Abstract
Multi-species microbial communities play a critical role in human health, industry, and waste remediation. Recently, the evolution of synthetic consortia in the laboratory has enabled adaptation to be addressed in the context of interacting species. Using an engineered bacterial consortium, we repeatedly evolved cooperative genotypes and examined both the predictability of evolution and the phenotypes that determine community dynamics. Eight Salmonella enterica serovar Typhimurium strains evolved methionine excretion sufficient to support growth of an Escherichia coli methionine auxotroph, from whom they required excreted growth substrates. Non-synonymous mutations in metA, encoding homoserine trans-succinylase (HTS), were detected in each evolved S. enterica methionine cooperator and were shown to be necessary for cooperative consortia growth. Molecular modeling was used to predict that most of the non-synonymous mutations slightly increase the binding affinity for HTS homodimer formation. Despite this genetic parallelism and trend of increasing protein binding stability, these metA alleles gave rise to a wide range of phenotypic diversity in terms of individual versus group benefit. The cooperators with the highest methionine excretion permitted nearly two-fold faster consortia growth and supported the highest fraction of E. coli, yet also had the slowest individual growth rates compared to less cooperative strains. Thus, although the genetic basis of adaptation was quite similar across independent origins of cooperative phenotypes, quantitative measurements of metabolite production were required to predict either the individual-level growth consequences or how these propagate to community-level behavior.
Introduction
Multi-species microbial communities critical to human health [1,2], industrial production [3], and waste remediation [4,5] are governed by complex social dynamics. Predicting how these communities might respond to environmental changes requires an understanding of how species interactions change the evolution of new traits. Interspecies interactions, like competition or cooperation, complicate environmental selective pressures and decrease the predictive power of monoculture behavior [6,7]. For example, a cooperative species that excretes a beneficial cellular product, or “public good,” may alter the environment and thus alter the selective pressure on other species [8]. When this cooperation is costly to individual growth, but rewarded by cooperative behavior, a complex tension arises in which the relative costs and benefits and the structure of the interactions determine whether the cooperation is driven to fixation, coexists, or is eliminated. From the point of view of adaptation, it is unclear whether evolution in communities should be highly variable between instances, owing to the presence of multiple partners, or highly similar, because there are often few phenotypes that primarily govern the interaction [7,9,10].
Parallelism at various levels is common in experimental evolution. Replicate microbial populations under selection for growth in liquid medium have often evolved parallel phenotypes [7,8] or genotypes [11–14]. For within-species interactions, examples have suggested that genetic parallelism can be common, despite a resulting range in phenotypic consequences from these mutants [15,16]. Here, we address the spectrum of mutations that gave rise to metabolic cooperation between species, and how the resulting metabolite production phenotypes translated to individual costs or community dynamics.
Our model system is a synthetic community, or consortium, comprised of two-members: an Escherichia coli methionine auxotroph (ΔmetB) and Salmonella enterica serovar Typhimurium [17]. Both members have a reciprocal requirement for the other to grow in lactose minimal media (Fig 1). E. coli excretes carbon byproducts that are consumed by S. enterica, as S. enterica cannot metabolize lactose. S. enterica must, in turn, evolve sufficient methionine excretion to support growth of the E. coli methionine auxotroph. By selecting in a spatially-structured environment, a cooperative methionine-producing S. enterica genotype was selected for, which could then sustain community growth either on petri dishes or in liquid media. In this initial instance of the evolution of cooperation methionine excretion by S. enterica was found to be individually costly, substantially decreasing the cooperator’s own growth rate compared to its ancestor [17].
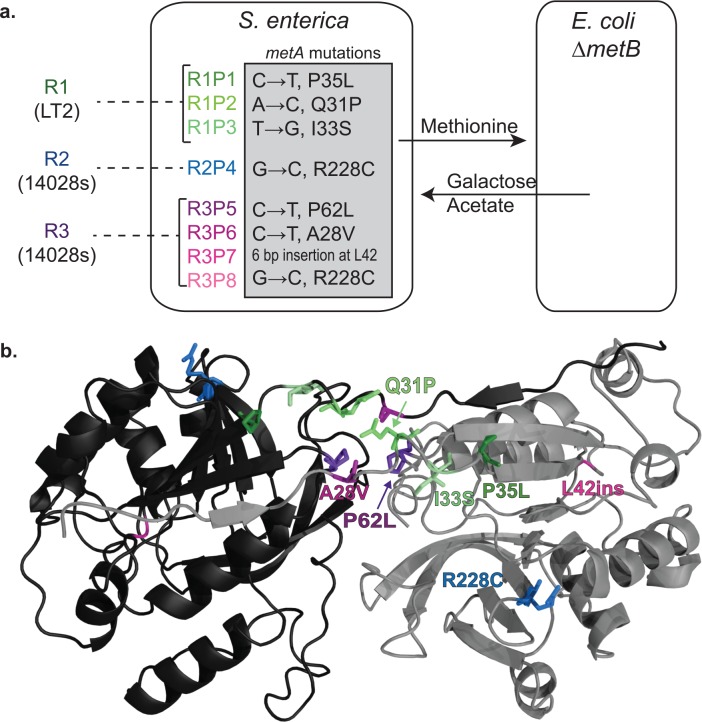
Fig 1. S. enterica cooperators evolved from ethionine resistant ancestors feature mutations in metA.
a) S. enterica ethionine resistant strains (R strains) were co-cultured with E. coli methionine auxotrophs on lactose minimal media. Adaptive methionine excretion by evolved cooperators enabled growth of E. coli ΔmetB, which in turn excretes usable carbon for S. enterica. Non-synonymous substitutions in the metA gene, encoding homoserine trans-succinylase (HTS), are listed next to producer strain name. b) A homology model of residues 2–297 of S. enterica HTS was created using Bacillus cereus HTS. Each evolved cooperator features a mutation at one highlighted residue. The active site is shown with its cognate substrate homoserine.
Here we uncover the genetic basis of methionine excretion in the previously evolved cooperative S. enterica, as well as repeat the use of a spatially-structured environment to select for a series of new cooperative strains from different, closely-related S. enterica backgrounds. We first examined the underlying molecular changes in our S. enterica mutants and found that all evolved strains featured mutations in the metA gene, which encodes the first step of methionine biosynthesis, homoserine trans-succinylase (HTS) [18]. We used molecular modeling to predict how these mutations modified HTS folding and homodimer formation and found that most mutations increased the binding affinity for dimerization. We determined the effect of the metA mutations upon growth of consortia in well-mixed, liquid medium. While growth in liquid is distinct from the spatially structured environment in which the S. enterica evolved, it represents an informative phenotype and is more experimentally tractable. Although we found genetic parallelism, the various metA alleles gave rise to a wide range of methionine excretion levels. We found that methionine excretion correlated negatively with individual growth of S. enterica, but positively with the growth rate of the consortia and with the proportion of E. coli partner that the given strains could maintain. The extent a given allele commits the cell to cooperation thus simultaneously determines the individual-level growth costs, the community growth rate and the species ratio in the consortium.
Results
Replicate S. enterica cooperators were evolved from multiple starting genotypes
To explore the genetic and physiological range of adaptations underlying the rise of cooperation in our consortium, we repeatedly evolved methionine-producing S. enterica strains by selecting for their ability to support growth of their auxotrophic E. coli partner (Fig 1). Repeated attempts to evolve WT S. enterica to support community growth failed. We thus turned to a classic route to generating methionine overproduction in S. enterica, selection of resistance to ethionine, a toxic methionine analog [19,20]. Methionine production in S. enterica is tightly regulated at the level of transcription and translation via end-product inhibition [18,21,22]. Ethionine represses methionine biosynthesis via the same mechanism [21]. A previously characterized ethionine resistant (EthR) strain [17] from S. enterica LT2 (Resistant strain, hereafter “R1”) did not excrete detectable levels of methionine, but was used as the ancestor for deriving an evolved cooperator that produced methionine (hereafter R1P1). We selected two new EthR backgrounds, “R2” and “R3,” from S. enterica 14028s. R1 is derived from the LT2 strain, while R2 and R3 were derived from 14028s; these two strains’ genome sequences differ by just 2% [23]. These three R strains were the ancestors for subsequent evolution of cooperation, and represent slightly distinct genetic backgrounds of the same serovar. None of these three strains excrete detectable amounts of methionine in the medium, but possess causative EthR mutations in methionine pathway transcriptional repressor metJ (Douglas et al., in preparation).
We evolved the new S. enterica cooperators by plating each R ancestor with E. coli ΔmetB on lactose agarose plates, and screened for regions of lactose utilization by the E. coli. Once lactose utilization was observed, that consortium “colony” was streaked to retrieve isolates of each species, allowing cooperative S. enterica to be isolated from their region of origin on the lactose plate. By repeatedly replaying the evolution of methionine excretion from each R ancestor we obtained seven new, independent cooperators that could support community growth like the original strain (here “R1P1”) isolated from R1 previously (Fig 1).
Genetic parallelism in cooperators evolved from different starting genotypes
In follow-up to preliminary genome sequencing of two of the S. enterica cooperators from different R ancestors (Fig 1; R1P1 and R2P4; sequence unpublished), targeted sequencing across the panel of evolved methionine-producing lineages revealed that they all contained mutations in metA, that encodes homoserine trans-succinylase (HTS) (Fig 1). All cooperators possessed either independent point mutations or an insertion in the coding region of HTS (Producer mutations; termed P1 to P8). Intriguingly, none appear within the active site (Fig 1). All but two substitutions fall within the first 62 amino acids of HTS. Each evolved metA allele is unique, with the exception of R228C in both metAP4 and metAP8 arising from an identical nucleotide substitution. Sequencing metA in the three ancestral R strains all showed the original metAWT allele, indicating that each metA mutation immediately preceded the emergence of the cooperative phenotype.
Molecular modeling suggests mutations tend to stabilize homodimer formation
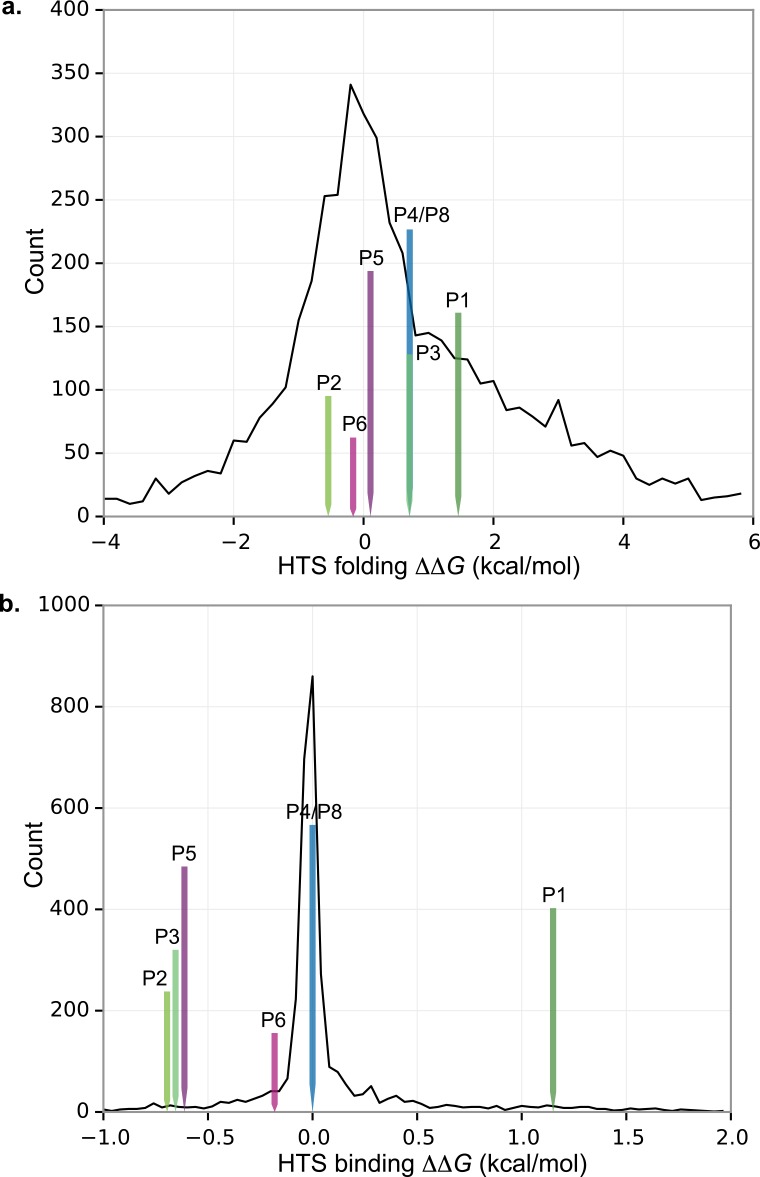
Molecular simulation suggests that the metA mutations are, at least in part, selected to stabilize homodimer formation of metA’s protein product, HTS, but not to stabilize folding of the monomer (Fig 2). Simulations were used to estimate the change in the protein folding and protein-protein binding stabilities for the six unique residue substitutions relative to wild type. To provide context for understanding the effects of these six mutations, all possible substitutions of HTS were calculated, that is, the changes in the stability were estimated for all 264 sites changing to the 19 other residue types (5016 mutations total). The figure shows histograms of the change in folding stability (Fig 2) and binding affinity (Fig 2) for all possible substitutions of HTS. The figure also indicates the six evolved substitutions seen in the experiments. The results show that two of the six evolved substitutions stabilize folding; this is consistent with chance since 38% of all possible mutations stabilize folding. The p-value for a Mann-Whitney U test of the effect of these six substitutions on folding stability was 0.38, consistent with the null hypothesis that they are random with respect to the distribution of all possible substitutions. By contrast, four evolved substitutions stabilize binding of the dimer, one (R228C) has no effect on stability since it is far from the binding interface, and one is destabilizing (P35L). The fraction of evolved substitutions that stabilize binding is much higher than one would expect by chance since only 29% of all possible mutations stabilize binding, and the corresponding p-value is 0.037.
Fig 2. Molecular modeling suggests evolved HTS substitutions increase dimer stability.
Stability changes were estimated using computer simulation. Histograms show stability changes ΔΔG for all possible amino acid changes for both folding and binding. Negative ΔΔG values correspond to stabilizing the folding of HTS or the dimer formation, and positive values destabilize folding or binding. The six single point mutations seen in this study are also indicated.
metA mutations are necessary for cooperative phenotype
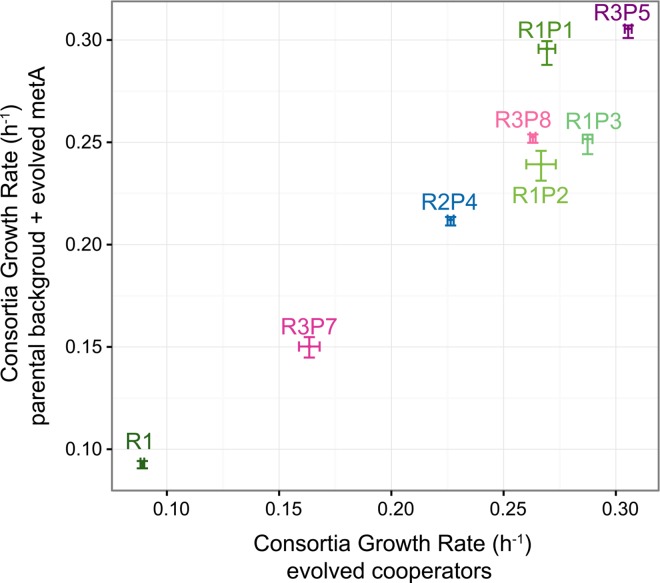
To test the necessity of the metA mutations for cooperation, the ancestral metAWT allele was substituted into each cooperative S. enterica mutant, and the ability to support growth in co-culture (consortia) with E. coli ΔmetB was measured by growth in minimal liquid medium. Evolved S. enterica strains with the metAWT allele reintroduced can no longer sustain growth in consortia with E. coli ΔmetB. Conversely, the evolved metA alleles were not sufficient for consortia growth in the wild-type S. enterica, as they could not restore cooperation when placed alone into wild-type LT2 background (S1 Fig, Welch’s t test, p < 0.05 for both RP strains versus their P strain, and both P strains versus WT). Direct substitution of any of the evolved metA alleles into their ancestral R background, however, restored consortia growth rates to the levels seen from the original evolved producers (R2 = 0.930, Pearson correlation, p < 0.001, throughout using each biological replicate separately) (Fig 3). While the metJ mutations that provide ethionine resistance are essential to consortia growth (Douglas et al. in preparation) the phenotypic diversity arising from different metA mutations within the same EthR background indicated that metA mutations are the primary driver of cooperative disparity between S. enterica strains.
Fig 3. Substituting evolved metA alleles into R strains recapitulates cooperative phenotype.
Consortia growth rates of R strains with evolved metA alleles correlates with growth rates of the original evolved strains (p-value < 0.001), including the control metAWT in the R1 background demonstrating the sufficiency of metAEVO alleles in the R backgrounds. Error bars indicate the standard error of three biological replicates.
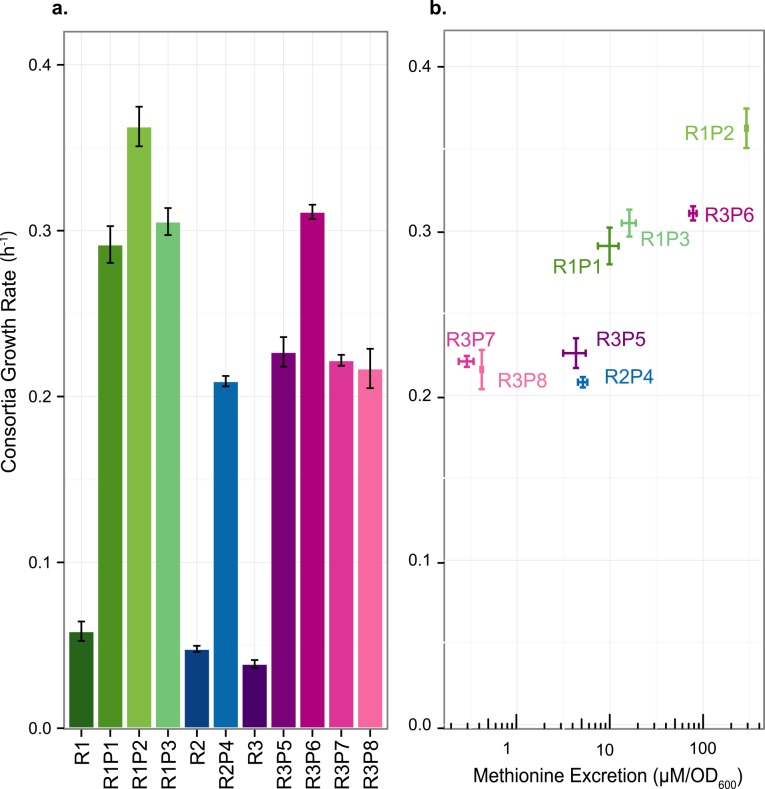
Methionine excretion by evolved cooperators is highly variable
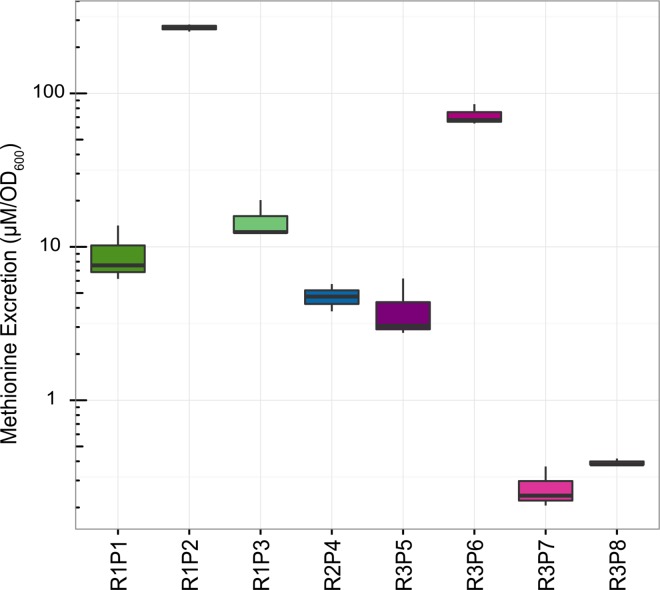
To quantify the variability of physiological consequences arising from parallel metA mutations in S. enterica evolved strains, methionine excretion was measured via gas chromatography/mass spectroscopy analysis of supernatant collected from mid-exponential phase S. enterica monocultures (Fig 4). Methionine excretion above the detection limit of 200 nM was not observed in conditioned media from the ancestral R strains. On the other hand, cooperators exhibited levels of production ranging from 0.272 to 268.2 μM in cultures normalized to an OD600 of 1.0 as a proxy for total biomass. This represents a 1000-fold range in methionine excretion despite similar genetic changes in the same locus.
Fig 4. Methionine excretion by S. enterica producers varies 1000-fold.
GC-MS quantification of methionine in conditioned media collected from evolved S. enterica strains at mid-log phase growth. Values are given as μM methionine per unit OD600. Each data point represents three biological replicates.
Methionine excretion correlates with group performance
In order to study the ecological consequences of the highly varied methionine excretion levels, consortia growth rates for ancestral and evolved S. enterica strains co-cultured with E. coli ΔmetB were measured in lactose minimal media. Although the measurements are of the dynamics of the total OD600 of the consortia, it should be noted that these dynamics were exponential, and involved consistent ratios of species through growth once they reach an equilibrium during the first growth cycle (S2 Fig). Thus, although the proportion of species differs across consortia (see below), both partner species grow at the same rate within each consortium. Consortia containing R strains show slow but measurable growth (0.038–0.054 h-1), suggesting either basal S. enterica cell death or methionine leakage by the ancestral backgrounds is sufficient to maintain some E. coli growth. Consortia growth rates when using evolved strains, however, were much faster and varied from 0.209 to 0.362 h-1 (Fig 5, Welch’s t test, p < 0.05 for all comparisons of RP strains to the corresponding R strain). These growth rates were positively correlated (R2 = 0.773, Pearson correlation, p < 0.05) in a log-linear fashion with the cooperators’ methionine excretion rates previously measured (Fig 5).
Fig 5. Increased consortia growth rate for evolved strains reflects increased S. enterica methionine excretion.
a) Growth rate of cooperative S. enterica co-cultured with E. coli ΔmetB in lactose minimal media. b) Growth rates of consortia containing evolved cooperators are positively correlated with methionine excretion by evolved S. enterica cooperators (p-value < 0.05). Error bars indicate standard error of three biological replicates.
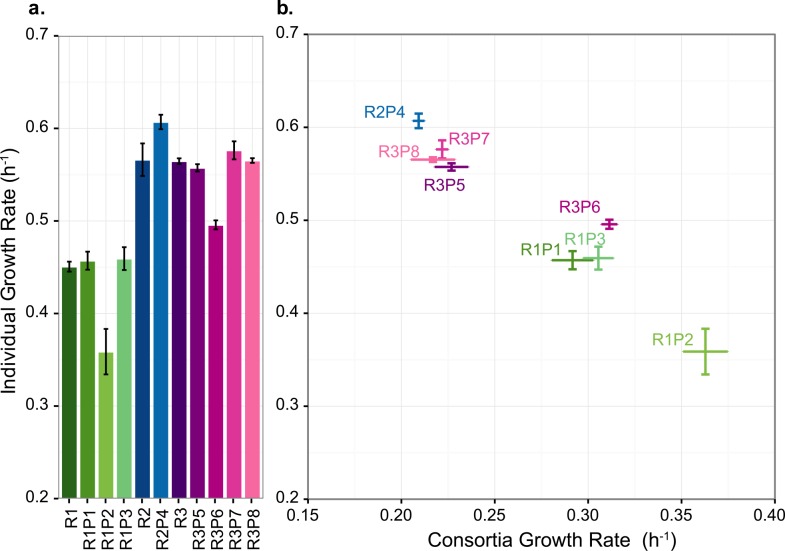
High methionine excretion is costly to individual growth
To assess the extent to which methionine production generated trade-offs between individual growth capacity and community performance, as was observed for the first S. enterica cooperator [17], growth of all individual isolates and reconstituted consortia with E. coli ΔmetB were characterized. S. enterica strain individual growth rates were measured in galactose minimal media (Fig 6). Galactose is one of the compounds excreted by the E. coli ΔmetB partner while growing in lactose minimal medium supplemented with methionine [WRH and CJM, unpublished data]. Consistent with the effects within each lineage, the R1 strain displays the slowest initial growth, and moderately cooperative R1-derived strains grow similarly to R1, with the most cooperative (R1P2) growing more slowly. The R2 and R3-derived strains exhibited a relatively similar pattern, whereby the most cooperative strain grows most slowly. Examining all evolved cooperators collectively, however, it becomes clear that there is a fairly tight, negative linear correlation (R2 = 0.927, Pearson correlation, p < 0.001) between individual growth on galactose and consortia growth on lactose when methionine production is required (Fig 6). Furthermore, for the most cooperative strain, R1P2, consortia growth rate was indistinguishable from its individual growth rate (Welch’s t test, p = 0.894). The other cooperators all grew faster individually than when partnered in co-dependence with E. coli. Even when excluding R1P2 as an outlier of cooperation, correlation between cooperators’ individual and consortia growth rates is maintained (R2 = 0.874, Pearson correlation, p < 0.05).
Fig 6. Decreases in individual growth of cooperators correlates with increases in consortia growth.
a) Growth rates of S. enterica strains in galactose minimal media show decreased individual fitness in the most cooperative S. enterica strains. b) A negative correlation (p-value < 0.001) between individual and consortia growth rates of evolved cooperators demonstrates a trade-off between individual and cooperative selective pressures in these consortia.
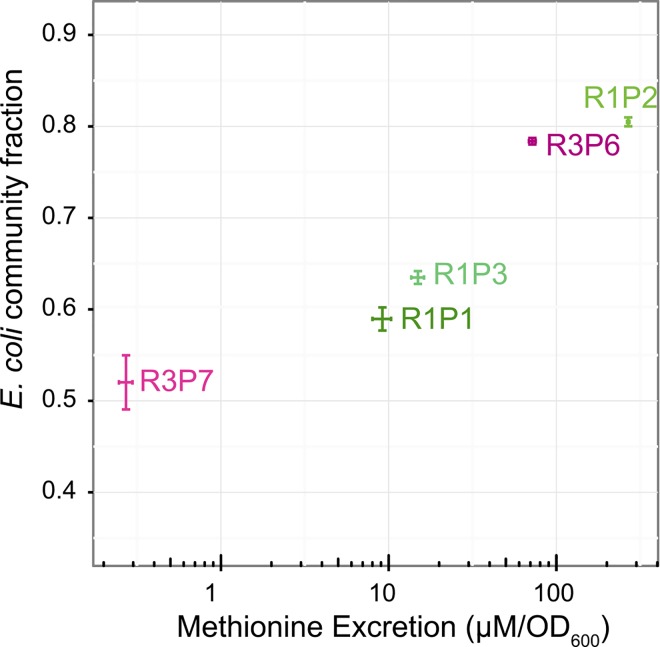
The most cooperative S. enterica strains support highest ratio of E. coli partner
Given the wide differences in methionine excretion, individual growth, and community performance, we further examined the consequence of each evolved metA allele upon the species ratio in our consortia. A subset of the S. enterica cooperators were fluorescently-labeled with yellow fluorescent protein, YFP, and co-cultured with cyan fluorescent protein, or CFP-labeled E. coli ΔmetB in both liquid and solid media. S. enterica methionine excretion correlates with E. coli percentages in liquid culture grown to stationary phase (R2 = 0.937, Pearson correlation, p < 0.05) (Fig 7), showing that individual behavior can predict this ecological trait.
Fig 7. Consortia composition in liquid communities correlates with S. enterica methionine excretion.
YFP-labeled S.enterica and CFP-labeled E. coli ΔmetB grown in liquid were sampled at completion of growth, and consortia composition determined via flow-cytometry. E. coli composition of each community correlates with methionine excretion of S. enterica within that community (p-value < 0.05). Error bars represent standard error of three biological replicates.
Discussion
By selecting for cooperative phenotypes multiple times with a synthetic, two-species consortium, we determined the degree of both the genetic and phenotypic parallelism for mutations that allowed between-species cooperation to emerge. Within the context of a multi-species, spatially-structured community, the environmental interactions are presumed to be more complex than those faced by individual, planktonic cells in well-mixed media. It might be expected that the range of adaptive mutations similarly increase just as biotic interactions often increase diversification. Because the initiation of cooperation was immediately observable on petri dishes due to the co-metabolic conversion of X-gal caused by lactose growth, our experimental design allowed us to obtain first step beneficial mutations as they occurred, before they competed with other possible mutations for fixation. By avoiding clonal interference [24] with other consortia-inducing mutations that generated separate colonies, it seems more likely that the genetic parallelism we observed was due to what mutations can occur, not just those whose selective coefficients are sufficiently large to outcompete others occurring in a given population size [25,26]. We also avoid the possible confounding effects of long-term positive interactions between genotypes such as cross-feeding that can lead to clonal reinforcement [27,28].
We found that the genetic basis for S. enterica to initiate a novel cooperative interaction was quite narrow, similar to previous work on within-species cooperation [29]. Why might natural selection have acted so narrowly? In this case, a relatively dramatic change in phenotype was required for consortia growth, and the ability of consortia to grow was limited by a single factor: methionine production. Given that regulation of methionine production (like many amino acid biosynthetic pathways) is strongly controlled at the first step of the five enzymes involved, HTS encoded by metA exerts a high degree of metabolic control [30] over this phenotype under these conditions. This suggests that when a single metabolic compound is the currency of exchange, changes at one critical point in central metabolism of one organism might be sufficient for large-scale ecological changes.
An additional factor that may have contributed to the observed genetic parallelism was that the evolved phenotype was likely a loss of function of an active negative regulation mechanism upon HTS levels. Previous work has demonstrated that the N-terminus region of the E. coli HTS destabilizes the protein via energy-dependent proteolysis [31]. Deletion or replacement of the first 68 amino acids of HTS in E. coli dramatically increased HTS half-life. Several of our P mutations were in HTS in S. enterica; the homologs in the two species share 95% amino acid identity. This suggests that the mutations we observed in S. enterica acted to increase protein stability. Consistent with this hypothesis, two independent mutations that stabilized the E. coli HTS and increased heat tolerance, S61T and I229T [32], neighbor amino acid substitutions in metAP4, metAP5, and metAP8. The R228C substitutions in metAP4 and metAP8 occur in the C-terminus region where other mutations have been found to desensitize HTS to allosteric inhibition of methionine [33]. Collectively, these prior experiments are consistent with the idea that any of a potentially wide range of mutations that increase HTS stability or reduce its sensitivity to end-product inhibition could lead to cooperation, and thus parallelism at the level of the gene.
While the probability and/or the magnitude of beneficial mutations in metA led to genetic parallelism, the resulting alleles conferred a wide spectrum of phenotypes. Genetic parallelism has been a fairly common finding from evolution of microbes in the laboratory, as well as environments such as chronic infections [34], but there are relatively few examples where consistency of phenotype between alleles has been tested [12,35–37]. Similar to work for within-species cooperation enabled by mutations affecting siderophore production [29] or a transcriptional regulator [15], we found that parallel mutations in metA resulted in variation in methionine production over three orders of magnitude. Furthermore, methionine production explained most of the variation in decreased growth rate on galactose caused by these alleles.
Though the link between parallel genetic changes and the resulting diversity in methionine excretion is unclear, this is one of few examples where the phenotypes of an individual microbial species have been shown to quantitatively correlate with community phenotypes in a complex assemblage [7,9,10,38]. The correlations between production, individual growth and community performance suggest that, at least in the absence of refinement through additional mutations, metabolite production itself governs the benefits and costs of cooperation: the more methionine you make, the slower you grow alone. It is also apparent, however, that methionine production was only substantially costly at high levels. The four isolates producing below 5 μM/OD600 grew alone with less than a 10% defect, whereas the four isolates producing 8 μM/OD600 had growth defects of 20–40%. A second general trend we found was that the more cooperative a strain is, the lower its observed equilibrium frequency in the community. This result parallels findings by Momeni et al 2013 in a single species [39]. In our work the most cooperative S. enterica strain (R1P2) is illustrative. Its consortia grows at a rate indistinguishable from its individual maximal growth rate, suggesting that it is not limited by growth substrate coming from E. coli, but that the E. coli is solely dependent upon a small, slow population of R1P2 for support. For all other S. enterica cooperators, they are capable of faster growth than their corresponding consortia with E. coli, suggesting that they are limited by growth substrate from E. coli. Since the same E. coli partner strain was present in all consortia, this in turn suggests that E. coli was limited in their own growth by the insufficient provisioning of methionine from their respective S. enterica partner strains. Simply by measuring the methionine output by the S. enterica partner, predictions about higher-level community characteristics within this community can be made with some accuracy.
The ability to correlate community composition with future behavior grows increasingly important as we come to rely on genetic sequencing of complex, multi-species microbial systems as a first pass estimator of community characteristics. Accurate inference of ecological behavior becomes more difficult as interactions between shifting selective pressures, genetic changes, and phenotypic expression within an organism are further complicated by species interactions. Metagenomic investigation of known interactions such as adaptive diversification have suggested that even time-series sequence data offer surprisingly few obvious clues of the underlying ecology [27,40]. We have shown here that even highly repeatable genetic adaptations in a dynamic community may translate into an unexpected array of phenotypes; our sequence data alone did not reflect the range of observed methionine excretion. Yet consistent evidence of physiological trade-offs and repeatable community dynamics gives some credence to the idea that the trajectory of even a species-rich evolving community might become predictable once the relevant phenotypes are characterized.
Materials and Methods
Growth media and strains
The experimental system consisted of an E. coli methionine auxotroph (E. coli ΔmetB) and an ethionine resistant S. enterica partner. An E. coli strain K12 BW25113 with a metB knockout was obtained from the Keio collection, with lactose metabolism restored as described [17]. Ethionine resistant S. enterica mutants from LT2 and 14028s backgrounds were selected as described [17]. Cultures were grown in 1X (liquid media) or 0.5X (solid media) “Hypho” minimal media containing C7 trace metal mix [41] and were supplemented with either 0.1% (liquid media) or 0.05% (solid media) galactose or lactose. Antibiotic concentrations used were: 50 μg/mL ampicillin, 25 μg/mL chloramphenicol. All antibiotics and chemicals were obtained from Sigma Aldrich (St. Louis, MO) unless otherwise noted. All strains are listed in S1 Table.
Evolution of methionine excreting S. enterica mutants
The two-step selection process for evolving a methionine-excreting S. enterica LT2 mutant is described in Harcombe 2010. Initial selection on ethionine, a competitive methionine analog, was again utilized to create new evolutionary ancestors from S. enterica serovar Typhimurium 14028s. Co-culturing of ethionine-resistant S. enterica and E. coli ΔmetB on lactose Hypho agarose plates to select for methionine excretion proceeded as described [17]. Briefly, 107 ethionine-resistant S. enterica cells grown overnight in Hypho galactose liquid media from a single colony were plated on lactose Hypho agarose plates with 107 E. coli ΔmetB cells grown overnight in Hypho lactose liquid media supplemented with 100 μM methionine. The bacteria were allowed to grow for 3 days at 37°C before the cells were scraped off, vortexed, and added to a fresh plate at 1/10 dilution. This second plate was allowed to grow for 3–5 days, or until colonies indicating co-growth of S. enterica and E. coli appeared. S. enterica strains were isolated from these colonies and tested for co-growth with E. coli ΔmetB in Hypho lactose liquid and solid media.
Genomic sequencing
Genomic DNA from S. enterica strains LT2, 14028s, and R2P4 was extracted from lysed cells using Wizard Genomic DNA Purification Kit (Promega, Madison, WI), and prepared for Illumina sequencing using the TrueSeq kit (Illumina, San Diego, CA). Samples were sent to The Microarray and Genomic Analysis Core facility at the University of Utah for sequencing on Illumina HiSeq 2000 sequencer, and aligned and analyzed using breseq, with all default users settings other than enabled polymorphism prediction [42] (http://www.barricklab.org/breseq).
Plasmids
Gene replacement amplicons utilized the chloramphenicol-containing pKD32 plasmid as a template, and pKD46 as a helper plasmid [43].
Structural analysis of HTS
Homology modeling of HTS (residues 2–297) from S. enterica was performed in SWISS-MODEL [44,45] using homoserine o-succinyltransferase from Bacillus cereus (PDB 2ghrA; 49% amino acid identity) as the template. Alignment of model to 2h2wA, as well as model visualization were performed in PyMOL [46].
Molecular modeling of metA mutations
Changes in protein folding stabilities and protein-protein binding stabilities were estimated using FoldX [47,48]. FoldX was chosen for this study to balance accuracy and speed [49,50]. Given the large number of substitutions studied here, it is not possible to use accurate statistical mechanical approaches such as all atom molecular dynamics simulation as done in previous studies [51,52]. Using an in-house script a total of 5016 mutations were generated and analyzed (each of the 264 residues in the protein structure mutated to one of other 19 types) for both the unbound monomer and bound dimer forms. The monomer and dimer structures were initially equilibrated six times in succession using "RepairPDB" to obtain a minimalized conformation, and mutant forms of both monomer and dimer were generated using “BuildModel”. Protein folding stabilities were then estimated using "Stability" on the monomer structures and protein-protein binding stabilities were estimated using “AnalyseComplex” on the dimer structures.
The p-values were calculated based on the null hypothesis that the folding and binding stabilities of the six experimental substitutions are random samples from the distribution of all possible substitutions. A non-parametric sum of ranks method (Mann-Whitney U test) was used for both folding and binding. The 5016 stability values for stability were first ranked in order of most stabilizing (smallest ΔΔG value) to least stabilizing (largest ΔΔG value). Tied values received a rank equal to the average of the ranks they span. One million random samples of six ΔΔG values were then drawn from the distribution. The sum of ranks were calculated for each draw and compared to the sum of ranks for the six experimental substitutions. The p-value was calculated as the number of draws where the sum of ranks for the random draw was less than or equal to the sum of ranks for the experimental substitutions. The calculation was performed five times for both binding and folding to ensure consistent results.
Gene Disruption
Gene disruptions were performed using the method of Datsenko and Wanner [43] with modifications described by Ellermeier et al. [53]. A selectable chloramphenicol marker (cat) flanked by 40 bp of the region surrounding the coding region of metA was constructed via PCR using plasmid pKD32 as template [38] and primers listed in S2 Table. PCR products were cleaned using QiaQuick PCR Purification kit (Qiagen) and electroporated into electrocompetent S. enterica cells carrying lambda Red helper plasmid pKD46 [43]. Cells were suspended in LB and recovered for 1 hr shaking at 37°C before being spread on selective media. Cells were purified once more selectively at 37°C before ΔmetA::cat insertion was verified via PCR.
P22 Transduction
To create lysates for P22 transduction, S. enterica donor strains were grown overnight, and then diluted 1:500 in 5mL LB+cat with 150 μL P22 HT int lysate stock and grown with shaking at 37°C for approximately 6 hours. After vortexing with 1 mL chloroform to kill remaining donor cells and centrifuging 10 minutes at 4550 x g to remove debris, lysate was stored at 4°C for up to 3 years. 200 μL overnight culture of recipient S. enterica strains were incubated with 100 μL lysate for 25 minutes at room temperature, rinsed twice with 100 mM sodium citrate LB, plated onto selective media, and grown overnight. After purifying once more selectively at 37°C, strains were cross-streaked against lytic P22 H5 lysate to test for remaining presence of phage.
Allele Replacement
Native metA loci were deleted via P22 transduction of ΔmetA::cat and selection on LB+ chloramphenicol. Cured ΔmetA::cat strains received replacement loci via P22 transduction of donor strains containing the desired new metA allele and selection on glucose minimal media. Amplification and sequencing of new metA locus confirmed no additional mutations were introduced.
Methionine measurements
Methionine measurements via GC-MS closely followed the method of Zamboni et al. [54]. To obtain conditioned media samples, overnight cultures of S. enterica strains were transferred at a dilution ranging from 50- to 200-fold into 30 mL galactose minimal media, and grown to mid-log phase in shaking flasks at 30°C. The E. coli release galactose and acetate, however, we chose to solely provide galactose to prevent the complexity of diauxic growth in batch culture. Galactose concentrations were also chosen to provide for sufficient growth of the S. enterica strains, rather than to imitate the levels experienced during co-culture experiments, and were kept the same across these strains. It should also be noted that we are using OD600 as a proxy for total biomass, rather than cell number per se, as it is the former that we wish to normalize production to. Samples were collected at mid-log phase to prevent depletion of exogenous methionine by S. enterica catabolism in stationary phase. Cultures were then pelleted for 10 minutes at 4°C and 4550 x g, and then filtered to complete removal of cells. After freezing at -80°C, thawed conditioned media was passed through solid phase extraction Chromaband Easy columns per manufacturer directions (Macherey-Nagel) and eluted in methanol. After removal of methanol in a vacuum centrifuge, and resuspension in 40 μL dimethylformamide, samples were placed into glass vials and derivatized for 1 hour at 85°C with 40 μL N-(tertbutyldimethylsilyl)-N-methyltrifluoroacetamide with 1% (wt/wt) tertbutyldimethyl-chlorosilane (Sigma). Derivatized samples were then immediately injected into a Shimadzu QP2010 GCMS (Columbia, MD). The injection source was 230°C. The oven was held at 160°C for 1 minute, increased to 310°C by 20°C min-1, and held at 310°C for 30 seconds. Column flow rate was 1.04 mL/min and the split ratio was 1.0. The column was a 30 m Rxi-1ms (Restek, Bellefonte, PA). Results were analyzed in GC-MS Postrun Analysis (Verison 2.70, Shimadzu, Kyoto, Japan). Methionine peak area was compared to an internal standard of 100 μM isoleucine added directly to the supernatants after thawing (S3 Fig). Experimental methionine concentrations were determined by a calibration curve, and then divided by the conditioned media culture’s final optical density as a proxy for total biomass. Methionine concentrations are given as μM/OD600 and represent a minimum of three biological replicates, each with three technical replicates.
Growth rate analysis
Strains were acclimated by inoculating single colonies into 640 μL medium in Costar 48-well cell culture plates (Product #3548, Corning Life Sciences, Tewksbury, MA) and placed in a humidified plate shaking tower (Liconic) at 30°C overnight. For growth rate measurements, strains were then transferred with a 1:1280 dilution into fresh medium and returned to the shaking tower. Optical densities were obtained every 30 minutes (individual growth) or 60 minutes (consortia growth) on a Wallac Victor 2 plate reader (Perkin-Elmer) until cultures reached saturation, using an automated measurement system [55]. Growth rates were quantified by fitting the data to an exponential growth model using custom analysis software [41] and averaging a minimum of three biological replicates.
Consortia Composition
S. enterica strains were labeled with a single chromosomal copy of Venus via transduction from LC1589 E. coli strain was labeled with a single chromosomal copy of Cerulean via P1 transduction from strain LC1511. Growth rates of the resulting fluorescent strains did not differ significantly from unlabeled strains. Hypho lactose liquid cultures were inoculated as described above, and 450 μL samples from replicate cultures in the same 48-well plate were collected at inoculation, mid-log, and stationary phase, and frozen at -80°C in 10% DMSO. 25 mL Hypho lactose agarose plates were inoculated with 50 μL of each YFP S. enterica strain and 50 μL of the CFP E. coli strain and incubated at 30°C for 72 hours before transfer. To transfer, 1 ml of Hypho added to the surface of plates, scraped, vortexed, and then 100 μL was transferred to new plates. Remaining culture from initial inoculation and each subsequent scraping were frozen at -80°C in 10% DMSO until analysis. Cytometry samples were diluted 1:1000 in PBS and analyzed on BD LSRFortessa II (BD, San Jose, CA). CFP-labeled E. coli and YFP-labeled S. enterica monoculture and non-fluorescent S. enterica were used determine cutoffs for 40,000 events gathered using the 405 nm and 488 nm lasers.
Supporting Information
Wild-type with the evolved alleles of metA cooperate more than with the original allele, but significantly less than the respective evolved isolates. Asterisks indicate presence of native allele of that background (i.e., no substitution).
(DOCX)
YFP-labeled S. enterica and CFP-labeled E. coli ΔmetB grown liquid media were sampled at various time points, and consortia composition determined via flow-cytometery. Each data point represents three biological replicates, each with three technical replicates.
(DOCX)
This sample is one biological replicate of R1P2, with the methionine and isoleucine peaks clearly visible. Fragment patterns at each peak match those of identified derivatized amino acids. Area under each peak corresponds to amino acid quantity.
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Lionello Bossi for pSUB11 and the members of the Marx lab, Stephen Lory, Andrew Murray, Karine Gibbs, and two anonymous reviewers for their manuscript suggestions and feedback.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded with an NIH (www.nih.gov) NRSA Award to WRH and a DOE (www.energy.gov) award to CJM (DE-SC0006731). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.The Human Microbiome Consortium (2012) Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3564958&tool=pmcentrez&rendertype=abstract. Accessed 19 February 2014. 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma B, Forney LJ, Ravel J (2012) The vaginal microbiome: rethinking health and diseases. Annu Rev Microbiol 66: 371–389. 10.1146/annurev-micro-092611-150157 The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smid EJ, Lacroix C (2013) Microbe-microbe interactions in mixed culture food fermentations. Curr Opin Biotechnol 24: 148–154. Available: http://www.ncbi.nlm.nih.gov/pubmed/23228389. Accessed 11 March 2014. 10.1016/j.copbio.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 4.Wagner M, Loy A, Nogueira R, Purkhold U, Lee N, Daims H. (2002) Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek 81: 665–680. Available: http://www.ncbi.nlm.nih.gov/pubmed/12448762. [DOI] [PubMed] [Google Scholar]
- 5.Paliwal V, Puranik S, Purohit HJ (2011) Integrated perspective for effective bioremediation. Appl Biochem Biotechnol 166: 903–924. Available: http://link.springer.com/10.1007/s12010-011-9479-5. Accessed 7 February 2014. 10.1007/s12010-011-9479-5 [DOI] [PubMed] [Google Scholar]
- 6.Liow LH, Van Valen L, Stenseth NC (2011) Red Queen: from populations to taxa and communities. Trends Ecol Evol 26: 349–358. Available: http://www.ncbi.nlm.nih.gov/pubmed/21511358. Accessed 20 March 2014. 10.1016/j.tree.2011.03.016 [DOI] [PubMed] [Google Scholar]
- 7.Lawrence D, Fiegna F, Behrends V, Bundy JG, Phillimore AB, Bell T, et al. (2012) Species interactions alter evolutionary responses to a novel environment. PLoS Biol 10: e1001330 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3352820&tool=pmcentrez&rendertype=abstract. Accessed 24 March 2014. 10.1371/journal.pbio.1001330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West S, Griffin AS, Gardner A, Diggle SP (2006) Social evolution theory for microorganisms. Nat Rev Microbiol 4: 597–607. Available: http://www.ncbi.nlm.nih.gov/pubmed/16845430. Accessed 20 February 2014. [DOI] [PubMed] [Google Scholar]
- 9.Hillesland KL, Stahl DA (2010) Rapid evolution of stability and productivity at the origin of a microbial mutualism. Proc Natl Acad Sci USA. 107(5): 2124–2129. 10.1073/pnas.0908456107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrade-Dominguez A, Salazar E, Vargas-Lagunas MC, Kolter R, Encarnacio S. (2014) Eco-evolutionary feedbacks drive species interactions. ISME. 8: 1041–1054. 10.1038/ismej.2013.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rainey PB, Travisano M (1998) Adaptive radiation in a heterogeneous environment. Nature 32: 69–72. [DOI] [PubMed] [Google Scholar]
- 12.Chou H-H, Marx CJ (2012) Optimization of gene expression through divergent mutational paths. Cell Rep 1: 133–140. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3407975&tool=pmcentrez&rendertype=abstract. Accessed 25 March 2014. 10.1016/j.celrep.2011.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE (2006) Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci U S A 103: 9107–9112. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1482574&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, et al. (2012) The molecular diversity of adaptive convergence. Science 335: 457–461. Available: http://www.ncbi.nlm.nih.gov/pubmed/22282810. Accessed 19 March 2014. 10.1126/science.1212986 [DOI] [PubMed] [Google Scholar]
- 15.Kim W, Racimo F, Schluter J, Levy SB, Foster KR (2014) Importance of positioning for microbial evolution. Proc Natl Acad Sci U S A 111: E1639–E1647. Available: http://www.ncbi.nlm.nih.gov/pubmed/24715732. 10.1073/pnas.1323632111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacLean RC, Buckling A (2009) The distribution of fitness effects of beneficial mutations in Pseudomonas aeruginosa. PLoS Genet 5: e1000406 Available: http://dx.plos.org/10.1371/journal.pgen.1000406. 10.1371/journal.pgen.1000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harcombe W (2010) Novel cooperation experimentally evolved between species. Evolution 64: 2166–2172. Available: http://www.ncbi.nlm.nih.gov/pubmed/20100214. Accessed 3 March 2014. 10.1111/j.1558-5646.2010.00959.x [DOI] [PubMed] [Google Scholar]
- 18.Old IG, Phillips SE, Stockley PG, Saint Girons I (1991) Regulation of methionine biosynthesis in the Enterobacteriaceae. Prog Biophys Mol Biol 56: 145–185. Available: http://www.ncbi.nlm.nih.gov/pubmed/1771231. [DOI] [PubMed] [Google Scholar]
- 19.Chubiz L, Douglas S, Harcombe W (2014) Combining engineering and evolution to create novel metabolic mutualisms between species Engingeering and Analzying Multicellular Systems. New York: Springer; pp. 39–49. [DOI] [PubMed] [Google Scholar]
- 20.Lawrence DA, Smith DA (1968) Regulation of methionine synthesis in Salmonella typhimurium: Mutants resistant to inhibition by analogues of methionine. 58: 473–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lawrence DA (1972) Regulation of methionine feedback-sensitive enzyme in mutants of Salmonella typhimurium. J Bacteriol 109: 8–11. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=247244&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hobson AC, Smith DA (1973) S-adenosylmethionine synthetase in methionine regulatory mutants of Salmonella typhimurium. Mol Gen Genet 126: 7–18. Available: http://www.ncbi.nlm.nih.gov/pubmed/4591373. [DOI] [PubMed] [Google Scholar]
- 23.Jarvik T, Smillie C, Groisman EA, Ochman H (2010) Short-term signatures of evolutionary change in the Salmonella enterica serovar typhimurium 14028 genome. J Bacteriol 192: 560–567. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2805332&tool=pmcentrez&rendertype=abstract. Accessed 26 March 2014. 10.1128/JB.01233-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerrish PJ, Lenski RE (1998) The fate of competing beneficial mutations in an asexual population. Genetica 102–103: 127–144. Available: http://www.ncbi.nlm.nih.gov/pubmed/9720276. [PubMed] [Google Scholar]
- 25.Rozen DE, de Visser JA, Gerrish PJ (2002) Fitness effects of fixed beneficial mutations in microbial populations. Curr Biol 12: 1040–1045. [DOI] [PubMed] [Google Scholar]
- 26.Hegreness M, Shoresh N, Hartl D, Kishony R (2006) An equivalence principle for the incorporation of favorable mutations in asexual populations. Science 311: 1615–1617. 10.1126/science.1122469 [DOI] [PubMed] [Google Scholar]
- 27.Herron MD, Doebeli M (2013) Parallel evolutionary dynamics of adaptive diversification in Escherichia coli. PLoS Biol 11: e1001490 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3576414&tool=pmcentrez&rendertype=abstract. Accessed 28 May 2014. 10.1371/journal.pbio.1001490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinnersley M, Wenger J, Kroll E, Adams J, Sherlock G, Rosenzweig F. (2014) Ex uno plures: clonal reinforcement drives evolution of a simple microbial community. PLoS Genet 10: e1004430 Available: 10.1371/journal.pgen.1004430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kümmerli R, Santorelli L a., Granato ET, Dumas Z, Dobay A, Griffin AS, et al. (2015) Co-evolutionary dynamics between public good producers and cheats in the bacterium Pseudomonas aeruginosa. J Evol Biol. Available: http://doi.wiley.com/10.1111/jeb.12751. [DOI] [PubMed] [Google Scholar]
- 30.Kascer H, Burns JA (1973) The control of flux. Symp Soc Exp Biol 27:65–104. [PubMed] [Google Scholar]
- 31.Biran D, Gur E, Gollan L, Ron EZ (2000) Control of methionine biosynthesis in Escherichia coli by proteolysis. Mol Microbiol 37: 1436–1443. [DOI] [PubMed] [Google Scholar]
- 32.Mordukhova EA, Lee H-S, Pan J-G (2008) Improved thermostability and acetic acid tolerance of Escherichia coli via directed evolution of homoserine O-succinyltransferase. Appl Environ Microbiol 74: 7660–7668. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2607180&tool=pmcentrez&rendertype=abstract. Accessed 3 March 2014. 10.1128/AEM.00654-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Usuda Y, Kurahashi O (2005) Effects of deregulation of methionine biosynthesis on methionine excretion in Escherichia coli. Appl Environ Microbiol 71: 3228–3234. 10.1128/AEM.71.6.3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman TD, Michel J-B, Aingaran M, Potter-Bynoe G, Roux D, Davis MR Jr, et al. (2011) Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 43: 1275–1280. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3245322&tool=pmcentrez&rendertype=abstract. Accessed 21 March 2014. 10.1038/ng.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper TF, Rozen DE, Lenski RE (2003) Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. PNAS 100: 1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woods RJ, Barrick JE, Cooper TF, Shrestha U, Kauth MR, Lenski RE. (2011) Second-order selection for evolvability in a large Escherichia coli population. Science 331: 1433–1436. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3176658&tool=pmcentrez&rendertype=abstract. Accessed 21 March 2014. 10.1126/science.1198914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plucain J, Hindré T, Le Gac M, Tenaillon O, Cruveiller S, Médigue C, et al. (2014) Epistasis and allele specificity in the emergence of a stable polymorphism in Escherichia coli. Science 343: 1366–1369. Available: http://www.ncbi.nlm.nih.gov/pubmed/24603152. Accessed 20 March 2014. 10.1126/science.1248688 [DOI] [PubMed] [Google Scholar]
- 38.Chubiz LM, Granger BR, Segrè D, and Harcombe WR 2015. Species interactions differ in their genetic robustness. Front Microbiol. 6:271 10.3389/fmicb.2015.00271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Momeni B, Brileya KA, Fields MW, Shou W. 2013. Strong inter-population cooperation leads to partner intermixing in microbial communities. eLife 2:e00230 10.7554/eLife.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marx CJ (2013) Can you sequence ecology? Metagenomics of adaptive diversification. PLoS Biol 11: e1001487 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3576389&tool=pmcentrez&rendertype=abstract. Accessed 23 May 2014. 10.1371/journal.pbio.1001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delaney NF, Kaczmarek ME, Ward LM, Swanson PK, Lee M-C, Marx CJ. (2013) Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS One 8: e62957 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3639900&tool=pmcentrez&rendertype=abstract. Accessed 24 April 2014. 10.1371/journal.pone.0062957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, et al. (2009) Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461: 1243–1247. Available: http://www.ncbi.nlm.nih.gov/pubmed/19838166. Accessed 19 March 2014. 10.1038/nature08480 [DOI] [PubMed] [Google Scholar]
- 43.Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS 97: 6640–6645. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=18686&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. Available: http://www.ncbi.nlm.nih.gov/pubmed/16301204. Accessed 20 March 2014. [DOI] [PubMed] [Google Scholar]
- 45.Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37: D387–D392. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2686475&tool=pmcentrez&rendertype=abstract. Accessed 25 March 2014. 10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.The PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC. (n.d.).
- 47.Guerois R, Nielsen JE, Serrano L (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320: 369–387. 10.1016/S0022-2836(02)00442-4 [DOI] [PubMed] [Google Scholar]
- 48.Schymkowitz JWH, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L. (2005) Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci U S A 102: 10147–10152. 10.1073/pnas.0501980102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller CR, Lee KH, Wichman HA, Ytreberg FM (2014) Changing folding and binding stability in a viral coat protein: a comparison between substitutions accessible through mutation and those fixed by natural selection. PLoS One 9: e112988 Available: http://dx.plos.org/10.1371/journal.pone.0112988. 10.1371/journal.pone.0112988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee KH, Miller CR, Nagel AC, Wichman HA, Joyce P, Ytreberg FM. (2011) First-step mutations for adaptation at elevated temperature increase capsid stability in a virus. PLoS One 6 10.1371/journal.pone.0025640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhan YA, Wu H, Powell AT, Daughdrill GW, Ytreberg FM (2013) Impact of the K24N mutation on the transactivation domain of p53 and its binding to murine double-minute clone 2. Proteins Struct Funct Bioinforma 81: 1738–1747. 10.1002/prot.24310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shyu C, Cavileer TD, Nagler JJ, Ytreberg FM (2011) Computational estimation of rainbow trout estrogen receptor binding affinities for environmental estrogens. Toxicol Appl Pharmacol 250: 322–326. Available: 10.1016/j.taap.2010.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ellermeier CD, Janakiraman A, Slauch JM (2002) Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290: 153–161. Available: http://www.ncbi.nlm.nih.gov/pubmed/12062810. [DOI] [PubMed] [Google Scholar]
- 54.Zamboni N, Fendt S-M, Rühl M, Sauer U (2009) (13)C-based metabolic flux analysis. Nat Protoc 4: 878–892. Available: http://www.ncbi.nlm.nih.gov/pubmed/19478804. Accessed 20 March 2014. 10.1038/nprot.2009.58 [DOI] [PubMed] [Google Scholar]
- 55.Delaney NF, Rojas Echenique JI, Marx CJ (2013) Clarity: an open-source manager for laboratory automation. J Lab Autom 18: 171–177. 10.1177/2211068212460237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild-type with the evolved alleles of metA cooperate more than with the original allele, but significantly less than the respective evolved isolates. Asterisks indicate presence of native allele of that background (i.e., no substitution).
(DOCX)
YFP-labeled S. enterica and CFP-labeled E. coli ΔmetB grown liquid media were sampled at various time points, and consortia composition determined via flow-cytometery. Each data point represents three biological replicates, each with three technical replicates.
(DOCX)
This sample is one biological replicate of R1P2, with the methionine and isoleucine peaks clearly visible. Fragment patterns at each peak match those of identified derivatized amino acids. Area under each peak corresponds to amino acid quantity.
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.