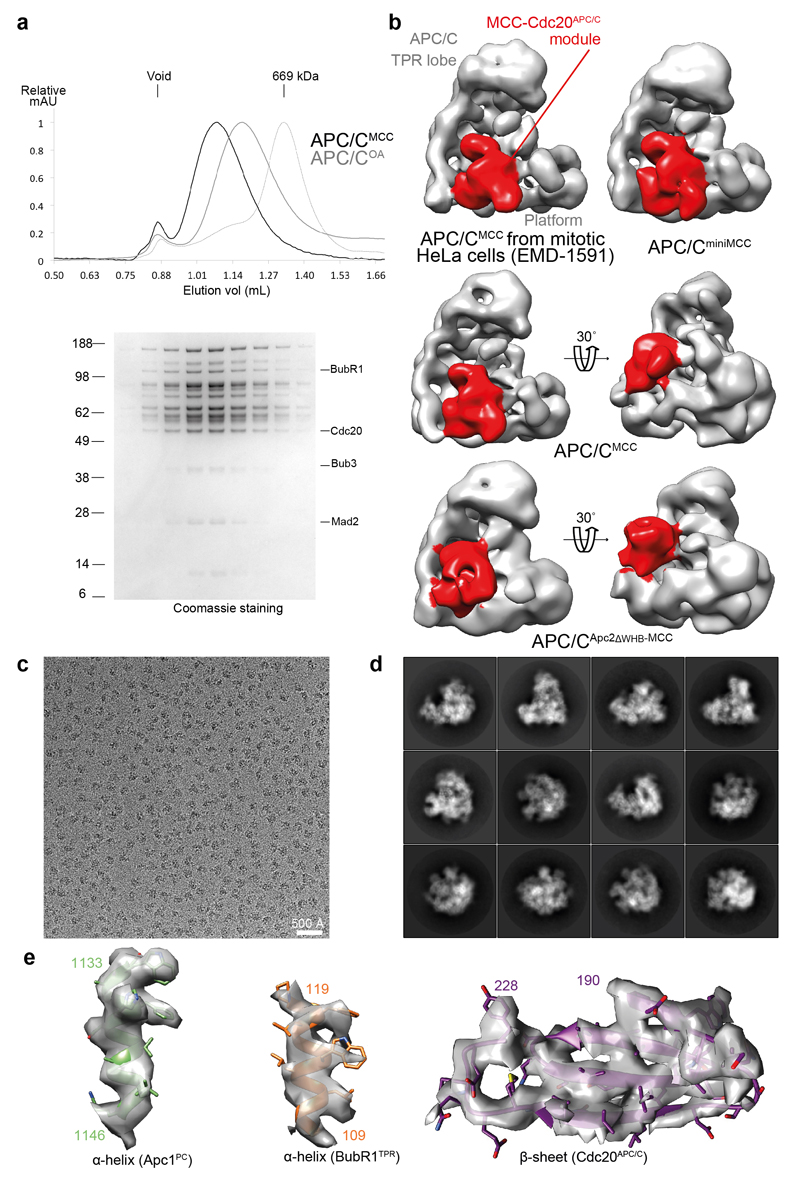
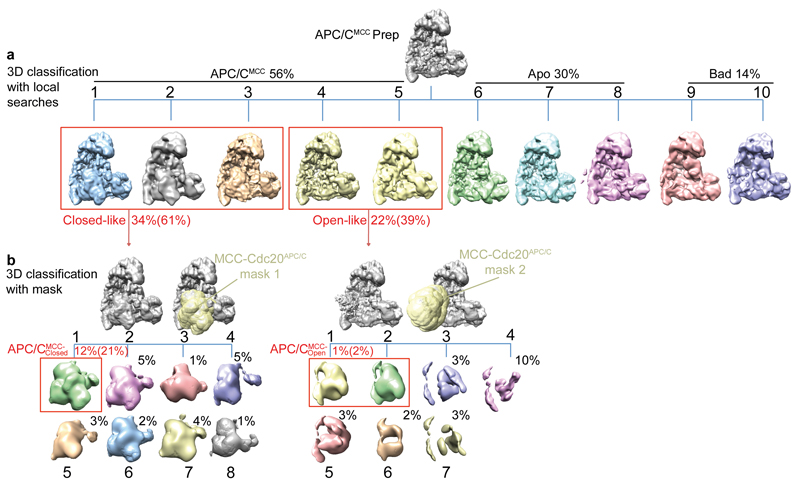
We reconstituted recombinant APC/CMCC using the insect cell/baculovirus expression system. Recombinant APC/CMCC incorporates two distinct Cdc20 subunits, termed Cdc20APC/C for the APC/CCdc20-associated subunit, and Cdc20MCC for the MCC-associated subunit (Extended Data Fig. 1a, j and Extended Data Table 1), consistent with 2,20. We determined negative stain and cryo-EM reconstructions of the APC/CMCC complex (Extended Data Table 2). The negative stain EM reconstruction of the recombinant APC/CMCC is essentially identical in structure to endogenous APC/CMCC isolated from checkpoint-arrested HeLa cells determined at a similar resolution 31 (Extended Data Fig. 2b). This substantiates the model that the physiological form of APC/CMCC includes two Cdc20 subunits 2,20. Both reconstructions feature a large density element termed the MCC-Cdc20APC/C module (MCC interacting with the Cdc20APC/C subunit of APC/CCdc20) occupying APC/C’s central cavity, extending from the “front” side of the platform domain (Extended Data Fig. 2b).
To understand quantitatively how the MCC interacts with APC/CCdc20, we determined a cryo-EM reconstruction of APC/CMCC at near-atomic resolution (Fig. 1a, b and Extended Data Figs 2c-e and 3 and Extended Data Table 2). Extensive 3D classification of APC/CMCC revealed conformational variability of the MCC-Cdc20APC/C module. This module adopts a stable rigid conformation in 21% of APC/CMCC particles (defined as APC/CMCC-Closed) (Extended Data Fig. 4b, class 1). A local resolution map of APC/CMCC-Closed shows that the central rigid regions are at 3.9 to 4.1 Å resolution, with the flexible outer regions at lower resolution (Extended Data Fig. 3c, d). We built an atomic model of APC/CMCC-Closed (Fig. 1 and Supplementary Video 1) guided by the cryo-EM structure of human APC/CCdh1.Emi1 (ref. 32) and the crystal structure of fission yeast MCC 18.
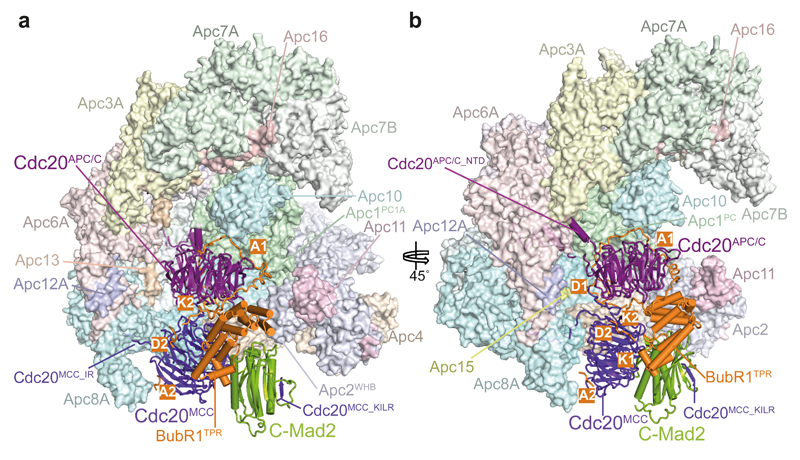
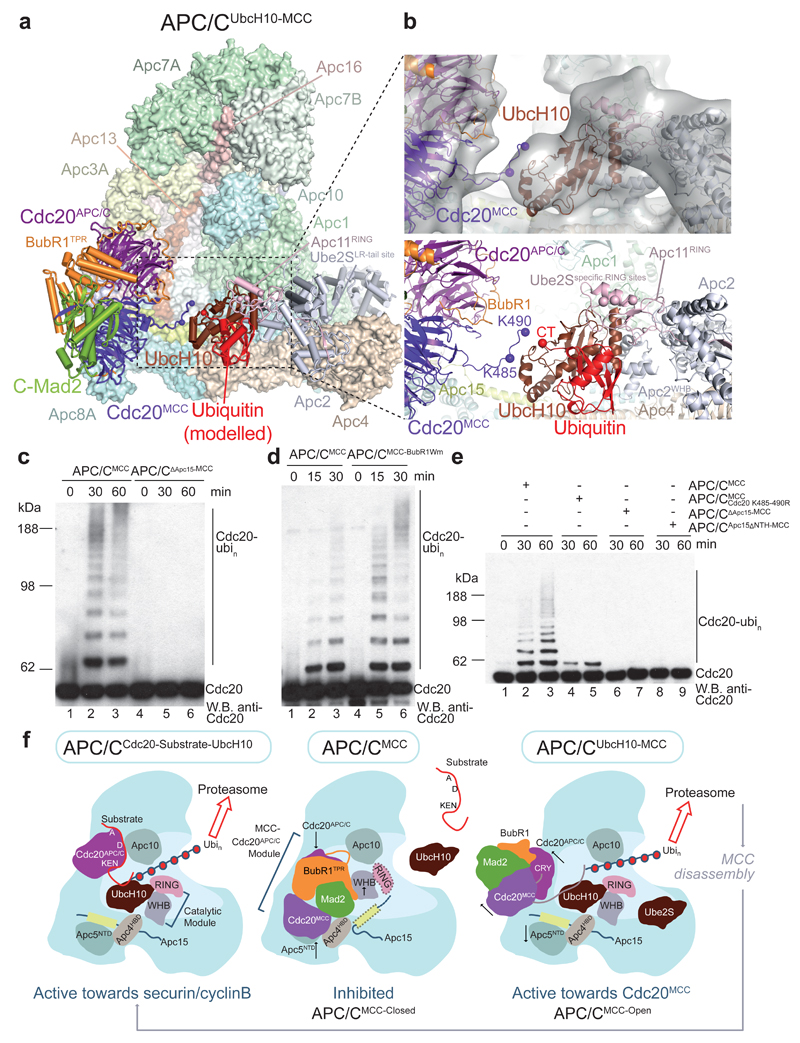
Figure 1. Overall structure of the APC/CMCC complex.
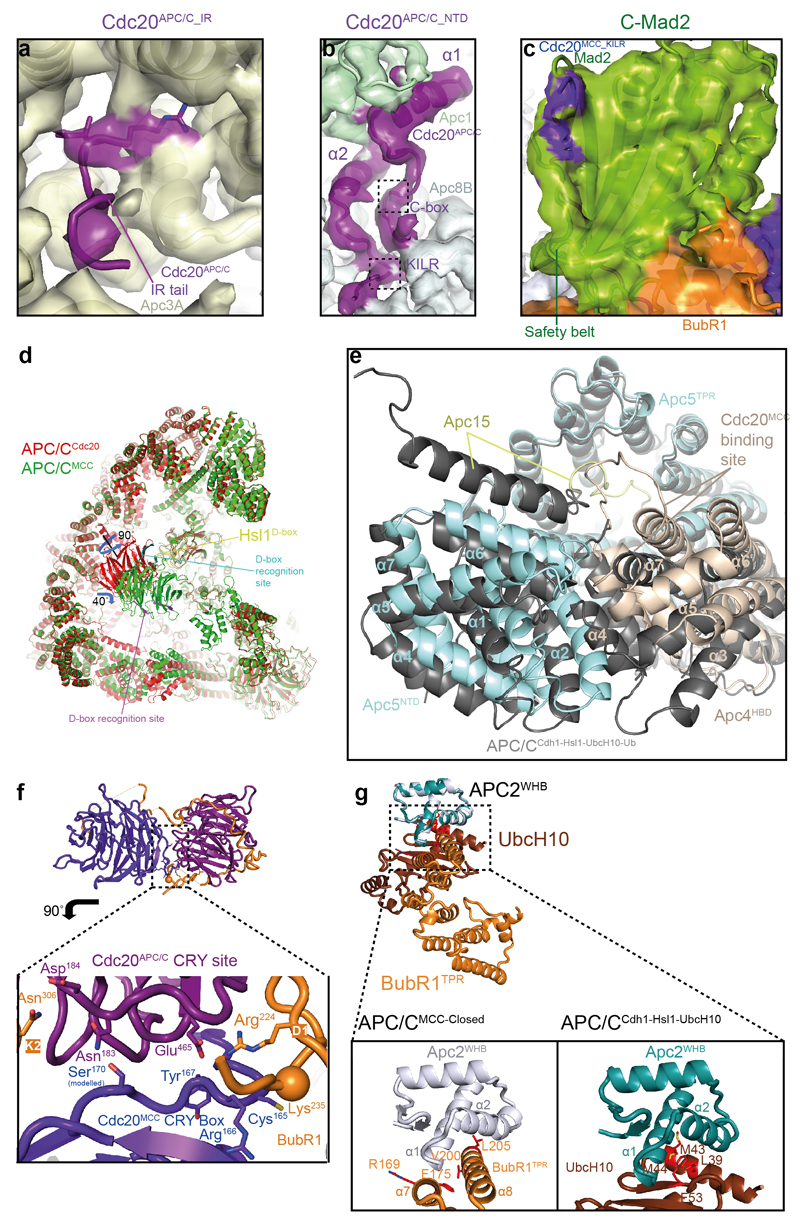
a, b. Two views of APC/CMCC. The MCC-Cdc20APC/C module is shown as a cartoon and the APC/C in a surface representation. BubR1 forms extensive contacts with Cdc20APC/C and Apc2. BubR1 inhibitory degrons visible in these views are highlighted.
In the APC/CMCC-Closed reconstruction, the secondary structure elements of the MCC-Cdc20APC/C module are clearly visible (Extended Data Fig. 2e), revealing that Cdc20APC/C, Cdc20MCC, Mad2 and the N-terminal and middle regions of BubR1, including its tetratricopeptide (TPR) domain (BubR1TPR) and its inhibitory degrons, are well defined (Figs 1a, b and 2a). However, despite their presence in reconstituted APC/CMCC (Extended Data Figs 1a and 2a), the C-terminal pseudo-kinase domain of BubR1 (Fig. 2a) and Bub3, were not visible in EM density, indicative of conformational variability. In agreement with this, both the structure and activity of APC/CMCC with Bub3 and the BubR1 C-terminus deleted (APC/CminiMCC) are indistinguishable from APC/CMCC (Extended Data Figs 1b, h, i and 2b). These data are consistent with the requirement of BubR1’s N-terminal ~363 residues (Fig. 2a) to sustain a SAC 33–36.
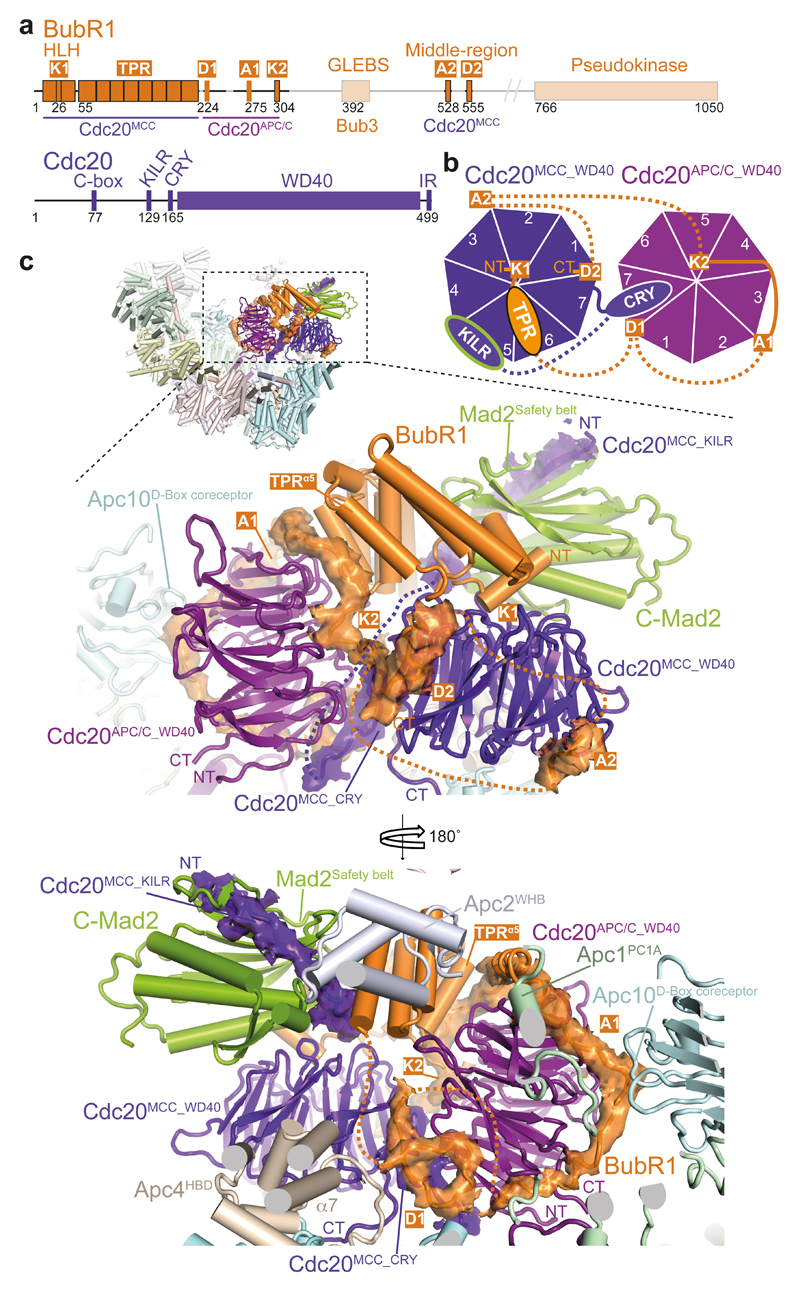
Figure 2. Interactions of BubR1 with Cdc20APC/C and Cdc20MCC.
a, Schematic of BubR1 and Cdc20. b, Schematic representation of the top views of the Cdc20APC and Cdc20MCC WD40 domains. WD40 domain blades are numbered and the positions of BubR1 inhibitory degrons (orange) are indicated. The CRY degron 49 mediates Cdc20MCC interactions with Cdc20APC/C (Extended Data Fig. 5f). c, Two views showing details of the MCC-Cdc20APC/C module. Cryo-EM density of the BubR1 inhibitory degrons, Cdc20MCC CRY box and KILR motif is shown. Interactions of the BubR1 A1 motif with the Apc10D-box coreceptor and Apc1; BubR1TPR with Apc2WHB; and Cdc20MCC with Apc4HBD are indicated (lower panel). Inset: overall view of APC/CMCC.
General architecture of APC/CMCC
In APC/CMCC, the MCC core elements comprising Cdc20MCC, BubR1TPR, and Mad2 resemble their counterparts in S. pombe MCC 18 (Fig. 1 and Supplementary Video 1). Mad2 adopts the closed conformation (C-Mad2) with its C-terminal segment (‘safety belt’) locking the tubular density of the Cdc20MCC KILR motif 10,11 (Figs 1 and 2c and Extended Data Fig. 5c). The MCC docks into the APC/CCdc20 cavity directly below Cdc20APC/C and interacts with Cdc20APC/C such that the two Cdc20 WD40 domains of APC/CMCC are arranged in an almost perpendicular fashion (Figs 1 and 2c). Cdc20MCC and BubR1 mediate interactions between the MCC and APC/CCdc20, with BubR1 dominating these interactions through its contacts to Cdc20APC/C and Apc2. C-Mad2 forms no direct contacts to APC/CCdc20 (Fig. 1). However, by stabilizing BubR1’s association with Cdc20MCC, C-Mad2 indirectly augments BubR1 interactions with APC/CCdc20 (refs 14,19,35). APC/CMCC-Closed is similar in structure to APC/C-coactivator complexes 22,32, with major conformational differences confined to Cdc20APC/C and the platform subunits Apc4, Apc5 and Apc15 (Extended Data Fig. 5d, e).
Contacts between the two Cdc20 subunits of APC/CMCC-Closed are mainly mediated by BubR1 that intertwines between them (Fig. 2c). In response to MCC binding, the WD40 domain of Cdc20APC/C is tilted by ~40° and rotated 90° about its central axis (Extended Data Fig. 5d) 22. This disrupts the D box-binding site formed by the interface of the D box co-receptors of Cdc20APC/C-WD40 and Apc10 (refs 21,32), and pulls the C-terminal IR tail of APC/CCdc20 away from the IR-tail binding site of Apc3A. A disengagement of the Cdc20APC/C IR tail from Apc3A is consistent with weak EM density at the IR tail-binding site (Extended Data Fig. 5a), and the finding that Cdc20 does not require Apc3 to bind the APC/C when the SAC is active 37. In contrast to the IR tail, the N-terminal domain of Cdc20APC/C maintains the same interactions with Apc8B and Apc1PC as seen in APC/CCdc20 (ref. 22) (Fig. 3a and Extended Data Fig. 5b).
Figure 3. Interactions of the MCC-Cdc20APC/C module with the APC/C and APC/C catalytic inhibition by the MCC.
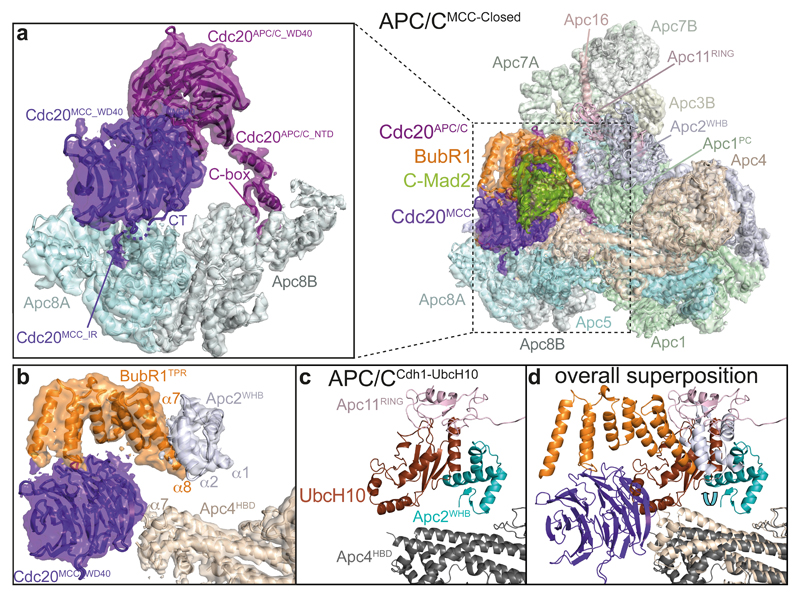
a, Right: an overview of the APC/CMCC model with the corresponding cryo-EM density. Left: segmented cryo-EM density of the Apc8 dimer and its two associated Cdc20 molecules. Cdc20APC/C interacts with Apc8B via its N-terminal domain (NTD). Cdc20MCC interacts with Apc8A through its C-terminal Ile Arg (IR) tail. b, c, d, Comparison of the binding mode of BubR1 and Cdc20MCC_WD40 in APC/CMCC with the binding mode of UbcH10 in APC/CCdh1-UbcH10-Ub (ref. 32). b, Segmented cryo-EM density of Cdc20MCC, BubR1TPR, Apc4HBD and Apc2WHB. (c) APC/CCdh1-UbcH10-Ub. (d) Both structures were superposed. BubR1TPR and Cdc20MCC_WD40 compete for the same binding surfaces on Apc2WHB and Apc4HBD that form the UbcH10 binding site in APC/CCdh1-UbcH10-Ub (Extended Data Fig. 5g). The Apc2WHB sub-domain of Apc2 is shifted in the APC/CMCC complex relative to APC/CCdh1-UbcH10-Ub and would clash with the UbcH10-binding site on Apc11RING.
Degron motifs of BubR1 interact with both Cdc20 subunits of APC/CMCC
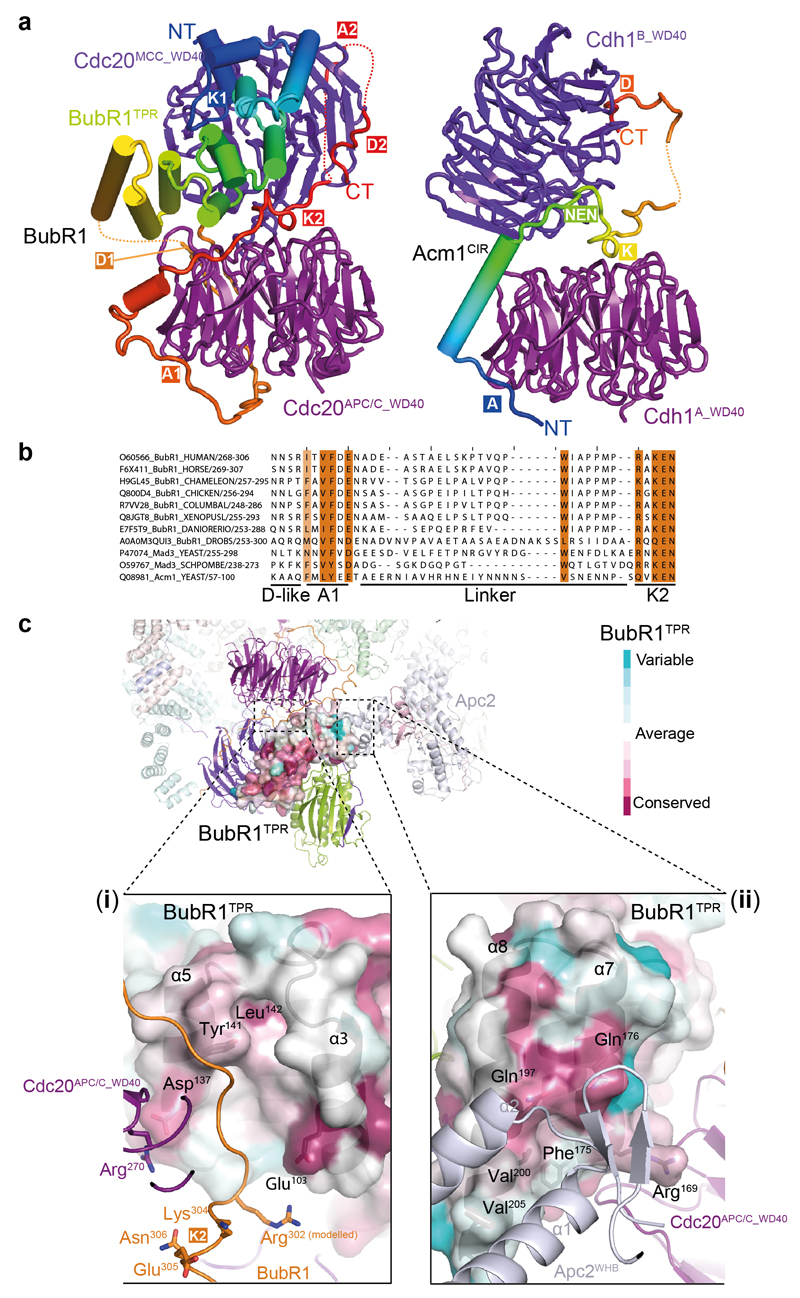
Similar to yeast MCC 18, in APC/CMCC, the N-terminal KEN-1 box (K1) (Fig. 2a and Supplementary Video 1), engages the KEN box-recognition site of the Cdc20MCC (Fig. 2b, c). Immediately following BubR1TPR and preceding KEN-2 (K2) is the N-terminal D box motif (D1) 20 (Fig. 2a). We assigned D1 to the loop-like density at the D-box recognition site of Cdc20APC/C, although due to the re-orientation of Cdc20APC/C in APC/CMCC, its position is diametrically opposed to its location in active APC/C-coactivator complexes (Extended Data Fig. 5d) 21,22,32. D1 and its flanking residues mediate the interface of Cdc20APC/C and Cdc20MCC (Figs 1 and 2c lower panel). This explains how mutating D1 disrupts MCC binding to a second Cdc20 subunit without affecting MCC integrity 20. By engaging the Cdc20APC/C D-box co-receptor in APC/CMCC, D1 would obstruct D box-dependent substrate recognition, consistent with its requirement for a checkpoint arrest 20.
EM density immediately C-terminal to D1 is disordered (Fig. 2c, lower panel) however, uninterrupted tubular density situated nearby and assigned to BubR1 is clearly defined wrapping around the opposite side of the Cdc20APC/C WD40 domain, connecting its bottom and top surfaces and contacting both the Cdc20APC/C KEN and ABBA motif-binding sites (labelled A1 and K2 in Fig. 2c). This BubR1-EM density feature bears a striking resemblance to the structure of Acm1 (APC/C-Cdh1-modulator 1), an inhibitor of yeast Cdh1 that utilizes ABBA, D box and KEN motifs to block the degron-recognition sites of Cdh1 (ref. 38) (Extended Data Fig. 6a). The BubR1 EM density contacting the top surface of Cdc20APC/C corresponds to the KEN-2 box (K2) (Fig. 2c, top panel). Guided by the Cdh1-Acm1 crystal structure 38, we modelled the KEN-2 box and the preceding ABBA motif (A1) (Fig. 2a and Extended Data Fig. 6a-c), to the KEN box and ABBA motif recognition sites of Cdc20APC/C (Fig. 2c).
The KEN-2 motif is invariant in BubR1 orthologs (Extended Data Fig. 6b), and although not required for MCC assembly 16,18,20,26,36,39, is essential for a spindle checkpoint arrest 16,26,36,39. Thus, similar to D1, a role for KEN-2 in mediating MCC interactions with APC/CCdc20 would explain its requirement in the SAC through stabilizing MCC interactions with Cdc20APC/C (ref. 20) and inhibiting degron recognition by APC/CCdc20.
A middle segment of metazoan BubR1 includes an ABBA motif and a D box (A2 and D2, respectively) (Fig. 2a) that mediate Cdc20-BubR1 interactions in a Mad2-independent manner 15,19,40–43. These motifs contribute weakly to sustaining the SAC 15,41,42, and play a role to recruit Cdc20 to unattached kinetochores 40–42. The tubular EM densities located at the ABBA motif and D-box recognition sites of Cdc20MCC were assigned to these motifs (Fig. 2c, top panel).
Cdc20MCC contacts to the APC/C
Although Cdc20MCC interactions with the APC/C are mainly mediated through BubR1 and APC/CCdc20, two additional contacts are notable. First, the C-terminal IR tail of Cdc20MCC binds to a site on Apc8A that is structurally equivalent to the Cdc20APC/C C-box binding site on Apc8B 32 (Fig. 3a and Supplementary Video 1). This interaction can be rationalized from the similarities in how the Apc3-IR tail and Apc8-C box binding sites interact with their cognate IR tail and C box motifs, respectively 32. Second, the lower surface of the Cdc20MCC WD40 domain interacts with the conserved acidic region of the Apc4 helix-bundle domain (Apc4HBD), close to the UbcH10 binding site 32,44 (Figs 2c and 3a, b).
The MCC suppresses APC/C E3 ligase activity
UbcH10 interacts with the APC/C through the RING domain of Apc11 and the winged-helix B (WHB) domain of Apc2 (Apc2WHB) 32,44. In APC/CMCC-Closed, BubR1TPR occludes both E2-binding sites (Figs 1 and 3). BubR1TPR interacts directly with the UbcH10 interface of Apc2WHB that repositions to contact BubR1TPR (Fig. 3b-d and Extended Data Figs 5g and 6c) 32,44. Thus, in APC/CMCC-Closed, the steric occlusion of UbcH10 binding would inhibit ubiquitin chain synthesis.
Apc15-dependent conformational flexibility of the MCC
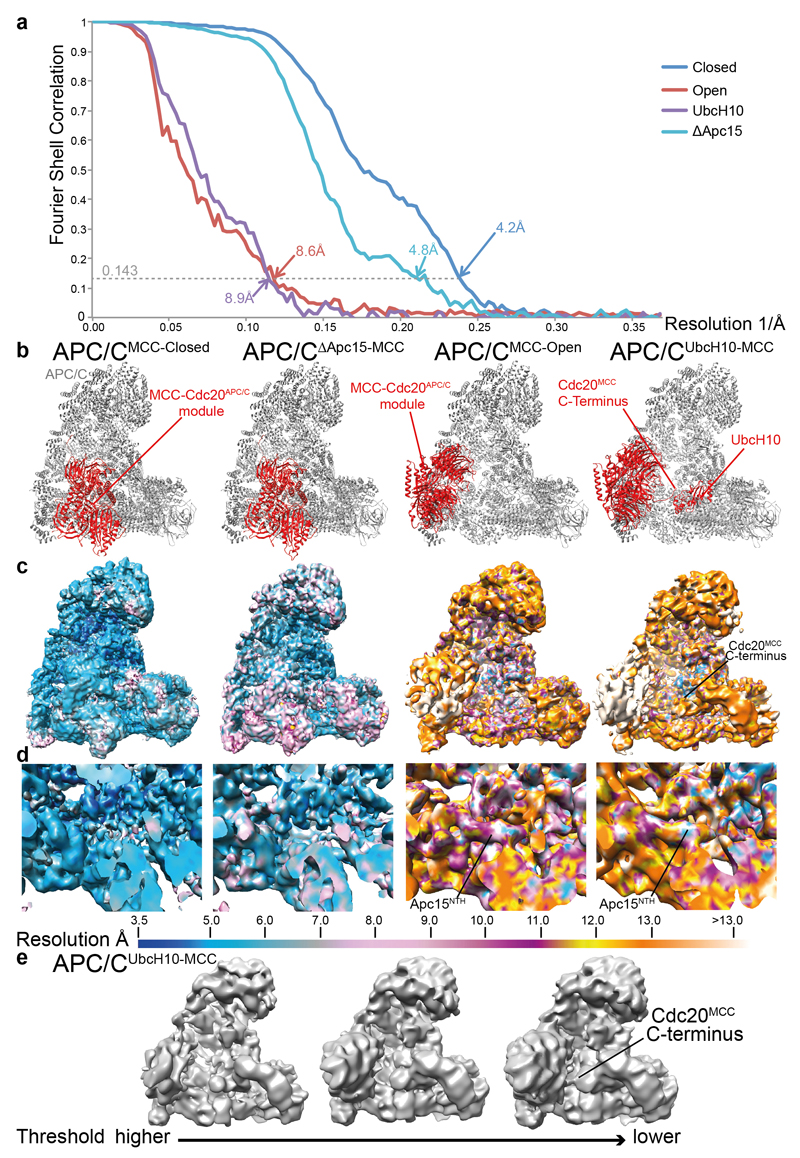
The APC/CMCC-Closed structure explains how the MCC inhibits APC/CCdc20 through obstructing substrate and UbcH10 interactions. However, Cdc20 auto-ubiquitination is the trigger for the spontaneous reactivation of the APC/C 23–29,45, dependent on the small APC/C subunit Apc15 (refs 28,29). In previously reported APC/C structures 22,32, Apc15 adopts an extended conformation, anchored to Apc5 by its N-terminus, and bridging Apc5 and Apc8A through its adjacent N-terminal helix (Apc15NTH). Strikingly, in APC/CMCC-Closed, Apc15NTH is structurally disordered. This results from Cdc20MCC shifting the tip of Apc4HBD that concomitantly repositions the adjacent N-terminal domain of Apc5 (Apc5NTD), disrupting its interaction with Apc15NTH (Fig. 4a, b and Extended Data Fig. 5d, e and Supplementary Video 2). The disorder of Apc15NTH and repositioning of Apc4HBD and Apc5NTD in APC/CMCC-Closed contrasts to their conformations in another structural state representing only 2% of APC/CMCC particles (termed APC/CMCC-Open) (Methods and Extended Data Fig. 4a, b and Extended Data Table 2). In APC/CMCC-Open, EM density assigned to the MCC-Cdc20APC/C module is weaker relative to APC/CMCC-Closed. However, EM density for Apc15NTH is clearly defined (Fig. 4e, f). Moreover, in APC/CMCC-Open, MCC-Cdc20APC/C is shifted towards APC/C’s TPR lobe, disrupting contacts between the MCC and the catalytic module, Apc4HBD and Apc10. The ‘open’ position of MCC-Cdc20APC/C is stabilized by Apc15NTH interacting with Apc5NTD that pushes onto the Cdc20MCC-binding site of Apc4HBD (Fig. 4f and Extended Data Fig. 5e). Thus, the stability of Apc15NTH influences the transition between open and closed conformations of the MCC-Cdc20APC/C module.
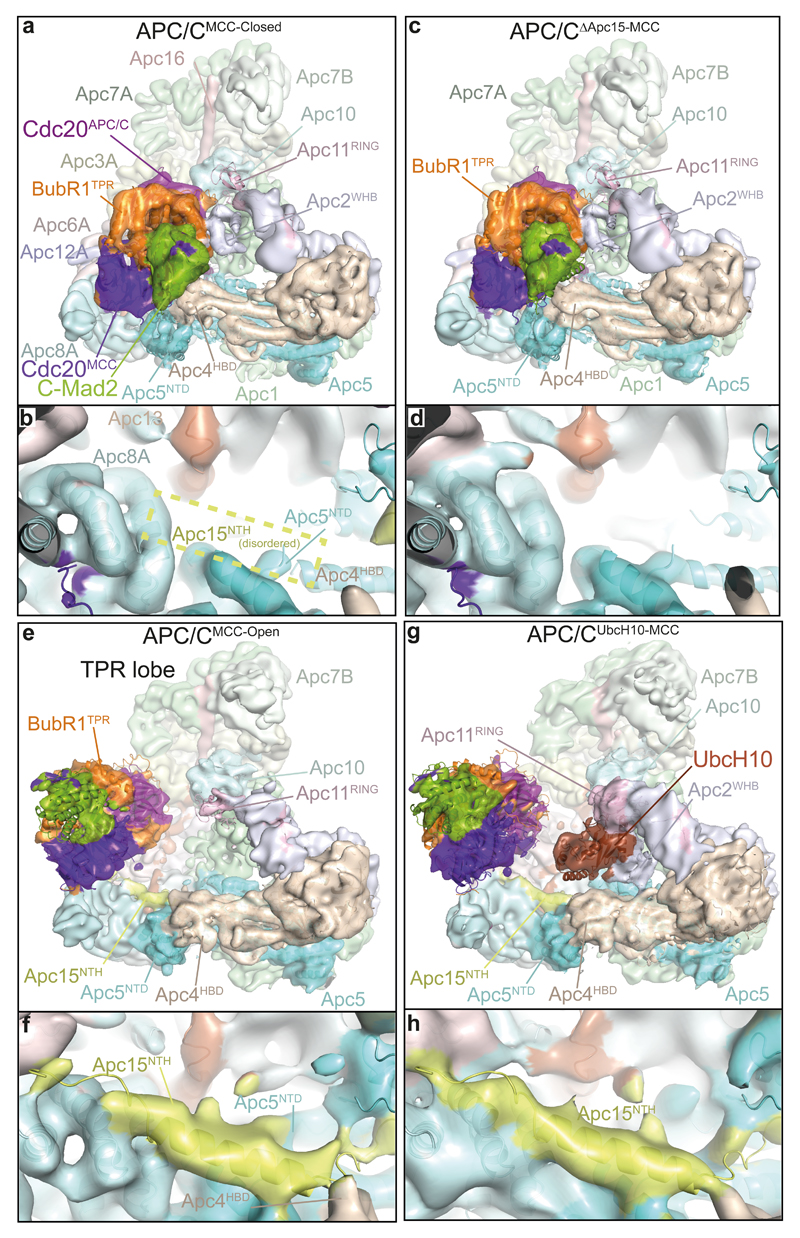
Figure 4. Cryo-EM structures of APC/CMCC-Open, APC/CΔApc15-MCC, APC/CUbcH10-MCC and comparison with APC/CMCC-Closed.
a, Top: overall view of the cryo-EM density of APC/CMCC-Closed and fitted coordinates for the MCC-Cdc20APC/C module, Apc2WHB and Apc11. The APC/CMCC subunits are coloured as in Figs 1 and 3. b, details of the Apc15NTH-binding site on Apc8A and Apc5. Apc8A and Apc5 are shown. The position of the disordered Apc15NTH is indicated by a box. c, d, APC/CΔApc15-MCC (Apc15 deleted). e, f, APC/CMCC-Open and g, h, the APC/CUbcH10-MCC complex. Shown in (a, b), in APC/CMCC-Closed, BubR1TPR interacts with Apc2WHB, and Apc15NTH is disordered. (c, d) In APC/CΔApc15-MCC the MCC-Cdc20APC/C adopts the closed conformation, blocking the catalytic module. Conversely in APC/CMCC-Open (e, f) and APC/CUbcH10-MCC (g, h), BubR1TPR no longer interacts with Apc2WHB, and Apc15NTH is ordered. (g) In APC/CUbcH10-MCC, Apc2WHB and Apc11RING interact with UbcH10. All cryo-EM reconstructions were filtered to 8.5 Å.
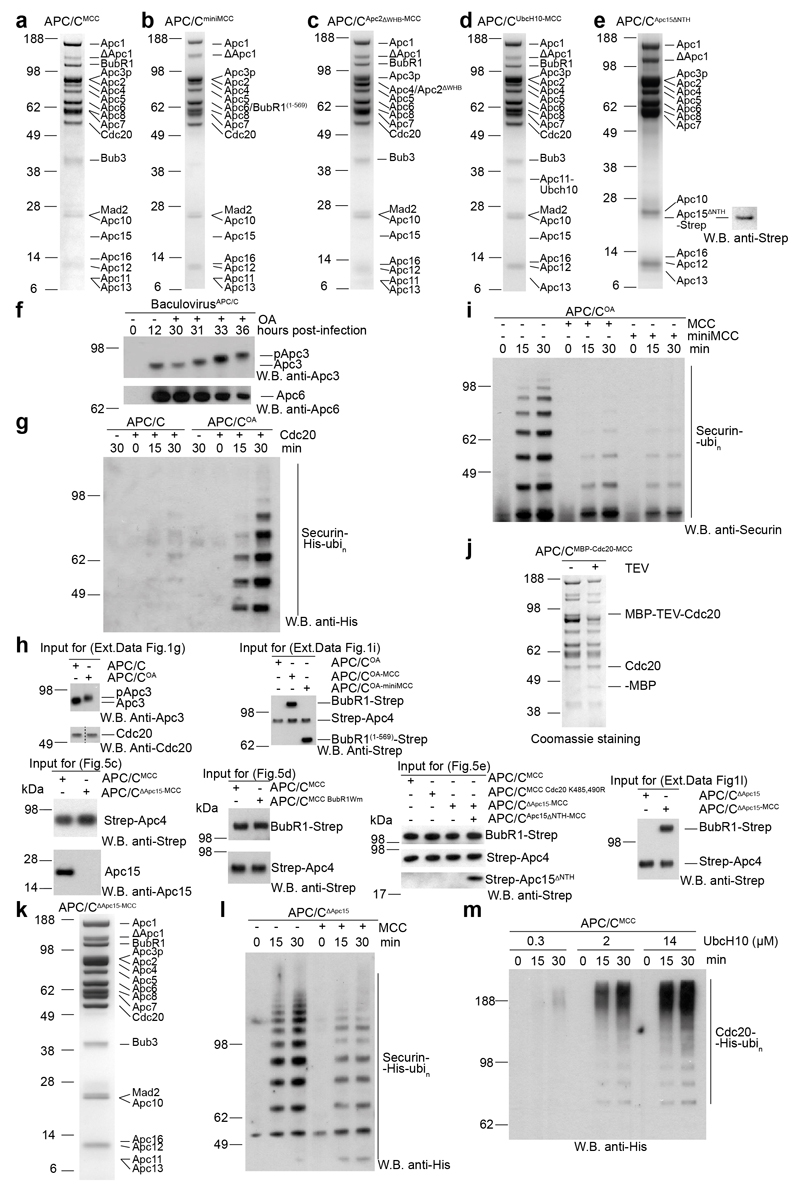
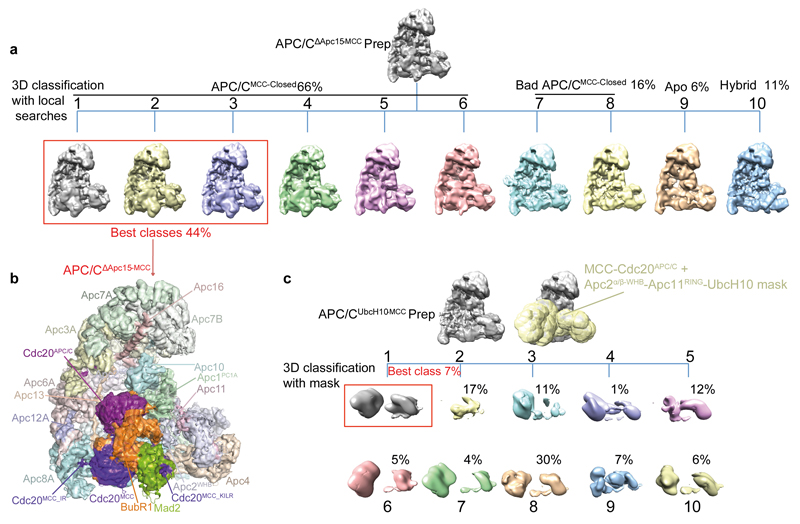
To further investigate how Apc15 modulates the position of MCC-Cdc20APC/C, we determined the structure of APC/CMCC with Apc15 deleted (APC/CΔApc15) (Extended Data Table 2 and Fig. 4c, d and Extended Data Fig. 3). APC/CΔApc15 assembled similarly to wild type APC/C and was catalytically active towards securin (Extended Data Fig. 1k-l). However, in contrast to APC/CMCC, the reconstituted APC/CΔApc15-MCC was defective for Cdc20 auto-ubiquitination, consistent with 28,29 (Fig. 5c and Extended Data Fig. 1h). 3D classification of the APC/CΔApc15-MCC cryo-EM data set showed that the MCC-Cdc20APC/C module adopts only the closed configuration (Fig. 4c, d and Extended Data Fig. 7b and Supplementary Video 2), with no evidence of APC/CMCC-Open (Extended Data Fig. 7a, classes 1 to 6). Thus, the absence of Apc15 promotes APC/CMCC-Closed, explaining why Apc15 is necessary for Cdc20 ubiquitination. In support of our structural data that Apc15NTH is required to stabilize APC/CMCC-Open, similar to APC/CΔApc15-MCC, APC/CMCC reconstituted with Apc15 lacking the Apc15NTH (Apc15ΔNTH), is defective in Cdc20 auto-ubiquitination (Fig. 5e, lanes 8 & 9 and Extended Data Fig. 1h).
Figure 5. Mechanism of Cdc20 auto-ubiquitination by APC/CUbcH10-MCC.
a, b, Model of a Cdc20MCC ubiquitination complex based on the APC/CUbcH10-MCC cryo-EM reconstruction. The UbcH10-ubiquitin conjugate is modelled in the closed conformation 50. b, Top: cryo-EM density and model of APC/CUbcH10-MCC. The EM map is filtered to 12 Å and displayed at slightly lower threshold than in Fig. 4g, see Extended Data Fig. 3e, for comparisons. Clear EM density connects Cdc20MCC with UbcH10. Bottom: The Cdc20MCC pre-ubiquitination model. Cdc20MCC residues Lys485 and Lys490, ubiquitinated in logarithmic and checkpoint-arrested cells, respectively 30, are in close proximity to the UbcH10 catalytic site (red sphere). c, Apc15 is required for Cdc20 ubiquitination by recombinant APC/CMCC. d, BubR1Wm mutations at the Apc2WHB interface (R169A, F175A,V200A, L205) (Extended Data Fig. 5g) stimulates Cdc20 ubiquitination. e, Cdc20 residues Lys485 and Lys490 are ubiquitinated by recombinant APC/CMCC (compare lanes 2,3 and 4,5). Apc15NTH is required for Cdc20 ubiquitination by recombinant APC/CMCC (lanes 8,9). f, cartoon illustrating reciprocal regulation of APC/C and MCC by APC/CMCC. In APC/CMCC-Closed, MCC inhibits substrate (e.g. securin and cyclin B) and UbcH10 recognition. Cyclin A and Nek2A can bypass the SAC. In APC/CMCC-Open, the UbcH10-binding site is exposed. Ube2S elongates Ub-conjugates initiated by UbcH10. Experiments in c-e were replicated three times. See Supplementary Fig. 1 for gel source data.
Since interactions between Apc2WHB and BubR1 stabilize APC/CMCC-Closed (Figs 1 and 3b), disrupting this interface should favour APC/CMCC-Open. Consistent with this idea, negative stain EM reconstructions of an APC/CMCC mutant with Apc2WHB deleted showed APC/CMCC adopting the open conformation, with no APC/CMCC-Closed (Extended Data Figs 1c and 2b). Importantly, in the complementary Apc2WHB-binding surface mutant of BubR1 (Extended Data Fig. 5g), Cdc20MCC auto-ubiquitination is stimulated (BubR1Wm, Fig. 5d and Extended Data Fig. 1h).
Mechanism of Cdc20MCC auto-ubiquitination from an APC/CUbcH10-MCC structure
Cdc20MCC auto-ubiquitination catalysed by APC/CMCC-UbcH10 is an intra-molecular process reliant on the APC/CMCC-Open conformation. To further explore this possibility, we determined the cryo-EM structure of APC/CMCC in complex with UbcH10 (APC/CUbcH10-MCC) (Extended Data Table 2). A 3D reconstruction using 7% of total particles showed clear EM density for both UbcH10 and the MCC-Cdc20APC/C module (Extended Data Fig. 7c). The the resultant EM map at 8.9 Å resolution allowed rigid body docking of the APC/C, UbcH10 and MCC-Cdc20APC/C coordinates. APC/CUbcH10-MCC resembles APC/CMCC-Open (Figs 4e-h and 5a and Extended Data Fig. 3 and Supplementary Video 2). Apc15NTH is ordered and MCC-Cdc20APC/C is rotated towards the TPR lobe leaving the catalytic module accessible to bind UbcH10 (Fig. 5a).
Strikingly, in APC/CUbcH10-MCC, UbcH10 induces an additional small rotation of MCC-Cdc20APC/C. The C-terminus of Cdc20MCC is visualized as an extended tubular density feature linking the Cdc20MCC WD40 domain with the catalytic site of UbcH10 (Fig. 5b & Extended Data Fig. 3e and Supplementary Video 2). Modelling the C-terminus of Cdc20MCC into this density shows that Lys485 and Lys490 are accessible to the UbcH10 catalytic site (Fig. 5b). Both residues are ubiquitinated in vitro (data not shown), and their replacement by Arg virtually eliminated Cdc20 auto-ubiquitination (Fig. 5e), indicating that these two residues are the major sites of Cdc20MCC ubiquitination in the context of APC/CMCC-UbcH10. Our model that the MCC competitively restricts access of UbcH10 to its binding site on the APC/C is consistent with the reduced binding of UbcH10 to endogenous APC/CMCC relative to APC/CCdc20 (ref. 31), and our finding that compared with securin ubiquitination, Cdc20 auto-ubiquitination required a 10-fold higher concentration of UbcH10 (Extended Data Fig. 1m), in agreement with 29. Although our data show that MCC suppresses APC/C - UbcH10 interactions, the effect of the MCC on modulating the affinity of the APC/C for the elongating E2 Ube2S, which contributes to checkpoint silencing 46–48, is less clear. The binding site for the Ube2S C-terminal LR tail at the Apc2-Apc4 interface 32, necessary for its association with the APC/C, is unaffected by the MCC (Fig. 5a), thus in principle allowing Ube2S to assemble ubiquitin chains onto the C-terminus of Cdc20MCC. However, the capacity of Cdc20 to stabilize APC/C – Ube2S complexes 48, may be affected by the interactions of BubR1 with the APC/CMCC.
In conclusion, our structural analysis of APC/CMCC and APC/CUbcH10-MCC provides a molecular explanation for the spontaneous APC/C reactivation resulting from SAC inactivation involving ubiquitin-mediated Cdc20MCC proteolysis. APC/C reactivation in response to the cessation of SAC signalling is facilitated by the reciprocal control of APC/C and MCC activities mediated by the conformational flexibility of APC/CMCC, influenced by Apc15. Modulation of this conformational change could allow for the regulation of Cdc20MCC auto-ubiquitination (Fig. 5f). A candidate for such a role is p31comet, a Mad2-binding protein proposed to stimulate Cdc20 ubiquitination in checkpoint-arrested cells 23,45.
Methods
Cloning and expression of human APC/C complex genes
The DNA coding sequences (CDSs) of the human APC/C subunits (wild type, mutant Apc2ΔWHB, Apc11-UbcH10 fusion and ΔApc15 were assembled by USER cloning into a modified version of the insect cell-baculovirus MultiBac expression system 32,51,52. All APC/C subunit CDSs were distributed in two recombinant vectors that were used for recombinant baculovirus generation. For APC/C expression, Hi-5 cells at a density of 2x106 cells/mL were co-infected with two pre-cultures of Sf9 cells each pre-infected with one of the two recombinant APC/C baculoviruses. APC/C expression (unphosphorylated) was performed for 30 h. To obtain APC/COA (phosphorylated APC/C), okadaic acid at a final concentration of 0.1 μM was added after 24 h of infection. Cells were harvested after 5 h of treatment.
Cloning and expression of human MCC complex genes and Cdc20
The CDSs of the human MCC subunits (Mad2, Cdc20, BubR1, Bub3) used for structural analysis were cloned into a pU2 plasmid 52 using the same method as for the APC/C. BubR1 was fused in frame with an N-terminal 3xFLAG tag. Cdc20 for individual expression was cloned into a pFastbac1HTA in frame with the His6-tag. In addition, an MBP tag followed by a TEV site between the starting codon of Cdc20 and the N-terminal His6 tag was added by restriction free cloning method (RF-cloning 53). To obtain a vector containing Mad2, Cdc20 and BubR1 (residues 1-569) CDSs (miniMCC construct), a Mad2 and Cdc20-containing expression cassette from a pU1 vector was shuttled (by the AvrII and PmeI sites) into a pFastbacDual vector (BstZ171 and SpeI sites) that contained 3xFLAG-BubR11-569 under the control of the p10 promoter. A C-terminal StrepIIx2 tag was added by RF-cloning into the BubR1 constructs used in ubiquitination assays. Expression of either the MCC or Cdc20 constructs was performed similarly to the APC/C (unphosphorylated) to avoid CDK-dependent inhibition of APC/C-Cdc20 interactions 54,55. Moreover, cells were harvested 48 h after infection. To express MCC complexes with the tagged versions of BubR1, virus containing the BubR1-StrepII constructs was co-infected with MCC virus. To express the MCC complex with the Cdc20K485R,R490R mutations, viruses containing the individual MCC subunits were used for co-infection. Apc15ΔNTH, a mutant form of Apc15 with a (Gly-Ser-Ala)6 linker substitution of the N-terminal helix (NTH: residues 23-57) was cloned into an E. coli pOPIN expression vector and purified using a C-terminal StrepIIx2 tag.
Reconstitution and purification of APCMCC complexes
To generate mitotic phosphorylated APC/C (APC/COA) we incubated APC/C expressing insect cells with the phosphatase inhibitor okadaic acid (OA) (as described above). The extent of APC/C phosphorylation was monitored by assessing the migration of the Apc3 subunit on SDS PAGE 56 (Extended Data Fig. 1a, f). The recombinant APC/COA was phosphorylated on ~110 sites (Extended Data Table 3), correlating closely with those previously identified in endogenous APC/C isolated from HeLa cells arrested by the mitotic checkpoint 56–58, and with sites phosphorylated in vitro by the mitotic APC/C activating kinases Cdk2-cyclinA2-Cks2 and Plk1 (ref. 22 (Extended Data Table 3). Compared with APC/C from untreated insect cells, and using Cdc20 as the coactivator, APC/COA readily ubiquitinates securin (Extended Data Fig. 1g, h).
The APC/CMCC complex was reconstituted by co-lysing APC/COA expressing cells with insect cells expressing separately MBP-tagged Cdc20 and the MCC (BubR1, Bub3, Mad2 and untagged Cdc20). Hi-5 cell pellets expressing either APC/COA or MBP-Cdc20 or MCC were mixed together in reconstitution buffer containing 50 mM Hepes (pH 8.2), 150 mM NaCl, 5% glycerol, 0.5 mM TCEP, 1 mM EDTA, 0.1 mM PMSF, 2 mM benzamidine, 5 units/ml benzonase (Novagen), Complete™ EDTA-free protease inhibitors (Roche), 50mM NaF, 20mM β-glycerophosphate and 0.1 μM okadaic acid. After complete mixing the cells were co-lysed by sonication and the lysate was centrifuged for 60 min at 20,000 rpm. The soluble fraction was loaded onto a Strep-Tactin Superflow Cartridge (Qiagen) for purification using the StrepIIx2 tag on Apc4 as described previously 21. The eluate was then applied to an anti-FLAG M2 Affinity Gel (A220, Sigma) column (directed against the N-terminal FLAG tag on BubR1) and incubated overnight. The APC/CMCC complex was eluted with a 3XFLAG peptide at a concentration of 50 μg/mL. The resulting elution was concentrated to around 1.4 mg/mL and run on a Superose™ 6 3.2/300 (GE Healthcare Life Sciences) gel filtration column pre-equilibrated with gel filtration buffer containing 20 mM Hepes (pH 8.0), 150 mM NaCl and 0.5 mM TCEP. The gel filtration was run on a ÄKTAmicro (GE Healthcare Life Sciences) with a flow rate of 50 μL/min.
An SDS-PAGE of purified APC/CMCC showed both versions of Cdc20, consistent with the incorporation of two distinct subunits of Cdc20 into APC/CMCC (refs 2,20) (Extended Data Fig. 1j). Reconstituted APC/CMCC is stable and homogeneous as shown by size-exclusion chromatography (Extended Data Fig. 2a).
The APC/CApc15ΔNTH complex was reconstituted by incubating recombinant APC/CΔApc15 with Apc15ΔNTH, at concentrations of 200 nM and 1 μM, respectively, followed by size exclusion chromatography. Anti-Apc15 antibodies were from Santa Cruz Biotechnology (sc-398448).
APC/C ubiquitination assay
To examine APC/C activity towards securin the ubiquitination assay was performed with 60 nM of recombinant human APC/C, 150 nM UBA1, 300 nM UbcH10, 300 nM Ube2S, 20 μM ubiquitin, 2 μM securin, 5 mM ATP, 0.25 mg/mL BSA and 7 nM of recombinant human Cdc20. The ubiquitination products of securin were detected by western blot with either an anti-His antibody (631212; Clontech) or an anti-securin antibody (700791; Invitrogen).
To test the activity of a pre-assembled APC/CMCC complex towards Cdc20MCC (Fig. 5c), ubiquitination reactions were performed with 250 nM of recombinant human APC/CCdc20-MCC and 10 μM of UbcH10 (40x excess). To test the activity of APC/C towards the Cdc20MCC from individually purified wild type and mutant MCCBubR1-StrepII (purification by StrepIIx2 affinity and gel filtration columns) ubiquitination reactions were performed with 200 nM of recombinant human APC/COA, 200 nM of recombinant human Cdc20 and either 300 or 600 nM of recombinant human MCCBubR1-StrepII (Fig. 5d, e). Either with a pre-assembled APC/CMCC complex or with a molar excess of MCC complex over free Cdc20 and APC/C only Cdc20MCC ubiquitination is promoted (data not shown)20. Cdc20 and the ubiquitination products of Cdc20MCC were detected by western blot with an anti-Cdc20 antibody (Cdc20 H-175 sc-8358; Santa Cruz Biotechnology, Inc.).
Electron microscopy
Freshly purified APC/CMCC samples were analysed by negative stain EM to check the sample quality and to obtain a low-resolution reconstruction. Micrographs were collected on a 2k*2k CCD camera fitted to a FEI Spirit electron microscope at an accelerating voltage of 120 kV, operated at a nominal magnification of 42,000 with a resulting pixel size of 2.46 Å/pixel at specimen level. Defocuses were set at approximately -2 μm. Particles were automatically selected using the autoboxer program implemented in EMAN 59. About 150 micrographs per sample were collected yielding ~10,000 particles. After 3D classification performed with RELION 60 only the prominent best class (30-40% of total amount of particles) was used for auto-refinement and final low-resolution structure determination.
Grid preparation for both negative stain and cryo-EM was performed as described previously 32,51. Cryo-EM micrographs were collected with an FEI Tecnai Polara electron microscope at an acceleration voltage of 300 kV and Falcon III direct detector. Micrographs were taken using EPU software (FEI) at a nominal magnification of 78,000, yielding a pixel size of 1.36 Å/pixel at specimen level. A total exposure time of 1.6 s were used at a dose rate of 27 electrons/pixel. Defocus range was set at -2.0 to -4.0 μm. Movie frames were recorded as described 32.
Image processing
Image processing was performed with RELION 1.4 (ref. 60). The initial steps including motion correction, CTF estimation, particle picking and particles sorting by Z-score and 2D classification were performed as described 32. Selected particles were used for a first round of 3D classification with global search and a sampling angular interval of 7.5°, using a 60 Å low-pass filtered APCCdh1-Emi1 EM map as a reference (ref. 32). Poorly characterized 3D classes, with poorly recognizable features, were discarded at this stage and the remaining particles were refined and corrected for beam-induced particle motion using particle polishing in RELION 61. Polished particles were used for another round of 3D classification with a local search within 15° and a smaller angular sampling interval of 3.7° (Extended Data Figs 4 and 7). The reconstruction generated from all the polished particles, low-pass filtered at 40 Å, was used as reference.
To isolate particles for the APC/CMCC-Closed state, classes showing closed-like features for the MCC-Cdc20APC/C module (e.g. proximity to Apc2, Apc4 and Apc10, Extended Data Fig. 4, classes 1-3) were combined and refined. The resultant map was used as reference for a subsequent 3D classification performed with a soft edge mask on the MCC-Cdc20APC/C module (Extended Data Fig. 4). The mask was created from a map converted from the fitted coordinates of the MCC-Cdc20 module, with three pixel extension and five pixels soft edge width. The MCC-Cdc20 module coordinates were created by fitting the MCC core coordinates and isolated Cdc20 (PDB ID: 4AEZ) 18, on the best MCC-Cdc20APC/C module density map (Extended Data Fig. 4, class 1).
To isolate particles for the APC/CMCC-Open state, classes showing open-like features for the MCC-Cdc20 module (e.g. proximity to TPR lobe and loss of contact with Apc2, Apc4 and Apc10, Extended Data Fig. 4, classes 4-5) were refined together. The obtained averaged class was used as a reference for a subsequent 3D classification performed with a larger mask (6 pixel extension and 6 pixel soft edge) created with the MCC-Cdc20APC/C module coordinates fitted into the corresponding density in the APC/CUbcH10-MCC reconstruction described below (Extended Data Figs 4 and 7).
To obtain the APC/CΔApc15-MCC structure the best classes from the 3D classification with local searches step were refined together (Extended Data Fig. 7a, classes 1-3).
To isolate the particles for the APC/CUbcH10-MCC reconstruction, instead of performing the 3D classification with local search steps, an initial classification with a large mask (similar to APC/CMCC-Open) was performed. The latter allowed the identification of a class that features both the MCC-Cdc20 module and the UbcH10-Apc11-Apc2WHB-Apc2α/β domain assembly 32. A large mask including the latter regions was created by fitting the MCC-Cdc20APC/C module coordinates and the UbcH10-Apc11-Apc2WHB-Apc2α/β domain assembly (PDB ID: 5A31) 32 in the preliminary APC/CUbcH10-MCC reconstruction. The latter mask was used for a re-classification of the initial particles and allowed the isolation of the final APC/CUbcH10-MCC particles (Extended Data Fig. 7c).
All resolution estimates were based on the gold standard Fourier Shell Correlation (FSC) = 0.143 criterion 62. Final FSC curves were calculated using a soft mask (five pixel extension and three pixel soft edge) of the two independent reconstructions. To visualize high-resolution details, all density maps were corrected for the modulation transfer-function of the detector and sharpened by applying negative B-factors, estimated using automated procedures.
Local resolution maps for all the cryo-EM reconstructions were calculated with RESMAP 63 using a resolution range between 3.5 and 15 Å and displayed with Chimera 64. For comparing structural features among the cryo-EM reconstructions, shown in Fig. 4 and Extended Data Fig. 3, which have different overall resolutions, a common filter of 8.5 Å was applied. This was selected based on the local resolution of the APC/CUbcH10-MCC map in the region assigned to Apc15 (the main region of relative comparison). APC/CUbcH10-MCC is the APC/C reconstruction with the lowest overall resolution. Filtering all the reconstruction to 8.5 Å resolution allowed a clear definition of the structural details of both Apc15 and other regions without the appearance of noise. To visualize the connecting density between UbcH10 and Cdc20 the APC/CUbcH10-MCC map was filtered to 12 Å resolution based on the local resolution of this area and the threshold was slightly lowered.
Model building
Initial fitting and superposition of coordinates was performed with Chimera 64. Model building of APC/CMCC was performed in COOT 65. APC/C platform, TPR lobe, Apc10 and accessory subunit coordinates from the atomic structure of APC/CCdh1-Emi1 (PDB: 4UI9) 32 were individually rigid body fit into the APC/CMCC-Closed cryo-EM density. A few regions such as Apc4HBD, Apc5NTD and Apc11 were also modified by flexible fitting. The Apc2WHB domain (PDB ID:4YII) 44 was rigid body fit into the corresponding density. Cdc20APC/C IR tail and NTD were rigid body fit from the coordinates of APC/CCdc20-Hsl1 cryo-EM structure 22. The Cdc20MCC IR tail was modelled by superposing the TPR domain of Apc3 including Cdc20IR from APC/CCdc20-Hsl1 to the TPR domain of APC/CMCC Apc8A. Two copies of human the Cdc20WD40 domain (PDB ID: 4GGA) 66, human C-Mad2 (PDB ID: 2V64) 8 and the human BubR1TPR domain (PDB ID: 3SI5) 67 were rigid body fit on the MCC-Cdc20 module density. Cdc20MCC CRY box, included in the human Cdc20WD40 domain crystal structure (PDB ID: 4GGA) 66 was modelled by flexible fitting. In addition, the Cdc20 KILR motif was modelled by rigid body fit of the MCC core crystal structure (PDB ID: 4AEZ) 18 into the corresponding density. A similar procedure was applied to model the first KEN1 and helix-loop-helix region of BubR1. BubR1 D1 and D2 were modelled by rigid body fit of Acm1 D box 3 (PDB ID: 3BH6) 38. Similarly BubR1 A1 and K2 were modelled by flexible fitting of the Acm1 region spanning the A-motif and KEN box as explained in the main text. BubR1 A2 was modelled as a rigid body fit of the Acm1 A-motif. Loop extensions were modelled as idealized polyalanine.
Model refinement was performed with REFMAC 5.8 (ref. 68). A REFMAC weight of 0.04 was defined by cross-validation using half reconstructions 69. A resolution limit of 4.0 Å was used. All available crystal structures or NMR structures were used for secondary structure restraints. The refinement statistics are summarized in Extended Data Table 3b.
Map visualization
Figures were generated using Pymol and Chimera 70. Structural conservation figures were generated using ConSurf 71.
Mass Spectroscopy
Purified proteins were prepared for mass spectrometric analysis by in solution enzymatic digestion, without prior reduction and alkylation. Protein samples were digested with trypsin or elastase (Promega), both at an enzyme to protein ratio of 1:20. The resulting peptides were analysed by nano-scale capillary LC-MS/MS using an Ultimate U3000 HPLC (ThermoScientific Dionex) to deliver a flow of approximately 300 nl/min. A C18 Acclaim PepMap100 5 µm, 100 µm x 20 mm nanoViper (ThermoScientific Dionex), trapped the peptides prior to separation on a C18 Acclaim PepMap100 3 µm, 75 µm x 250 mm nanoViper (ThermoScientific Dionex, San Jose, USA). Peptides were eluted with a 90 min gradient of acetonitrile (2% to 50%). The analytical column outlet was directly interfaced via a nano-flow electrospray ionization source, with a hybrid quadrupole orbitrap mass spectrometer (Q-Exactive Plus Orbitrap, ThermoScientific). LC-MS/MS data were then searched against an in house LMB database using the Mascot search engine (Matrix Science) 72, and the peptide identifications validated using the Scaffold program (Proteome Software Inc.) 73. All data were additionally interrogated manually.
Sequence alignment
Sequence alignment was performed using Jalview 74.
Extended Data
Extended Data Figure 1. Biochemical characterization of recombinant APC/CMCC complex and preparations of wild type and mutant complexes.
a,b,c,d,e SDS PAGE gels (stained with coomassie) of gel filtration peak fraction from wild type and mutant APC/CMCC complexes preparations used in this study. Western blot against Strep tag in (e) confirms the presence of Apc15ΔNTH-Strep.construct. f, Top: Western blot performed with anti-Apc3 antibody to monitor the time-dependent phosphorylation of this subunit induced by okadaic acid (OA) treatment in the APC/C-expressing insect cells. Bottom: Western blot against was used as a loading control and reflects the decrease in cell viability after addition of OA (data not shown). g, Western blot against His6-tagged ubiquitin of in vitro securin ubiquitination assays performed with either APC/C or APC/COA in either the presence or absence of Cdc20. h, the input sample for the ubiquitination assays performed in this study is shown. i, Western blot against securin of in vitro securin ubiquitination assays performed with APC/COA and Cdc20 with or without either MCC or miniMCC. j, SDS PAGE of APC/CMCC reconstituted with MBP-TEV-Cdc20APC/C and untagged Cdc20MCC. MBP-TEV-Cdc20APC/C TEV cleavage products are indicated. k, SDS PAGE gels of reconstituted APC/CΔApc15-MCC complex. l, Western blot against His6-tagged ubiquitin of in vitro securin ubiquitination assays performed with APC/C ΔApc15 and Cdc20 with or without MCC. m, Western blot against His6-tagged ubiquitin of in vitro Cdc20 ubiquitination assays performed with APC/CMCC and increasing concentrations of UbcH10. Experiments g, l, m were replicated three times and i four times. See Supplementary Fig. 1 for gel source data.
Extended Data Figure 2. Stability of APC/CMCC complex, negative staining-EM reconstructions of APC/CMCC wild type and mutant complexes and Cryo-EM analysis.
a, Top: chromatogram showing the elution profile of the APC/CMCC complex run on a Superose 6 column. Apo APC/COA and thyroglobulin standard molecular weight marker (669 kDa) are indicated. Bottom: SDS PAGE of the eluted fractions. APC/CMCC elutes earlier than APC/COA. b, Negative stain-EM reconstructions performed for this study and EMD-1591 (ref. 31) are shown. APC/C (grey) and MCC-Cdc20APC/C module (red) are highlighted. The APC/CMCC and APC/CApc2ΔWHB-MCC reconstructions are also shown in the same orientation as in Figure 4 to facilitate comparisons. c, A typical cryo-EM micrograph of APC/CMCC-Closed representative of 20,234 micrographs. d, Gallery of two-dimensional class averages of APC/CMCC-Closed showing different views representative of 50 two-dimensional classes. e, density quality for secondary structures. The APC/CMCC-Closed map was filtered to 4.0 Å.
Extended Data Figure 3. Resolution and other Cryo-EM features of APC/CMCC complexes.
a, Fourier Shell Correlation (FSC) curves, (b, c, d) local resolution maps calculated with RESMAP 63 are shown for all the cryo-EM reconstructions determined in this study, b, ribbon representations of structures shown, c, overall views of local resolution maps, d, close up of platform region (Apc4, Apc5, Apc15). All the maps shown in (c) and (d) are filtered to 8.5 Å. Local resolution colour scheme is indicated in the bar at the bottom of (d). e, the APC/CUbcH10-MCC reconstruction filtered at 12 Å and shown at different threshold levels. The lowest threshold is the same as in Fig. 5b.
Extended Data Figure 4. Three-dimensional classification of APC/CMCC.
a, b, 3D class averages obtained by classification with local searches (see Methods) are shown in (a). Classes 1-5 (56%) show density for the MCC-Cdc20APC/C module with classes 1-3 and 4-5 having a closed-like and an open-like conformation respectively (framed in red, see Methods). Particles from classes 1-3 and 4-5 were separately refined and re-classified using a mask (yellow, see Methods) shown in (b) to isolate the best quality particles for APC/CMCC-Closed (mask 1: left) and APC/CMCC-Open (mask 2: right). Percentages for each of the classes relative to the total number of selected particles are indicated. The percentages relative to the total number of APC/CMCC particles are indicated in parenthesis.
Extended Data Figure 5. Features of the APC/CMCC structure.
a,b,c, Cryo-EM density and fitted coordinates for Cdc20APC/C_IR (a), Cdc20APC/C_NTD (b), C-Mad2 (c) are shown. Colours for each subunit are as for Fig. 1. d, Overall superposition of the APC/CCdc20-Hsl1 structure (red) with the APC/CMCC-closed structure (green). The Cdc20WD40 change of position is illustrated and the blades forming the D-box (yellow) binding pocket is highlighted. e, Superposition of Apc4, Apc5 and Apc15 between APC/CCdh1-Hsl1-UbcH10-Ub structure (grey) and APC/CMCC (subunit colours as in Fig. 1) shows the dramatic conformational change of Apc4HBD, Apc5NTD and Apc15NTH induced by Cdc20MCC binding to its indicated binding site on Apc4HBD. f, close up view of the Cdc20MCC CRY box recognition site of Cdc20APC/C. The CRY box also contacts BubR1 in proximity to D1. Colours for each subunit are as for Fig. 1. g, Superposition of the Apc2WHB domains from APC/CMCC and APC/CCdh1-Hsl1-UbcH10-Ub structures and the corresponding interacting regions of BubR1TPR and UbcH10 are shown. Bottom left: the residues mutated in BubR1Wm that contact Apc2WHB and used in the ubiquitination assay shown in Fig. 5d are indicated (red). Bottom right: Residues of UbcH10 (red) that contact the corresponding site on Apc2WHB ablate APC/C UbcH10-dependent ubiquitination activity 44.
Extended Data Figure 6. Conservation analysis on BubR1A1-K2 and BubR1TPR regions.
a, Similarities in modes of binding of BubR1 to two Cdc20 subunits of APC/CMCC (left) and Acm1 to two Cdh1 subunits in the Acm1-Cdh1 heterotrimer 38 (right). D box, KEN box, NEN box and ABBA motif are labelled as D, K, NEN and A. BubR1 (colour-ramped from blue to red indicating N to C-terminus) mediates Cdc20 dimer interface, whereas Acm1 mediates a Cdh1 dimer interface. b, Local sequence alignment performed with BubR1A1-K2 region sequences from several species (described on the left as: sequence identifier_protein name_species/residue number) and the S. cerevisiae Acm1A-KEN region. A D-box-like feature (corresponding to Emi1D-box 7-10 positions) 32 precedes the first ABBA motif (A1). A 21-33 residues long linker connects the A1 to the second KEN-box (K2). Conserved positions are highlighted in orange. c, ConSurf analysis of the BubR1TPR region highlighting conserved residues on the Cdc20APC/C binding pocket (i) and on the Apc2WHB domain pocket (ii). The Cdc20APC/C binding pocket is required for a functional SAC 67. This site interacts with residues of BubR1 immediately N-terminal to KEN-2, thereby reinforcing their contacts with Cdc20APC/C. Residues conservation is indicated in a gradient from cyan to purple. BubR1, Cdc20APC/C and Apc2WHB are coloured as in Fig. 1.
Extended Data Figure 7. Three-dimensional classification of APC/C ΔApc15-MCC and APC/C UbcH10-MCC.
a, 3D class averages obtained by classification with local searches (see Methods) are shown for APC/CΔApc15-MCC. Particles from classes 1-3 were refined together for obtaining the final APC/CΔApc15-MCC reconstruction shown in (b). The APC/CΔApc15-MCC map was filtered to 4.8 Å. c, 3D class averages obtained by classification using a mask (yellow, see Methods) are shown for APC/C UbcH10-MCC. Class 1 was used for the final reconstruction. Percentages relative to the total amount of particles are indicated for each of the classes.
Extended Data Table 1. Summary of phosphorylation sites in APC/C complexes used in this study.
| Protein | Phospho-sites | untreated | OA | Reference |
|---|---|---|---|---|
| Apc1 | LWS51DGA | 57 | ||
| QEVT65IHE | 57 | |||
| PPGS202PRE | 56,57,58 | |||
| RVKS276EEE | 56 | |||
| QGGT291PQN | 56,57,58 | |||
| NVAT297SSS | 56,57,58 | |||
| HLRS307LSK | 57 (Interphase) | |||
| RSLS309KGD | 57 | |||
| KGDS313PVT | 57 | |||
| SPVT316SPF | 57 | |||
| HSRS341PSI | 56,57,58 | |||
| RSPS343ISN | 56,57,58 | |||
| PSIS345NMA | 56,57 | |||
| AALS351RAH | 56,57,58 | |||
| RAHS355PAL | 56,57,58 | |||
| GVHS362FSG | 56,57,58 | |||
| HSFS364GVQ | 56,57 | |||
| FNIS372SHN | 56,57 | |||
| NISS373HNQ | 56,57 | |||
| HNQS377PKR | 56,57,58 | |||
| ISHS386PNS | 57 (Interphase) | |||
| SNGS394FLA | 57 | |||
| RPST530PLD | 57,58 | |||
| DGVS536TPK | 56 | |||
| GVST537PKP | 56,57 | |||
| LLGS547LDE | 57 | |||
| VLLS555PVP | 57,58 | |||
| LHDS569LYN | 57 | |||
| SLY571NED | 56 | |||
| GSLS688PVI | 56,57,58 | |||
| ARPS699ETG | 57 | |||
| ETGS703DDD | 57 | |||
| LCLS731PSE | 58 | |||
| RHST921SVS | 57 | |||
| HSTS922VSS | 57 | |||
| KHKS1347PSY | 57 | |||
| AHVY1463IIA | ||||
| Apc2 | ELDS205RYA | 58 | ||
| RPAS314PEA | 56,57,58 | |||
| GQDS470EDD | 57 | |||
| RRSS499DII | ||||
| HQFS532FSP | 58 | |||
| FSFS534PER | 56,58 | |||
| LIDS732DDE | N.D. | 57 (Interphase) | ||
| DDES736DSG | N.D. | 57 (Interphase) | ||
| ESDS738GMA | N.D. | 57 (Interphase) | ||
| Apc3 | TVLT203ETP | 58 | ||
| LTET205PQD | 56,57,58 | |||
| PQDT209IEL | 56 | |||
| SKYS225LNT | 56,57,58 | |||
| AVIS241PDT | 56,57 | |||
| ETPS291PGD | 56 | |||
| GDGS296YLQ | 56 | |||
| LQNY301TNT | 56 | |||
| QNYT302NTP | 56 | |||
| YTNT304PPV | 56 | |||
| SVFS334QSG | 58 | |||
| SGNS339REV | 58 | |||
| REVT343PIL | 57,58 | |||
| QTST358TPQ | 58 | |||
| TSTT359PQV | 58 | |||
| QVLS364PTI | 56,57 | |||
| LSPT366ITS | 56,58 | |||
| PTIT368SPP | 56,58 | |||
| TITS369PPN | 56,57,58 | |||
| LFTS384DSS | 58 | |||
| SDSS387TTK | 57,58 | |||
| INDS426LEI | 56,57,58 | |||
| LEIT430KLD | 56 | |||
| KLDS434SII | 56,57 | |||
| LDSS435IIS | 56,57,58 | |||
| SIIS438EGK | 57,58 | |||
| STIT446PQI | 56,57,58 | |||
| SSMT809DAD | ||||
| ADDT814QLH | ||||
| AAES821DEF | ||||
| Apc4 | KGKY469FNV | 56 | ||
| EVLS777ESE | 57,58 | |||
| LSES779EAE | 56 | |||
| Apc5 | MELT178SRD | 57 | ||
| ELTS179RDE | 57 | |||
| LDVS195VRE | 56,58 | |||
| KALT232PAS | 58 | |||
| Apc6 | FEKY107LKD | 56,58 | ||
| KDES112GFK | 56,57 | |||
| NIIS559PPW | 56,57,58 | |||
| TGLT580PLE | 56,57,58 | |||
| LETS585RKT | 57,58 | |||
| SRPS595LEE | 56 | |||
| Apc7 | VRPS119TGN | 56 | ||
| TGNS123AST | 56,57 | |||
| NSAS125TPQ | 56,57 | |||
| SAST126PQS | 57,58 | |||
| DQKS545LEG | ||||
| KEES557PTD | ||||
| Apc8 | VAVT13AAV | |||
| QGET556PTT | 56,57,58 | |||
| TPTT559EVP | 56,58 | |||
| ETPT564TEV | 56,57,58 | |||
| NTPT584RRV | 56,57 | |||
| RRVS588PLN | 56,57,58 | |||
| LNLS593SVT | 57 | |||
| NLSS594VTP | 57 | |||
| SSVT596P | 57,58 |
The red shadowing shows the presence of phosphorylation sites and white indicates its absence
Extended Data Table 2. Glossary of terms and abbreviations used in this study.
| Abbreviation | Full name |
|---|---|
| APC/C | Anaphase-promoting complex/cyclosome (Apc1-Apc8, Apc10-Apc13, Apc15, Apc16) |
| MCC | Mitotic checkpoint complex (Cdc20, Mad2, BubR1, Bub3) |
| SAC | Spindle assembly checkpoint |
| Cdc20 | Mitotic APC/C coactivator subunit |
| Cdh1 | Late mitotic APC/C coactivator subunit |
| UbcH10 | Initiating E2 |
| Ube2S | Elongating E2 |
| APC/CCdc20 | APC/C – Cdc20 complex |
| APC/CCdh1 | APC/C – Cdh1 complex |
| APC/CMCC | Inhibited APC/CCdc20 – MCC complex |
| APC/CMCC-Closed | Closed conformation of APC/CMCC |
| APC/CMCC-Open | Open conformation of APC/CMCC |
| APC/CUbcH10-MCC | APC/CMCC – UbcH10 complex |
| APC/CCdh1-Emi1 | APC/CCdh1 – Emi1 complex |
| APC/CCdh1,substrate,UbcH10-Ub | APC/CCdh1 – substrate-UbcH10 complex |
| APC/C∆Apc15 | APC/C with Apc15 deleted |
| APC/COA | Reconstituted phosphorylated APC/C isolated from BV/insect cells cultures incubated with okadaic acid |
| Cdc20APC/C | Cdc20 subunit of APC/CCdc20 |
| Cdc20MCC | Cdc20 subunit of MCC |
| Cdc20APC/C-WD40 | WD40 domain of Cdc20 subunit of APC/CCdc20 |
| C-Mad2 | Closed conformation of Mad2 |
| C-Mad2SB | C-mad2 safety belt |
| O-Mad2 | Open conformation of Mad2 |
| IR tail | Apc3 and Apc8 interacting C-terminal Ile-Arg motif of Cdc20 (Cdh1 and Apc10) |
| C box | Apc8 interacting motif of Cdc20 and Cdh1 (DRYIPxR) |
| KILR motif | C-Mad2-interaction motif of Cdc20MCC and an Apc8-interacting motif of Cdc20APC/C |
| LR tail | Leu-Arg-Arg-Leu C-terminal motif common to Ube2S and Emi1 |
| D box | APC/C degron: RxxLxx[IV]xN |
| KEN box | APC/C degron: KEN |
| ABBA motif (A motif) | APC/C degron: Fx[ILV][FHY]x[DE] |
| CRY motif | APC/C degron: CRY |
| D1, D2 | D-1 box of BubR1, D-2 box of BubR1 |
| A1, A2 | ABBA-1 motif of BubR1, ABBA-2 motif of BubR1 |
| K1, K2 | KEN-1 box of BubR1, KEN-2 box of BubR1 |
| Apc4HBD | Helix bundle domain of Apc4 |
| Apc5NTD | N-terminal domain domain of Apc5 |
| Apc15NTH | N-terminal helix of Apc15 |
| BubR1TPR | BubR1 tetratricopeptide (TPR) domain |
Extended Data Table 3. Summary of cryo-EM data and statistics.
| a. Statistics of all cryo-EM reconstructions | |||||
| Samples | Micrographs collected | Particles used for final reconstruction | Resolution (Å) | ||
| APC/CMCC-Closed | 20,234 | 155,263 | 4.2 | ||
| APC/CMCC-Open | 6,188 | 8.6 | |||
| APC/CUbcH10-MCC | 2,340 | 8,491 | 8.9 | ||
| APC/CΔApc15-MCC | 5,054 | 163,308 | 4.8 | ||
| b. Statistics of all cryo-EM structure determination | |||||
| Data collection | |||||
| EM | FEI Polara, 300K eV | ||||
| Detector | FEI Falcon III | ||||
| Pixel size (Å) | 1.36 | ||||
| Defocus range (µm) | 2.0-4.0 | ||||
| Reconstruction | APC/CMCC-Closed | APC/CMCC-Open | APC/CUbcH10-MCC | APC/CΔApc15-MCC | |
| Software | RELION 1.4 | RELION 1.4 | RELION 1.4 | RELION 1.4 | |
| Accuracy of rotation (degrees) | 1.318 | 2.594 | 3.235 | 1.61 | |
| Accuracy of translations (pixels) | 0.843 | 1.763 | 2.09 | 1.08 | |
| Final resolution (Å) | 4.2 | 8.6 | 8.9 | 4.8 | |
| Refinement | APC/CMCC-Closed | ||||
| Software | RefMac 5.8 | ||||
| Refmac weight | 0.04 | ||||
| Resolution limit (Å) | 4.0 | ||||
| Residue number | 9,014 | ||||
| Average Fourier shell correlation | 0.761 | ||||
| R factor | 0.2505 | ||||
| Rms bind length (Å) | 0.0126 | ||||
| Rms bond angle (°) | 1.746 | ||||
| Validation | |||||
| Ramachandran plot | |||||
| Preferred | 8427 (93.49%) | ||||
| Allowed | 353 (3.92%) | ||||
| Outliers | 234 (2.60%) | ||||
Supplementary Material
Acknowledgments
This work was supported by a Cancer Research UK grant (C576/A14109) and the Medical Research Council (MRC_UP_1201/6) to DB and a Long Term EMBO Fellowship to C.A. We thank members of the Barford group for discussions, X. Bai and S. Scheres for their help with RELION; C. Savva and S. Chen for EM facilities; J. Grimmett and T. Darling for computing and W. Zachariae and J. Pines for their invaluable advice and JP for communicating data prior to publication.
Footnotes
Author contributions. Z.Z. cloned an initial mutant MCC construct used by C.A. to generate other MCC constructs. C.A. cloned Cdc20, purified proteins, performed the protein complex reconstitutions and biochemical analysis. C.A. and L.C. prepared grids, collected and analysed EM data and determined the 3D reconstructions. C.A. fitted coordinates, built models and made the figures with help of L.C. Z.Z. together with J.Y. generated APC/C viruses. J.Y. provided E1, E2 and ubiquitin for ubiquitination assays. S.M. and M.S. performed mass spectrometry. D.B. directed the project and designed experiments with C.A. C.A. and D.B. wrote the manuscript.
Author information. EM maps are deposited with EM-DB with accession code EMD-4037. Protein coordinates of APC/CMCC are deposited with RCSB with PDB code 5LCW.
The authors declare no competing financial interests.
References
- 1.Meyer HJ, Rape M. Processive ubiquitin chain formation by the anaphase-promoting complex. Seminars in cell & developmental biology. 2011;22:544–550. doi: 10.1016/j.semcdb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Primorac I, Musacchio A. Panta rhei: the APC/C at steady state. The Journal of cell biology. 2013;201:177–189. doi: 10.1083/jcb.201301130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Current biology: CB. 2012;22:R966–980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Musacchio A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Current biology : CB. 2015;25:R1002–1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 5.Hoyt MA, Totis L, Roberts BTS. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 6.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 7.De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Current biology : CB. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 8.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 9.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Developmental cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Molecular cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 11.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint. The EMBO journal. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. The Journal of cell biology. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. The Journal of cell biology. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Molecular biology of the cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312:1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes & development. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nature cell biology. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 19.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Developmental cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 20.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517:631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–393. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, et al. Molecular mechanism of APC/C activation by mitotic phosphorylation. Nature. 2016;533:260–264. doi: 10.1038/nature17973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 24.Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446:876–881. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 25.Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes & development. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PloS one. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ge S, Skaar JR, Pagano M. APC/C- and Mad2-mediated degradation of Cdc20 during spindle checkpoint activation. Cell cycle. 2009;8:167–171. doi: 10.4161/cc.8.1.7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster SA, Morgan DO. The APC/C subunit Mnd2/Apc15 promotes Cdc20 autoubiquitination and spindle assembly checkpoint inactivation. Molecular cell. 2012;47:921–932. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uzunova K, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nature structural & molecular biology. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nature cell biology. 2011;13:1234–1243. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–454. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malureanu LA, et al. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Developmental cell. 2009;16:118–131. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suijkerbuijk SJ, et al. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Developmental cell. 2012;22:1321–1329. doi: 10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 35.Han JS, et al. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Molecular cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lara-Gonzalez P, Scott MI, Diez M, Sen O, Taylor SS. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. Journal of cell science. 2011 doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izawa D, Pines J. How APC/C-Cdc20 changes its substrate specificity in mitosis. Nature cell biology. 2011;13:223–233. doi: 10.1038/ncb2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He J, et al. Insights into Degron Recognition by APC/C Coactivators from the Structure of an Acm1-Cdh1 Complex. Molecular cell. 2013;50:649–660. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elowe S, et al. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. Journal of cell science. 2010;123:84–94. doi: 10.1242/jcs.056507. [DOI] [PubMed] [Google Scholar]
- 40.Lischetti T, Zhang G, Sedgwick GG, Bolanos-Garcia VM, Nilsson J. The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nature communications. 2014;5:5563. doi: 10.1038/ncomms6563. [DOI] [PubMed] [Google Scholar]
- 41.Di Fiore B, et al. The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Developmental cell. 2015;32:358–372. doi: 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diaz-Martinez LA, et al. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. The Journal of biological chemistry. 2015;290:2431–2443. doi: 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaisari S, Sitry-Shevah D, Miniowitz-Shemtov S, Hershko A. Intermediates in the assembly of mitotic checkpoint complexes and their role in the regulation of the anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:966–971. doi: 10.1073/pnas.1524551113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown NG, et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:5272–5279. doi: 10.1073/pnas.1504161112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Molecular cell. 2011;44:710–720. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 46.Garnett MJ, et al. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nature cell biology. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williamson A, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires e3-dependent tracking of the emerging conjugate. Molecular cell. 2014;56:232–245. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 49.Reis A, Levasseur M, Chang HY, Elliott DJ, Jones KT. The CRY box: a second APCcdh1-dependent degron in mammalian cdc20. EMBO reports. 2006;7:1040–1045. doi: 10.1038/sj.embor.7400772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, et al. Recombinant expression, reconstitution and structure of human anaphase-promoting complex (APC/C) The Biochemical journal. 2013;449:365–371. doi: 10.1042/BJ20121374. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Yang J, Barford D. Recombinant expression and reconstitution of multiprotein complexes by the USER cloning method in the insect cell-baculovirus expression system. Methods. 2016;95:13–25. doi: 10.1016/j.ymeth.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 53.van den Ent F, Lowe J. RF cloning: a restriction-free method for inserting target genes into plasmids. Journal of biochemical and biophysical methods. 2006;67:67–74. doi: 10.1016/j.jbbm.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 54.Yudkovsky Y, Shteinberg M, Listovsky T, Brandeis M, Hershko A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem Biophys Res Commun. 2000;271:299–304. doi: 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]
- 55.Labit H, et al. Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. The EMBO journal. 2012;31:3351–3362. doi: 10.1038/emboj.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kraft C, et al. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. The EMBO journal. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hegemann B, et al. Systematic phosphorylation analysis of human mitotic protein complexes. Science signaling. 2011;4:rs12. doi: 10.1126/scisignal.2001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steen JA, et al. Different phosphorylation states of the anaphase promoting complex in response to antimitotic drugs: a quantitative proteomic analysis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:6069–6074. doi: 10.1073/pnas.0709807104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 60.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bai XC, Fernandez IS, McMullan G, Scheres SH. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife. 2013;2:e00461. doi: 10.7554/eLife.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian W, et al. Structural analysis of human Cdc20 supports multisite degron recognition by APC/C. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:18419–18424. doi: 10.1073/pnas.1213438109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bolanos-Garcia VM, et al. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding Site. Structure. 2011;19:1691–1700. doi: 10.1016/j.str.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell. 2014;157:823–831. doi: 10.1016/j.cell.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z, et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. Journal of structural biology. 2012;179:269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Landau M, et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic acids research. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 73.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 74.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.