Abstract
Activation of inflammatory gene expression is regulated, among other factors, by post-translational modifications of histone proteins. The most investigated type of histone modifications are lysine acetylations. Histone deacetylases (HDACs) remove acetylations from lysines, thereby influencing (inflammatory) gene expression. Intriguingly, apart from histones, HDACs also target non-histone proteins. The nuclear factor κB (NF-κB) pathway is an important regulator in the expression of numerous inflammatory genes, and acetylation plays a crucial role in regulating its responses. Several studies have shed more light on the role of HDAC 1-3 in inflammation with a particular pro-inflammatory role for HDAC 3. Nevertheless, the HDAC-NF-κB interactions in inflammatory signalling have not been fully understood. An important challenge in targeting the regulatory role of HDACs in the NF-κB pathway is the development of highly potent small molecules that selectively target HDAC iso-enzymes. This review focuses on the role of HDAC 3 in (NF-κB-mediated) inflammation and NF-κB lysine acetylation. In addition, we address the application of frequently used small molecule HDAC inhibitors as an approach to attenuate inflammatory responses, and their potential as novel therapeutics. Finally, recent progress and future directions in medicinal chemistry efforts aimed at HDAC 3-selective inhibitors are discussed.
Keywords: Lysine acetylation, histone deacetylase (HDAC), inflammation, nuclear factor κB (NF-κB), epigenetics, HDAC inhibitors
Introduction
Inflammatory responses involve differential expression of hundreds of genes and are frequently driven by well-defined stimulus-regulated transcription factors such as nuclear factor-κB (NF-κB) [1]. Activation of inflammatory gene expression encompasses a wide variety of co-regulatory mechanisms acting at different levels of the transcription activation process. Among these mechanisms the regulation of gene expression by post-translational modifications of histone proteins has gained significant attention in recent years. Exploration of histone-modifying enzymes is currently a major topic in drug discovery programs in academia and pharmaceutical companies. Nevertheless, it proves to be difficult to define specific post-translational modifications and specific histone-modifying enzymes as molecular targets for drug discovery in particular diseases. The diversity in types and functions of histone post-translational modifications illustrates the complexity of signal transduction cascades.
The increasing number of post-translational histone modifications that is being discovered illustrates the potential complexity on this level [2]. The most investigated type of histone modifications are lysine acetylations, which are regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) [3]. These enzymes influence the way in which DNA is wrapped around histones and packaged into chromatin, a highly organized and dynamic protein-DNA complex [4]. HATs enable the transfer of acetyl groups from acetyl coenzyme A (Ac-CoA) to lysine residues, whereas HDACs, on the other hand, deacetylate lysine residues. In general, increased levels of histone acetylation are associated with increased transcriptional activity, whereas decreased levels of acetylation are associated with repression of gene expression.
Steady-state levels of acetylation of the histones (and other non-histone proteins) result from the balance between the opposing activities of HATs and HDACs [5]. When the equilibrium between HATs and HDACs is altered, dysregulation of (disease-associated) genes could result in diseases like cancer and chronic inflammation. For example, neutrophilic airway inflammation is associated with increased HAT and reduced HDAC activity in peripheral blood monocytes, which is not associated with alterations in gene expression of the individual enzymes [6]. Similarly, a decreased activity and mRNA expression was observed of HDAC 1 and HDAC 2 in patients suffering from asthma [7].
Many different types of cellular proteins are subject to lysine acetylations, which therefore are expected to co-regulate multiple cellular functions. High-resolution mass spectrometry for the proteome-wide identification and quantitation of acetylation sites provided an in-depth view of the in vivo acetylome identifying 3600 acetylations sites [8]. Using this method, a global view of the lysine acetylome and its changes upon HDAC inhibition can be acquired. Recently, a comprehensive study demonstrated the specificities of 19 HDAC inhibitors towards modulation of the acetylation levels of multiple lysine acetylations in human cells [9]. In addition, the specificities of HDAC inhibitors have been extensively determined at the enzyme level using in vitro HDAC inhibition assays with recombinant (human) deacetylases [10] or proteomic-bases approaches [11,12]. Strategies taking into account the cellular environment in the assessment of the iso-enzyme selectivity of HDAC inhibitors reveal surprising selectivity levels for compounds previously denoted pan-HDAC inhibitors based on selectivity studies using purified recombinant HDAC catalytic domains This indicates that the cellular context is important for assessment of the selectivity of HDAC inhibitors.
Apart from histones and macromolecular complexes also transcription factors like NF-κB, activator protein 1 (AP-1), signal transducers and activators of transcription (STATs) and nuclear receptors [13] are modified by histone-modifying enzymes. The NF-κB transcription factor is an important regulator in the expression of numerous (inflammatory) genes, which is for that reason, an interesting target for anti-inflammatory drug development. The NF-κB pathway is relatively well investigated and several regulatory mechanisms have been described to control the NF-κB response to specific stimuli. It has been reported that post-translational modifications, i.e., acetylation of (regulatory) proteins like NF-κB play a crucial role in regulating the intensity, length and specificity of inflammatory responses [14,15]. NF-κB can be (de)acetylated at various lysine residues resulting in NF-κB activation or deactivation [16].
Considering the importance of lysine acetylations in the NF-κB pathway, development of small molecules interfering with the activity of specific HDAC iso-enzymes has great potential to suppress inflammation. To support further development of small molecule inhibitors for HDACs for applications in inflammatory diseases, here, we provide an overview of the role of HDAC 3 and lysine deacetylation in NF-κB-mediated inflammation. In addition, we discuss applications of frequently used small molecule HDAC inhibitors as an approach to attenuate inflammatory responses and address recent progress in medicinal chemistry efforts aimed at the development of HDAC 3-selective inhibitors.
Classes of HDACs
Mammalian HDAC enzymes can be classified into four main groups, based on their homology with orthologues identified in yeast [17]. Class I HDACs, including HDAC 1, 2 and 8 are predominantly found within the nucleus, due to the presence of a nuclear localization sequence and the absence of a nuclear export signal sequence within HDAC 1, 2, and 8. However, HDAC 3 has both a nuclear import and export signal, which enables its localization in both the cytoplasm and nucleus [17,18]. Class I HDACs have a ubiquitous tissue distribution [17]. Class II HDACs are subdivided into two groups, IIA (HDAC 4, 5, 7, 9) and IIB (HDAC 6 and 10), and are predominantly found in the cytoplasm [19]. Class II HDACs are able to shuttle between the cytoplasm and the nucleus, and have a more tissue-specific distribution than class I HDAC [17]. Then, sharing similarities with both Class I and II HDACs, there is HDAC 11, which is the only member of class IV. Class III HDACs are also called sirtuins (SIRT1-SIRT7) and localize in the cytoplasm, nucleus and mitochondria. These HDACs act via different mechanisms than class I and II and require a co-factor NAD+ for activation [20].
HDAC 3 in Inflammation
Several roles for HDAC 1-2 have been described in inflammatory diseases [21–25] (summarized in table 1) with a particular pro-inflammatory role for HDAC 3. HDAC 3 has been reported to be an important player in monocyte recruitment to sites of inflammation, and in macrophage cytokine production. Similarly, siRNA knockdown of HDAC 3 in Human Umbilical Vein Endothelial Cells (HUVEC) was shown to reduce vascular cell adhesion molecule 1 (VCAM-1) expression and hence suppress monocyte adhesion [26]. Furthermore, in an allergic skin inflammation mouse model, a pro-inflammatory role for HDAC 3 was found [27]. When mice were injected with HDAC 3 siRNA at the same day as the induction of skin inflammation, disease progression was less severe. Acute smoke exposure has been shown to reduce HDAC 3 activity in human alveolar macrophages resulting in increased inflammatory responses [28]. Lower nuclear content of HDAC 3, in the context of equivalent total HDAC protein levels following smoke exposure, may reflect increased nuclear export of HDAC 3, allowing increased NF-κB driven cytokine expression, which can contribute to inflammation. In addition, siRNA-mediated knockdown of HDAC 3 in human macrophages was shown to increase the production of Interleukin 8 (IL-8) and Interleukin 1β (IL-1β) in response to lipopolysaccharide (LPS) stimulation [28]. Similar to these results, it was demonstrated that macrophages lacking HDAC 3 were hyperresponsive to Interleukin 4 (IL-4) stimulation [29]. Taken together these studies indicate an important role for HDAC 3 in inflammatory responses.
Table 1. HDAC 1-3 in (NF-κB-mediated) inflammation.
| HDAC | Activity/Level | Observed Effects | Refs |
|---|---|---|---|
| HDAC 1 | ↑ RA synovial fluid | ↑ TNFα levels | [21] |
| ↓ Degradation | ↑ TNFα and IL-1β levels | [22] | |
| ↓ T cell-specific loss | ↑ inflammatory response in vivo allergic airway inflammation model | [23] | |
| HDAC 2 | ↓ Reduced levels | Observed in COPD patients | [24] |
| ↓ Reduced levels | Observed in smoking asthmatics compared to non-smoking asthmatics | [25] | |
| HDAC 3 | ↓ | ↓ expression 50% of studied inflammatory genes in macrophages | [38] |
| ↓ siRNA knockdown | ↓ vascular cell adhesion molecule-1; suppresses monocyte adhesion | [26] | |
| ↓ siRNA knockdown | ↓ disease progression in allergic skin inflammation mouse model | [27] | |
| ↓ Activity after smoke | ↑ inflammatory responses in alveolar macrophages | [28] | |
| ↓ siRNA knockdown | ↑ IL-8 and IL-1β production after LPS stimulation | [28] | |
| ↓ | Macrophages hyperresponsive to IL-4 stimulation | [29] |
Role of NF-κB Acetylation in Inflammation
The NF-κB transcription factors are a family of inducible transcription factors that play a central role in the expression of various cytokines, chemokines and adhesion molecules, which are involved in inflammatory and immune responses and in cell survival [30]. NF-κB transcription factors exist in homo- or hetero-dimeric complexes consisting of different members of the Rel family of proteins. The most prevalent and best studied of these complexes is the p50-p65 heterodimer. In quiescent cells, the p50-p65 complex is present in the cytoplasm in an inactive form, bound to inhibitory proteins known as IκBs. The NF-κB pathway plays an important role in chronic inflammatory diseases like inflammatory bowel disease, rheumatoid arthritis, chronic obstructive pulmonary disease (COPD) and asthma [31,32].
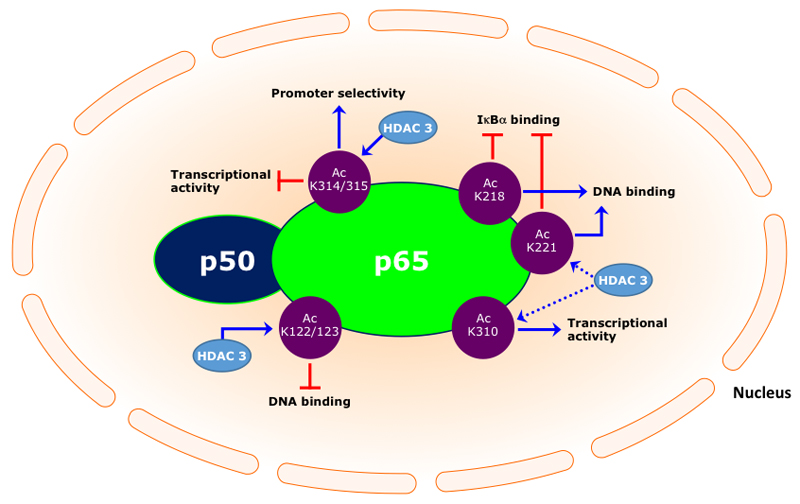
Acetylations of specific lysine residues in the NF-κB p65 transcription factor play a key role in the regulation of its transcriptional capacity, its DNA binding ability and duration of its action [33]. Seven lysine residues of p65 have been described to be subjected to acetylation, respectively, 122, 123, 218, 221, 310, 314 and 315. Acetylation of lysines 122 and 123 decreases DNA binding, whereas acetylation at lysines 218 and 221 increases binding to κB enhancers. Acetylation at lysine 310 is required for full transcriptional activity of p65, however, does not affect the DNA binding or its assembly with IκBα [34]. Acetylation at lysine 314 and 315 does not affect the general transcriptional activity of the NF-κB complex, but rather increase promoter selectivity [35].
This demonstrates that direct acetylation or deacetylation of specific lysine residues of NF-κB plays a crucial role in the regulation of NF-κB-mediated gene expression, which raises the idea to attenuate inflammatory responses by modulating NF-κB acetylation levels using HDAC inhibitors. Consequently, a challenge lies ahead in targeting specific HDACs that are involved in lysine deacetylation of NF-κB p65.
HDAC 3-NF-κB Interactions in Inflammation
A number of studies have shown that HDAC 3 deacetylates the NF-κB p65 subunit, leading to repression of its transcriptional activity [34]. An early study on the HDAC-NF-κB interaction demonstrated that HDAC 3 plays an essential role in the deacetylation of p65 (promoting effective binding to IκBα), thereby negatively regulating Tumour Necrosis Factor α (TNFα)-induced luciferase activity [15], which was abolished in the presence of Trichostatin A (TSA). Similarly, it has been demonstrated that HDAC 3-dependent NF-κB deacetylation promotes interaction with the inhibitor protein IκBα resulting in nuclear export of NF-κB and hence repression of inflammatory gene expression [36]. In contrast, Ziesché et al reported that HDAC 3 is required for Interleukin 1 (IL-1)-induced (human IL-8 or murine Cxcl2) gene expression [37]. The positive regulatory role of HDAC 3 involves binding to the NF-κB p65 subunit and concomitantly deacetylation at various lysines. Interestingly, by using mutagenesis it was demonstrated that acetylation at lysine 314 and 315, in addition to acetylation of lysine 122 and 123, negatively regulate NF-κB activity. Moreover, it has been shown that HDAC 3 is involved in the removal of the inhibitory NF-κB p65 acetylations at lysine 122, 123, 314 and 315 [37]. Therefore, HDAC 3 is an important positive regulator in IL-1-induced inflammatory signalling by deacetylating four specific lysines in the NF-κB p65 subunit. Another paper describing a co-activatory role of HDAC 3 for NF-κB-dependent gene expression came from the Natoli lab [38]. This study demonstrated that HDAC 3-deficient macrophages are unable to activate almost half of the LPS-induced inflammatory genes expression. HDAC 3 ensures that NF-κB is kept in a primarily deacetylated and thus active state. Similar to these results, it was demonstrated that HDAC 3 promotes TNFα expression in LSP-stimulated cardiomyocytes [39]. In addition, a recent study demonstrated that a decrease in nuclear accumulation of NF-κB in MDA-MB-231 cells was accompanied by a decrease in HDAC 3 [40], though the role of HDAC 3 in the reduction of NF-κB accumulation was not studied in detail. A schematic representation of the regulation of NF-κB activity by acetylation and the role of HDAC 3 is shown in figure 1. Taken together, HDAC 3 can be considered as an important activator of NF-κB-mediated inflammatory gene expression by keeping NF-κB in a primarily deacetylated (active) state.
Figure 1.
HDAC Inhibitors as Anti-Inflammatory Drugs for the Treatment of (NF-κB-Mediated) Inflammation
After their introduction in cancer therapy, HDAC inhibitors (HDACi) are gaining attention for application in other diseases as well. Their potential applications range from neurodegenerative (Alzheimer’s and Huntington’s disease) to inflammatory diseases such as asthma, rheumatoid arthritis, and also viral infections [41–44]. Of particular interest are their anti-inflammatory properties, which are observed at 10-100 fold lower doses than those used for treatment of cancer [45]. Sever al HDACi including TSA, phenylbutyrate, suberoylanilide hydroxamic acid (SAHA) and ITF2357 have demonstrated anti-inflammatory effects both in vitro and in vivo [46–50]. For example, it has been demonstrated that pan-HDAC inhibitors such as TSA and SAHA delay NF-κB nuclear translocation, thereby reducing gene expression upon TNFα stimulation [51]. Also, TSA repressed inducible Nitric Oxide Synthase (iNOS) expression in IL-1β/LPS/Interferon γ (IFNγ)-stimulated murine macrophages [52]. Furthermore, inhibition of class I/II HDACs by TSA and nicotinamide effectively blocked the TNFα and Interleukin 6 (IL-6) production by macrophages in patients with rheumatoid arthritis [53]. Furthermore, cytokine array analysis revealed that HDAC 3 inhibition by TSA resulted in decreased monocyte chemoattractant protein-1 (MCP-1) secretion [27]. Recently it has also been demonstrated that TSA suppresses induction of inflammatory cytokines in an acute-on-chronic liver failure model in rats through regulating the acetylation of NF-κB [54]. In contrast, however, in OP9 preadipocytes it was shown that HDAC inhibition by TSA enhanced the expression of inflammatory proteins and NF-κB-dependent transcriptional activity, which might be caused by the increase in the acetylation of NF-κB p65 at lysine 310 and duration of the nuclear translocation of NF-κB [55]. The latter effect was presumably due to acetylation of p65 at lysine 218 or 221 thereby attenuating NF-κB interaction with IκBα. These studies indicate that both anti-inflammatory and pro-inflammatory effects have been observed upon treatment with pan-HDACi.
Several studies show that inhibition of deacetylases extends NF-κB transcriptional activity [56,57], whereas other studies demonstrate that inhibition of deacetylation decreases NF-κB transcriptional activity [58,59]. This indicates that the resulting effect of HDAC inhibition depends on the selectivity for specific NF-κB acetylation sites, which suggests that selective inhibition of HDACs might result in specific effects on inflammatory gene expression. The utility of small molecule inhibitors of HDACs that affected inflammation (in vitro and/or in vivo) by regulating NF-κB p65 lysine acetylation have been reported [60–65], and are summarized in table 2. These studies demonstrate that selective HDAC inhibitors provide chances for further elucidation of the role of HDACs in inflammatory diseases, which provides a perspective towards anti-inflammatory drug discovery.
Table 2. Small molecule inhibitors of HDACs and their effects on (NF-κB-mediated) inflammation.
| HDAC inhibitor | Selectivity | Effects in disease models | Refs |
|---|---|---|---|
| Trichostatin A (TSA) | pan-HDAC | Suppression of TNFα in vitro | [60] |
| SAHA (Vorinostat) | pan-HDAC | Reduces LPS-induced cytokine release in vitro and in vivo Suppresses LPS-stimulated NF-κB nuclear accumulation |
[50,68] |
| LBH589 (Panabinostat) | pan-HDAC | Promotes NF-κB activation | [57] |
| ITF2357 (Givinostat) | pan-HDAC | Inhibition TNFα expression in peripheral blood mononuclear cells Therapeutic benefit in patients suffering from juvenile idiopathic arthritis |
[58,62] |
| LAQ824 (Dacinostat) | pan-HDAC | Inhibition of IL-10 expression | [63] |
| MS-275 (Entinostat) | HDAC 1-3 | Promotes NF-κB activation Suppresses LPS-stimulated NF-κB nuclear accumulation |
[64,68] |
| MGCD0103 (Mocetinostat) | HDAC 1-3, 11 | - | [61,65] |
| MI192 | HDAC 2-3 | Suppresses IL-6 production in primary PBMCs | [69] |
| KBH-A42 | HDAC 1-2 | Suppression of TNFβ production in vitro and in vivo | [70] |
| RGFP966 | HDAC 3 | Supresses TNFα-induced CCL2 expression | [71] |
| Inhibition of pro-inflammatory genes in vitro and ex vivo; ↑ IL-10 | [72] |
HDAC inhibitors with improved selectivity have demonstrated anti-inflammatory effects both in vitro and in vivo. For example, the benzamide type HDACi MS-275, with selectivity for HDAC 1-3, demonstrated anti-inflammatory effects in several inflammation models, e.g. a rat experimental autoimmune prostatitis [66] and mouse models of arthritis [67]. MS-275 also demonstrated anti-inflammatory effects in human rheumatoid arthritis synovial fibroblastic E11 cells [68]. Moreover, it was shown that MS-275 inhibited LPS-induced NF-κB nuclear accumulation (up to ~75%), and increased the association between NF-κB and the HAT p300. Less acetylated NF-κB was observed in the nucleus when cells were treated with MS-275, which confirms the inhibitory effect of MS-275 on NF-κB nuclear accumulation. In addition, IL-6 and Interleukin 18 (IL-18) were reduced in a concentration-dependent manner in E11 cells [68]. Moreover, in THP-1 cells, MS-275 suppressed LPS-induced IL-1β, IL-6, IL-18 and TNFα expression [68]. A recent study demonstrated that HDAC 2-3-selective inhibitor MI192 reduced the TNFα production and dose dependently suppressed IL-6 production in primary Peripheral blood mononuclear cells (PBMCs) isolated from rheumatoid arthritis patients [69]. Moreover, HDAC 1-2 inhibitor KBH-A42 reduced the TNFα production in vitro and in vivo [70]. RGFP966, a selective and potent inhibitor of HDAC 3 with nanomolar activity and no effective inhibition of any other class I HDAC at low micromolar concentrations, reduced the TNFα-induced expression of chemokine (C-C motif) ligand 2 (CCL2) in vitro [71]. Selective pharmacological inhibition of HDAC 3 using RGFP966 demonstrated anti-inflammatory effects in response to LPS/IFNγ in RAW 264.7 macrophages and ex vivo in mouse precision-cut lung slices (PCLS) [72]. Remarkably, upon RGFP966 treatment, the expression of the anti-inflammatory gene IL-10 was upregulated in PCLS. In addition, the transcriptional activity of NF-κB p65 was significantly reduced. The finding presented in these studies justify further optimization and investigation of pharmacological HDAC 3-selective inhibitors, such as RGFP966, and provide an interesting starting point for further development of anti-inflammatory drugs for application in inflammatory diseases.
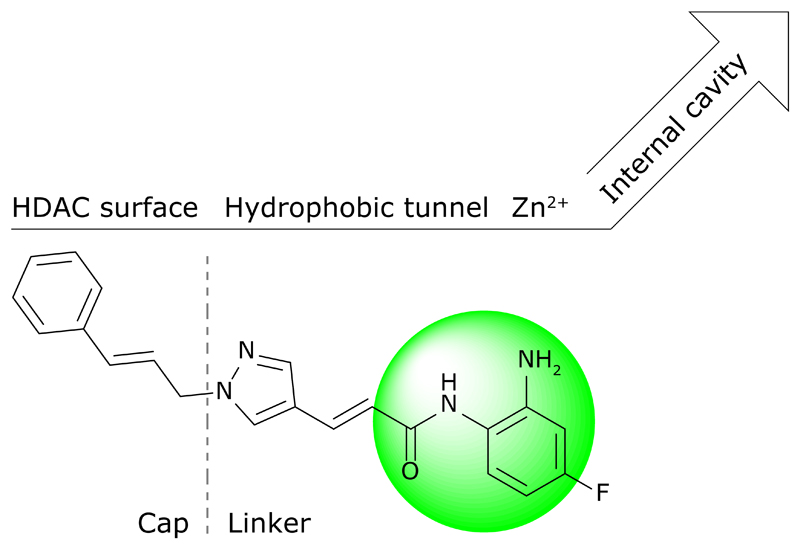
Currently, besides RGFP966 and some other 2-amino benzamide-type inhibitors [73], most newly developed compounds only exhibit class I or II HDAC selectivity and are not truly selective for HDAC iso-enzyme not to mention HDAC 3 [74]. Of note here is that the current literature often expresses selectivity as a significant difference in the half maximal inhibitory concentration (IC50), while measuring and comparing equilibrium inhibitory constants (Ki) or kinetic inhibitory constants (ki) might be more appropriate [75]. Consequently, the selectivity profile of many new inhibitors remains to be accurately mapped. Lack of selectivity partially stems from the high homology between class I HDAC iso-enzymes, but has most likely persisted due to the ubiquitous use of hydroxamic acid as metal-binding group in inhibitor designs. Following the “cap-linker-chelator” model many efforts were focused on making novel linker and capping groups, keeping hydroxamic acid as the preferred chelator. Examples include pyrazoles [76] and spirochromanes [77], and yet none were shown to be isoform selective. Additionally, hydroxamate-based compounds have poor pharmacokinetic properties [78], which severely limit their use.
An interesting strategy to obtain HDAC 3-selective inhibitors would be exploration of the HDAC 3 14 Å-internal cavity. This cavity is expected to function as an acetate releasing channel and may harbour strong ionic interaction sites for novel inhibitors [79]. Although all class I HDACs have a similar internal cavity there is enough sequence variation that can be exploited to achieve selectivity, as has been shown with biarylbenzamides [80]. Benzamides are in that sense especially good starting points because of the possibility to easily append many different groups that may target this internal cavity.
Lastly, the ongoing search for new metal-binding groups [81], and therefore new ways to target the internal binding domains, has resulted in the discovery of potent compounds with non-chelating zinc-binding groups, such as trifluoromethyloxadiazoles [82] and peptide-based thiols [83] and ketones [84], thereby challenging the traditional cap-linker-chelator model. Continuous medicinal chemistry efforts may further defy this model and will most likely yield ever more selective HDAC inhibitors, which are highly desired in the treatment of and in research on inflammation.
Concluding Remarks
Post-translational modifications like lysine acetylations play a crucial role in regulation of NF-κB mediated signalling. The importance of lysine acetylations in the regulation of the NF-κB pathway raises the idea to attenuate inflammatory responses by modulating NF-κB acetylation levels. Small molecules interfering with the activity of histone-modifying enzymes like HDACs have great potential to regulate the NF-κB signalling pathway. Pan-HDACi TSA and SAHA have already demonstrated to suppress the secretion of cytokines in vitro, and attenuate disease severity in animal models of inflammatory diseases, which directed interest towards small molecule compounds that selectively target specific HDACs.
Since many HDACi applied in literature lack selectivity for particular HDAC iso-enzymes, the assignment of specific effects/functions to specific HDACs is hampered. Therefore, new inhibitors targeting HDAC iso-enzymes more selectively need to be developed as tools for these kinds of studies. It is encouraging that recent literature demonstrates the development of HDACi with improved selectivity profiles, which generates chances for further elucidation of the role of HDACs in inflammatory diseases, providing a better perspective towards anti-inflammatory drug discovery and reducing the risk of unwanted side effects.
HDAC 3 has been reported as an important player in monocyte recruitment to sites of inflammation, and in expression of inflammatory genes. In addition, HDAC 3 deficient macrophages stimulated with LPS demonstrated reduced production of inflammatory cytokines. This implies that HDAC 3 is an important player in stimulating inflammation. Therefore, based on current literature on the activating role of HDAC 3 in pro-inflammatory gene expression via the NF-κB pathway, we argue that selective inhibition of HDAC 3 inhibits pro-inflammatory gene expression in model systems for inflammatory diseases by inhibition of the activation of the NF-κB pathway. This justifies further development and investigation of pharmacological HDAC 3-selective inhibitors with the perspective toward development of anti-inflammatory drugs for application in inflammatory diseases.
Highlights.
HDAC 3 plays a key role in NF-κB-mediated signaling
HDAC inhibitors demonstrate promising anti-inflammatory effects.
Selective HDAC 3 inhibitors have potential for treatment of inflammatory diseases.
Figure 2.
Acknowledgments
We are grateful for financial support by the European Research Council with an ERC starting grant (309782) to FJD. Further support was obtained from the Netherlands Organization of Scientific Research (NWO) by a VIDI grant to FJD (723.012.005). We acknowledge COST action ‘epigenetics from bench to bedside (TD0905)’ and ‘epigenetic chemical biology (CM1406)’ for building a European network on epigenetics.
References of Interest
• of special interest
•• of outstanding interest
•• Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G: Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A 2012, 109:E2865-74.
Describing a coactivating function of HDAC 3 for NF-κB-dependent gene expression
•• Ziesche E, Kettner-Buhrow D, Weber A, Wittwer T, Jurida L, Soelch J, Muller H, Newel D, Kronich P, Schneider H et al.: The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-kappaB. Nucleic Acids Res 2013, 41:90-109.
Shows a coactivatory role of HDAC 3 for transcription of the majority of IL-1-induced human and murine genes by three different loss-of-function approaches for HDAC 3
• Kim Y, Kim K, Park D, Lee E, Lee H, Lee YS, Choe J, Jeoung D: Histone deacetylase 3 mediates allergic skin inflammation by regulating expression of MCP1 protein. J Biol Chem 2012, 287:25844-25859.
Identified HDAC 3 as a mediator of allergic skin inflammation in mouse models
• Morioka N, Tomori M, Zhang FF, Saeki M, Hisaoka-Nakashima K, Nakata Y: Stimulation of nuclear receptor REV-ERBs regulates tumor necrosis factor-induced expression of proinflammatory molecules in C6 astroglial cells. Biochem Biophys Res Commun 2016, 469:151-157.
Study applying HDAC 3-selective inhibitor RGFP966 demonstrating that TNFα-induced expression of CCL2 was affected by HDAC 3 inhibition
•• Wambua MK, Nalawansha DA, Negmeldin AT, Pflum MK: Mutagenesis studies of the 14 A internal cavity of histone deacetylase 1: insights toward the acetate-escape hypothesis and selective inhibitor design. J Med Chem 2014, 57:642-650.
Well-conducted investigation of the internal cavity of HDAC 1, providing inspiration for novel selective inhibitor designs
References
- 1.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 3.Dekker FJ, van den Bosch T, Martin NI. Small molecule inhibitors of histone acetyltransferases and deacetylases are potential drugs for inflammatory diseases. Drug Discov Today. 2014;19:654–660. doi: 10.1016/j.drudis.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 6.Gunawardhana LP, Gibson PG, Simpson JL, Powell H, Baines KJ. Activity and expression of histone acetylases and deacetylases in inflammatory phenotypes of asthma. Clin Exp Allergy. 2014;44:47–57. doi: 10.1111/cea.12168. [DOI] [PubMed] [Google Scholar]
- 7.Ito K, Caramori G, Lim S, Oates T, Chung KF, Barnes PJ, Adcock IM. Expression and activity of histone deacetylases in human asthmatic airways. Am J Respir Crit Care Med. 2002;166:392–396. doi: 10.1164/rccm.2110060. [DOI] [PubMed] [Google Scholar]
- 8.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 9.Scholz C, Weinert BT, Wagner SA, Beli P, Miyake Y, Qi J, Jensen LJ, Streicher W, McCarthy AR, Westwood NJ, et al. Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol. 2015;33:415–423. doi: 10.1038/nbt.3130. [DOI] [PubMed] [Google Scholar]
- 10.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 12.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci U S A. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 14.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 16.Ito K. Impact of post-translational modifications of proteins on the inflammatory process. Biochem Soc Trans. 2007;35:281–283. doi: 10.1042/BST0350281. [DOI] [PubMed] [Google Scholar]
- 17.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parra M, Verdin E. Regulatory signal transduction pathways for class IIa histone deacetylases. Curr Opin Pharmacol. 2010;10:454–460. doi: 10.1016/j.coph.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 20.Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J. Sirtuins: the 'magnificent seven', function, metabolism and longevity. Ann Med. 2007;39:335–345. doi: 10.1080/07853890701408194. [DOI] [PubMed] [Google Scholar]
- 21.Horiuchi M, Morinobu A, Chin T, Sakai Y, Kurosaka M, Kumagai S. Expression and function of histone deacetylases in rheumatoid arthritis synovial fibroblasts. J Rheumatol. 2009;36:1580–1589. doi: 10.3899/jrheum.081115. [DOI] [PubMed] [Google Scholar]
- 22.Gopal YN, Van Dyke MW. Depletion of histone deacetylase protein: a common consequence of inflammatory cytokine signaling? Cell Cycle. 2006;5:2738–2743. doi: 10.4161/cc.5.23.3522. [DOI] [PubMed] [Google Scholar]
- 23.Grausenburger R, Bilic I, Boucheron N, Zupkovitz G, El-Housseiny L, Tschismarov R, Zhang Y, Rembold M, Gaisberger M, Hartl A, et al. Conditional deletion of histone deacetylase 1 in T cells leads to enhanced airway inflammation and increased Th2 cytokine production. J Immunol. 2010;185:3489–3497. doi: 10.4049/jimmunol.0903610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barnes PJ. Role of HDAC2 in the pathophysiology of COPD. Annu Rev Physiol. 2009;71:451–464. doi: 10.1146/annurev.physiol.010908.163257. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad T, Barnes PJ, Adcock IM. Overcoming steroid insensitivity in smoking asthmatics. Curr Opin Investig Drugs. 2008;9:470–477. [PubMed] [Google Scholar]
- 26.Inoue K, Kobayashi M, Yano K, Miura M, Izumi A, Mataki C, Doi T, Hamakubo T, Reid PC, Hume DA, et al. Histone deacetylase inhibitor reduces monocyte adhesion to endothelium through the suppression of vascular cell adhesion molecule-1 expression. Arterioscler Thromb Vasc Biol. 2006;26:2652–2659. doi: 10.1161/01.ATV.0000247247.89787.e7. [DOI] [PubMed] [Google Scholar]
- 27.Kim Y, Kim K, Park D, Lee E, Lee H, Lee YS, Choe J, Jeoung D. Histone deacetylase 3 mediates allergic skin inflammation by regulating expression of MCP1 protein. J Biol Chem. 2012;287:25844–25859. doi: 10.1074/jbc.M112.348284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winkler AR, Nocka KN, Williams CM. Smoke exposure of human macrophages reduces HDAC3 activity, resulting in enhanced inflammatory cytokine production. Pulm Pharmacol Ther. 2012;25:286–292. doi: 10.1016/j.pupt.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 29.Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ, Feng D, Steger DJ, Schug J, Artis D, Lazar MA. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. 2011;25:2480–2488. doi: 10.1101/gad.175950.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisastra R, Dekker FJ. Inflammation, Cancer and Oxidative Lipoxygenase Activity are Intimately Linked. Cancers (Basel) 2014;6:1500–1521. doi: 10.3390/cancers6031500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghizzoni M, Haisma HJ, Maarsingh H, Dekker FJ. Histone acetyltransferases are crucial regulators in NF-kappaB mediated inflammation. Drug Discov Today. 2011;16:504–511. doi: 10.1016/j.drudis.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B, Yang XD, Lamb A, Chen LF. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal. 2010;22:1282–1290. doi: 10.1016/j.cellsig.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buerki C, Rothgiesser KM, Valovka T, Owen HR, Rehrauer H, Fey M, Lane WS, Hottiger MO. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziesche E, Kettner-Buhrow D, Weber A, Wittwer T, Jurida L, Soelch J, Muller H, Newel D, Kronich P, Schneider H, et al. The coactivator role of histone deacetylase 3 in IL-1-signaling involves deacetylation of p65 NF-kappaB. Nucleic Acids Res. 2013;41:90–109. doi: 10.1093/nar/gks916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proc Natl Acad Sci U S A. 2012;109:E2865–74. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu H, Shan L, Schiller PW, Mai A, Peng T. Histone deacetylase-3 activation promotes tumor necrosis factor-alpha (TNF-alpha) expression in cardiomyocytes during lipopolysaccharide stimulation. J Biol Chem. 2010;285:9429–9436. doi: 10.1074/jbc.M109.071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohapatra DK, Reddy DS, Ramaiah MJ, Ghosh S, Pothula V, Lunavath S, Thomas S, Valli SN, Bhadra MP, Yadav JS. Rugulactone derivatives act as inhibitors of NF-kappaB activation and modulates the transcription of NF-kappaB dependent genes in MDA-MB-231cells. Bioorg Med Chem Lett. 2014;24:1389–1396. doi: 10.1016/j.bmcl.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 41.Fischer A. Targeting histone-modifications in Alzheimer's disease. What is the evidence that this is a promising therapeutic avenue? Neuropharmacology. 2014;80:95–102. doi: 10.1016/j.neuropharm.2014.01.038. [DOI] [PubMed] [Google Scholar]
- 42.Royce SG, Karagiannis TC. Histone deacetylases and their inhibitors: new implications for asthma and chronic respiratory conditions. Curr Opin Allergy Clin Immunol. 2014;14:44–48. doi: 10.1097/ACI.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 43.Grabiec AM, Tak PP, Reedquist KA. Function of histone deacetylase inhibitors in inflammation. Crit Rev Immunol. 2011;31:233–263. doi: 10.1615/critrevimmunol.v31.i3.40. [DOI] [PubMed] [Google Scholar]
- 44.Wightman F, Ellenberg P, Churchill M, Lewin SR. HDAC inhibitors in HIV. Immunol Cell Biol. 2012;90:47–54. doi: 10.1038/icb.2011.95. [DOI] [PubMed] [Google Scholar]
- 45.Dinarello CA, Fossati G, Mascagni P. Histone deacetylase inhibitors for treating a spectrum of diseases not related to cancer. Mol Med. 2011;17:333–352. doi: 10.2119/molmed.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halili MA, Andrews MR, Labzin LI, Schroder K, Matthias G, Cao C, Lovelace E, Reid RC, Le GT, Hume DA et al. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J Leukoc Biol. 2010;87:1103–1114. doi: 10.1189/jlb.0509363. [DOI] [PubMed] [Google Scholar]
- 47.Nasu Y, Nishida K, Miyazawa S, Komiyama T, Kadota Y, Abe N, Yoshida A, Hirohata S, Ohtsuka A, Ozaki T. Trichostatin A, a histone deacetylase inhibitor, suppresses synovial inflammation and subsequent cartilage destruction in a collagen antibody-induced arthritis mouse model. Osteoarthritis Cartilage. 2008;16:723–732. doi: 10.1016/j.joca.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 48.Grabiec AM, Korchynskyi O, Tak PP, Reedquist KA. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann Rheum Dis. 2012;71:424–431. doi: 10.1136/ard.2011.154211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Furlan A, Monzani V, Reznikov LL, Leoni F, Fossati G, Modena D, Mascagni P, Dinarello CA. Pharmacokinetics, safety and inducible cytokine responses during a phase 1 trial of the oral histone deacetylase inhibitor ITF2357 (givinostat) Mol Med. 2011;17:353–362. doi: 10.2119/molmed.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leoni F, Zaliani A, Bertolini G, Porro G, Pagani P, Pozzi P, Dona G, Fossati G, Sozzani S, Azam T et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc Natl Acad Sci U S A. 2002;99:2995–3000. doi: 10.1073/pnas.052702999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Imre G, Gekeler V, Leja A, Beckers T, Boehm M. Histone deacetylase inhibitors suppress the inducibility of nuclear factor-kappaB by tumor necrosis factor-alpha receptor-1 down-regulation. Cancer Res. 2006;66:5409–5418. doi: 10.1158/0008-5472.CAN-05-4225. [DOI] [PubMed] [Google Scholar]
- 52.Yu Z, Zhang W, Kone BC. Histone deacetylases augment cytokine induction of the iNOS gene. J Am Soc Nephrol. 2002;13:2009–2017. doi: 10.1097/01.asn.0000024253.59665.f1. [DOI] [PubMed] [Google Scholar]
- 53.Grabiec AM, Krausz S, de Jager W, Burakowski T, Groot D, Sanders ME, Prakken BJ, Maslinski W, Eldering E, Tak PP, Reedquist KA. Histone deacetylase inhibitors suppress inflammatory activation of rheumatoid arthritis patient synovial macrophages and tissue. J Immunol. 2010;184:2718–2728. doi: 10.4049/jimmunol.0901467. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Q, Yang F, Li X, Wang LW, Chu XG, Zhang H, Gong ZJ. Trichostatin A Protects Against Experimental Acute-on-Chronic Liver Failure in Rats Through Regulating the Acetylation of Nuclear Factor-kappaB. Inflammation. 2015;38:1364–1373. doi: 10.1007/s10753-014-0108-7. [DOI] [PubMed] [Google Scholar]
- 55.Sato T, Kotake D, Hiratsuka M, Hirasawa N. Enhancement of inflammatory protein expression and nuclear factor Kappab (NF-Kappab) activity by trichostatin A (TSA) in OP9 preadipocytes. PLoS One. 2013;8:e59702. doi: 10.1371/journal.pone.0059702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khan N, Jeffers M, Kumar S, Hackett C, Boldog F, Khramtsov N, Qian X, Mills E, Berghs SC, Carey N et al. Determination of the class and isoform selectivity of small-molecule histone deacetylase inhibitors. Biochem J. 2008;409:581–589. doi: 10.1042/BJ20070779. [DOI] [PubMed] [Google Scholar]
- 57.Rosato RR, Kolla SS, Hock SK, Almenara JA, Patel A, Amin S, Atadja P, Fisher PB, Dent P, Grant S. Histone deacetylase inhibitors activate NF-kappaB in human leukemia cells through an ATM/NEMO-related pathway. J Biol Chem. 2010;285:10064–10077. doi: 10.1074/jbc.M109.095208. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Leoni F, Fossati G, Lewis EC, Lee JK, Porro G, Pagani P, Modena D, Moras ML, Pozzi P, Reznikov LL, et al. The histone deacetylase inhibitor ITF2357 reduces production of pro-inflammatory cytokines in vitro and systemic inflammation in vivo. Mol Med. 2005;11:1–15. doi: 10.2119/2006-00005.Dinarello. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faraco G, Pittelli M, Cavone L, Fossati S, Porcu M, Mascagni P, Fossati G, Moroni F, Chiarugi A. Histone deacetylase (HDAC) inhibitors reduce the glial inflammatory response in vitro and in vivo. Neurobiol Dis. 2009;36:269–279. doi: 10.1016/j.nbd.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 60.Chung YL, Lee MY, Wang AJ, Yao LF. A therapeutic strategy uses histone deacetylase inhibitors to modulate the expression of genes involved in the pathogenesis of rheumatoid arthritis. Mol Ther. 2003;8:707–717. doi: 10.1016/s1525-0016(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 61.Marson CM, Matthews CJ, Yiannaki E, Atkinson SJ, Soden PE, Shukla L, Lamadema N, Thomas NS. Discovery of potent, isoform-selective inhibitors of histone deacetylase containing chiral heterocyclic capping groups and a N-(2-aminophenyl)benzamide binding unit. J Med Chem. 2013;56:6156–6174. doi: 10.1021/jm400634n. [DOI] [PubMed] [Google Scholar]
- 62.Vojinovic J, Damjanov N. HDAC inhibition in rheumatoid arthritis and juvenile idiopathic arthritis. Mol Med. 2011;17:397–403. doi: 10.2119/molmed.2011.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H, Cheng F, Woan K, Sahakian E, Merino O, Rock-Klotz J, Vicente-Suarez I, Pinilla-Ibarz J, Wright KL, Seto E, et al. Histone deacetylase inhibitor LAQ824 augments inflammatory responses in macrophages through transcriptional regulation of IL-10. J Immunol. 2011;186:3986–3996. doi: 10.4049/jimmunol.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai Y, Rahmani M, Dent P, Grant S. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol. 2005;25:5429–5444. doi: 10.1128/MCB.25.13.5429-5444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat Struct Mol Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang ZY, Schluesener HJ. HDAC inhibitor MS-275 attenuates the inflammatory reaction in rat experimental autoimmune prostatitis. Prostate. 2012;72:90–99. doi: 10.1002/pros.21410. [DOI] [PubMed] [Google Scholar]
- 67.Lin HS, Hu CY, Chan HY, Liew YY, Huang HP, Lepescheux L, Bastianelli E, Baron R, Rawadi G, Clement-Lacroix P. Anti-rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen-induced arthritis in rodents. Br J Pharmacol. 2007;150:862–872. doi: 10.1038/sj.bjp.0707165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choo QY, Ho PC, Tanaka Y, Lin HS. Histone deacetylase inhibitors MS-275 and SAHA induced growth arrest and suppressed lipopolysaccharide-stimulated NF-kappaB p65 nuclear accumulation in human rheumatoid arthritis synovial fibroblastic E11 cells. Rheumatology (Oxford) 2010;49:1447–1460. doi: 10.1093/rheumatology/keq108. [DOI] [PubMed] [Google Scholar]
- 69.Gillespie J, Savic S, Wong C, Hempshall A, Inman M, Emery P, Grigg R, McDermott MF. Histone deacetylases are dysregulated in rheumatoid arthritis and a novel histone deacetylase 3-selective inhibitor reduces interleukin-6 production by peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Rheum. 2012;64:418–422. doi: 10.1002/art.33382. [DOI] [PubMed] [Google Scholar]
- 70.Choi Y, Park SK, Kim HM, Kang JS, Yoon YD, Han SB, Han JW, Yang JS, Han G. Histone deacetylase inhibitor KBH-A42 inhibits cytokine production in RAW 264.7 macrophage cells and in vivo endotoxemia model. Exp Mol Med. 2008;40:574–581. doi: 10.3858/emm.2008.40.5.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morioka N, Tomori M, Zhang FF, Saeki M, Hisaoka-Nakashima K, Nakata Y. Stimulation of nuclear receptor REV-ERBs regulates tumor necrosis factor-induced expression of proinflammatory molecules in C6 astroglial cells. Biochem Biophys Res Commun. 2016;469:151–157. doi: 10.1016/j.bbrc.2015.11.086. [DOI] [PubMed] [Google Scholar]
- 72.Leus NG, van der Wouden PE, van den Bosch T, Hooghiemstra WT, Ourailidou ME, Kistemaker LE, Bischoff R, Gosens R, Haisma HJ, Dekker FJ. HDAC 3-selective inhibitor RGFP966 demonstrates anti-inflammatory properties in RAW 264.7 macrophages and mouse precision-cut lung slices by attenuating NF-kappaB p65 transcriptional activity. Biochem Pharmacol. 2016;108:58–74. doi: 10.1016/j.bcp.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suzuki T, Kasuya Y, Itoh Y, Ota Y, Zhan P, Asamitsu K, Nakagawa H, Okamoto T, Miyata N. Identification of highly selective and potent histone deacetylase 3 inhibitors using click chemistry-based combinatorial fragment assembly. PLoS One. 2013;8:e68669. doi: 10.1371/journal.pone.0068669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Micelli C, Rastelli G. Histone deacetylases: structural determinants of inhibitor selectivity. Drug Discov Today. 2015;20:718–735. doi: 10.1016/j.drudis.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 75.Chou CJ, Herman D, Gottesfeld JM. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J Biol Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao Y, Liao C, Li Z, Wang Z, Sun Q, Liu C, Yang Y, Tu Z, Jiang S. Design, synthesis, and biological evaluation of 1, 3-disubstituted-pyrazole derivatives as new class I and IIb histone deacetylase inhibitors. Eur J Med Chem. 2014;86:639–652. doi: 10.1016/j.ejmech.2014.09.024. [DOI] [PubMed] [Google Scholar]
- 77.Thaler F, Moretti L, Amici R, Abate A, Colombo A, Carenzi G, Fulco MC, Boggio R, Dondio G, Gagliardi S, et al. Synthesis, biological characterization and molecular modeling insights of spirochromanes as potent HDAC inhibitors. Eur J Med Chem. 2016;108:53–67. doi: 10.1016/j.ejmech.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 78.Fraczek J, Vanhaecke T, Rogiers V. Toxicological and metabolic considerations for histone deacetylase inhibitors. Expert Opin Drug Metab Toxicol. 2013;9:441–457. doi: 10.1517/17425255.2013.754011. [DOI] [PubMed] [Google Scholar]
- 79.Wambua MK, Nalawansha DA, Negmeldin AT, Pflum MK. Mutagenesis studies of the 14 A internal cavity of histone deacetylase 1: insights toward the acetate-escape hypothesis and selective inhibitor design. J Med Chem. 2014;57:642–650. doi: 10.1021/jm401837e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE, Harrington P, et al. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1:2) Bioorg Med Chem Lett. 2008;18:973–978. doi: 10.1016/j.bmcl.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 81.Chen K, Xu L, Wiest O. Computational exploration of zinc binding groups for HDAC inhibition. J Org Chem. 2013;78:5051–5055. doi: 10.1021/jo400406g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol. 2013;9:319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 83.Bertino EM, Otterson GA. Romidepsin: a novel histone deacetylase inhibitor for cancer. Expert Opin Investig Drugs. 2011;20:1151–1158. doi: 10.1517/13543784.2011.594437. [DOI] [PubMed] [Google Scholar]
- 84.Ueda H, Nakajima H, Hori Y, Fujita T, Nishimura M, Goto T, Okuhara M. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. I. Taxonomy, fermentation, isolation, physico-chemical and biological properties, and antitumor activity. J Antibiot (Tokyo) 1994;47:301–310. doi: 10.7164/antibiotics.47.301. [DOI] [PubMed] [Google Scholar]