Abstract
The nuclear envelope (NE) physically separates the cytoplasmic and nuclear compartments. While this barrier provides advantages, it also presents a challenge for the nuclear export of large ribonucleoprotein (RNP) complexes. Decades-old dogma holds that all such border-crossing is via the nuclear pore complex (NPC). However, the diameter of the NPC central channel limits the passage of large cargos. Here, we review evidence that such large RNPs employ an endogenous NE-budding pathway, previously thought to be exclusive to the nuclear egress of Herpes viruses. We discuss this and other models proposed, the likelihood that this pathway is conserved, and the consequences of disrupting NE-budding for synapse development, localized translation of synaptic mRNAs, and laminopathies inducing accelerated aging.
Introduction
Envelopment of genomic material by a double membrane, a hallmark of eukaryotic cells, was first described over a hundred years ago [1] and likely first occurred some 3 billion years ago. The segregation of the nucleoplasm from the cytosol enabled the spatial-temporal separation of translation from transcription [2]. This, in turn, allowed the evolution of additional regulatory levels necessary for the development of complex multi-cellular organisms, e.g., RNA export and cytoplasmic trafficking, or spatially restricted post-translational control, such as phosphorylation, ubiquitinylation and sumoylation. The nuclear envelope (NE) is composed of an outer nuclear membrane (ONM), which is continuous with the endoplasmic reticulum (ER), and an inner nuclear membrane (INM) that faces the nucleoplasm. Beneath the INM there is a dense meshwork, the lamina, primarily comprising intermediate filament proteins, the Lamins. Far from representing a passive component of the nucleoskeleton, the Lamins play dynamic roles in nuclear mechanical stability, chromatin organization, regulation of gene expression, genome stability, and cell division and differentiation [3]. Lamins often exist in two flavors, A-type Lamins (encoded by the LMNA gene in mammals, which generates two isoforms, LMNA and LMNC, and the lamC gene in Drosophila), and B-type Lamins (encoded by LMNB1 and LMNB2 in mammals and lamDm0 in flies). While B-type Lamins are expressed from the earliest stages of cellular differentiation and are required for cell viability, Atype Lamins are usually present in differentiated cells and are not required for cell survival. Indeed, animals such as C. elegans lack A-type Lamin, expressing only B-type Lamin. In humans a very large number of mutations (300+) in the LMNA gene have been identified, causing tissue-specific dystrophies and early aging, and collectively known as laminopathies [4].
Until recently it had been assumed that, except during early phases of mitosis, when the NE and lamina break down to be subsequently reassembled during telophase, the Nuclear Pore Complex (NPC) was the only gateway in and out of the nucleus. The NPC, first described over 50 years ago [5], is a ~125MDa protein complex, with ~3000 of them spanning each higher eukaryotic cell NE. It consists of some 500 proteins, primarily multiple copies of ~30 distinct nucleoporins that form a central channel through which cargo is transported in and out of the nucleus in a highly-regulated fashion [6, 7]. Both passive diffusion and facilitated transport, enabled by nuclear localization and export sequences within proteins and RNAs, as well as NPC-associated elements, are supported by the NPC.
A major problem with postulating the NPC as the sole route of nucleo-cytoplasmic transport is the relatively small size of molecular complexes that can transverse through its central channel. Studies using gold particles complexed to nuclear-localizing cargo of various sizes, used as molecular calipers to measure the size of the NPC central channel, placed its diameter at ~39 nm [8]. Moreover, recent structural studies indicate that the NPC channel is relatively flexible, being able to stretch up to 52 nm in diameter [9, 10]. There has been, however, a significant number of studies in diverse eukaryotic cells and tissues indicating the existence of much larger (100–700nm) RNPs in the cytoplasm [11–15]. Given the NPC size constrains it has been proposed that these super-large RNPs are assembled in the cytoplasm and that RNAs exit the nucleus one by one through the NPC. A potential mechanism to explain the nuclear export of RNPs larger than the maximum diameter of the NPC central channel has been suggested in the study of Balbiani ring RNPs which are synthesized in the nucleus of larval salivary gland cells of the dipteran Chironomus tentans, with a ~50 nm in diameter. Within the nucleus, these RNPs form a ribbon which assumes a ring-like conformation [16]. During RNP extrusion through the NPC, they adopt an elongated conformation, enabling their passage through the NPC [16]. The transit of other “oversized” RNPs from the nucleus to the cytoplasm might also be accounted for by the physical properties of RNPs, in which components appear to exist in a condensed liquid phase [17], enabling a degree of deformability. Thus, flexibility of either NPC and/or granule structure allows some larger RNPs to pass through the NPC.
An alternative, NPC-independent mechanism for nuclear exit, has been uncovered from studies of the nuclear egress of Herpes viruses (HVs). HV nucleocapsids, themselves large nucleoproteins, are assembled in the nucleus and must escape to the cytoplasm for maturation and release from the cell. This mechanism, NE-budding, appears to be a conserved mechanism for the nuclear export of very large RNPs which are assembled in the nucleus.
HV egress from the nucleus
HVs are clinically-important double-stranded DNA viruses endemic in human and animal populations and induce infections such as oral and genital herpes caused by Herpes simplex, chicken pox and shingles, caused by Varicella zoster, mononucleosis, caused by Barr-Epstein virus, and Kaposi’s sarcoma caused by Human herpesvirus 8. As is typical of viral replication, both viral and cellular proteins are employed through all phases of infection, replication, maturation and shedding from the host cell. Given the large size of HV nucleocapsids (~ 125 nm) they escape from the nucleus though a process of NE membrane remodelling, involving envelopment and de-envelopment of the nucleocapsid at the NE [18–24]. A widely accepted model of this process is that nucleocapsids disrupt the nuclear lamina, gaining access to the INM. Subsequently, they undergo primary envelopment by deforming the INM, thus budding into the perinuclear space (between INM and ONM; Fig. 1A). In this way, perinuclear nucleocapsids become enveloped by an INM coat. During de-envelopment, the capsid INM coat fuses with the ONM, liberating a naked capsid into the cytoplasm (Fig. 1A) for secondary envelopment at the Golgi and release from the cell.
Figure 1. Nuclear egress of HV nucleocapsids.
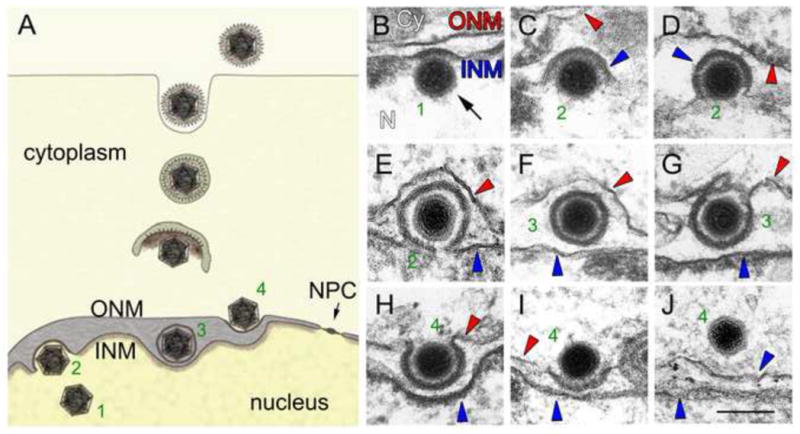
A diagrammatic representation of the envelopment and de-envelopment process at the NE (A). Secondary envelopment of capsids in the cytoplasm and release of mature viral particles are also shown. Transmission electron micrographs of different stages of HV nuclear egress, showing nucleocapsid docking (B), primary envelopment at the INM (C, D, E), residence at the perinuclear space (F), de-envelopment at the ONM (G–I) and escape to the cytoplasm (J). Panels A and B–J are from Figure 1C and Figure 2, respectively, from [21] and are used with permission. The numbers shown in (B–J) correspond to the stages similarly numbered in (A).
The so-called Nuclear Egress Complex (NEC), required for primary envelopment at the INM, has been well characterized [20, 22–26]. For simplicity, here we use the Herpes Simplex Virus-1 (HSV-1) protein nomenclature, although similar viruses encode orthologs which may bear different names. The HSV-1 NEC requires a complex of two virally-encoded proteins, pUL34 and pUL31. In the absence of the NEC, nuclear egress of viral particles ceases and capsids in various states of maturity accumulate in the nucleus.
The NEC also recruits viral and cellular kinases which locally phosphorylate INM-associated lamina proteins, leading to a disruption of the lamina at these sites [22, 23, 26]. There is some specificity to the Lamin-kinase interactions, as viral kinases, pUS3 and pUL13 primarily phosphorylate the A/C-Lamins, while cellular Protein Kinase C (PKC) phosphorylates the A- and B-Lamins [26, 27].
In addition to PKC, the cellularly-encoded AAA+ superfamily member TorsinA (TorA) also appears to be involved in HV nuclear egress in some cells [28]. TorA plays roles in maintaining NE architecture and ER function [29] and mutations in TorA underlie early onset torsion dystonia in humans, a movement disorder [30–32].
Overexpressing wild type TorA in HSV-1-infected cells reduced viral replication leading to the accumulation of membrane-bound granules that retain pUL34 [28]. These data indicate that TorA might play a role early in the NE-budding process, possibly during the invagination of the INM-covered particle into the perinuclear space or in its dissociation from the INM. In contrast, complete elimination of the TorA/B coding regions in double knockout Hela cell lines resulted in minimal effects on viral production [33]. This discrepancy between the overexpression and knockout studies remains to be resolved.
A flurry of recent structural studies of the different NECs have provided mechanistic insights as to how pUL34 and pUL31 interact with each other, the immature virion and the lamina to effect NE-budding [34–42]. Together with earlier work, they support a model where tightly-interacting pUL34 and pUL31dimers bind simultaneously to the surface of the nucleocapsid and an unknown partner(s) on the nucleoplasmic side of the INM, to dock the capsid. These dimers multimerize, eventually completely coating the capsid, and inducing the changes in INM curvature required for budding into the perinuclear space. Strikingly, expression of the two NEC components in uninfected cells results in the formation of “empty”, but correctly sized, vesicles in the perinuclear space, between the IMN and ONM [43, 44]. This indicates that at least the initial steps of NE-budding do not require docking of a viral particle. Similarly, studies using artificial membranes or liposomes show that pUL31 and pUL34 are sufficient to deform membranes to induce vesiculation (reviewed in [23]). In a giant unilamellar vesicle in vitro model, pUL34, which has been shown to recruit pUL31 to the membrane, facilitates membrane budding and abscission [45]. The requirement for pUL34 can be bypassed by tethering pUL31 to the membrane, suggesting that membrane-localized pUL31 alone is sufficient for budding and abscission.
The Fz2 nuclear import pathway and the nuclear egress of large RNPs
The first clues suggesting the presence of an endogenous cellular NE-budding pathway similar to HV nuclear egress came from an unexpected direction: the study of the roles of trans-synaptic Wnt signalling at the developing Drosophila neuromuscular junction (NMJ). Wnts are secreted glycoproteins, with myriad of roles in development throughout the animal kingdom, which bind and signal through various receptors, including the Frizzled (Fz) family of receptors [46].
Wingless (Wg; mammalian Wnt-1 ortholog) is secreted by motor-neuron presynaptic terminals and binds to Fz2 receptors on the opposing muscle membrane [47, 48], a ruffled membrane called the subsynaptic reticulum (SSR; Fig. 2A, B). Reductions in signalling through this pathway result in aberrant NMJ growth, and incomplete differentiation of synaptic boutons (components of the NMJ that carry out neurotransmission), particularly the differentiation of postsynaptic structures. These processes rely on the so-called Frizzled Nuclear Import (FNI) Wg pathway. In the FNI pathway, Fz2 is endocytosed from the muscle cell surface and proteolytically-cleaved to yield a small cytoplasmic carboxyterminal fragment (Fz2C), which is imported into the muscle nuclei and incorporated into large nuclear Fz2C foci [47]. Fz2 endocytosis and proteolysis was suggested to be constitutive, while the nuclear translocation of Fz2C required Wg [47], the multiple PDZ domain adaptor, GRIP [49] and two members of the importin family [50], adaptors involved in importing proteins through the NPC.
Figure 2. The Frizzled Nuclear Import pathway and NE-budding at the Drosophila NMJ.
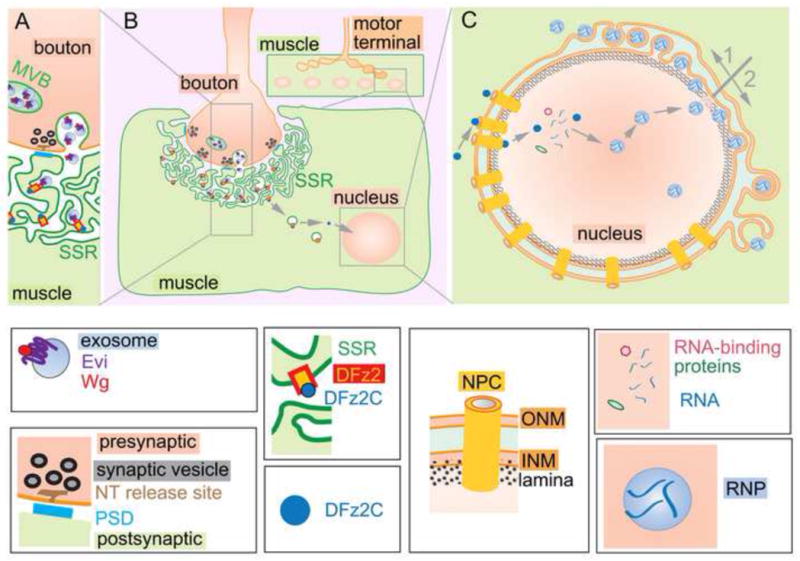
A high magnification view of the corresponding box in (B) schematically showing the release of Wg via exosomes by presynaptic boutons and the binding of Wg to Fz2 receptors at the ruffled surface of the muscle cell (the subsynaptic reticulum; SSR) is shown (A). The FNI pathway, in which Fz2 receptors are endocytosed at the SSR of the muscle, a cytoplasmic fragment of the receptor (DFz2C) is cleaved, and this fragment is imported into the nucleus (B). A schematic representation of the proposed endogenous NE-budding model, based on EM observations in Drosophila and mouse (C). DFz2C fragments are imported into the nucleus by a mechanism that require nucleoporins and become associated with large RNPs. These RNPs dock at the NE and disrupt the nuclear lamina though a mechanism involving aPKC. Subsequent remodeling of the INM and the ONM, either by pathway 1 (indicated in grey; arrow up) or by pathway 2 (arrow down). The key to the different proteins and structures depicted is shown below panels A–C. MVBs are multivesicular bodies and PSD is the postsynaptic density.
Examination of nuclear Fz2C foci by confocal microscopy followed by image reconstruction through deconvolution software revealed that the foci were composed of individual Fz2C granules associated with the NE, and were usually present in clusters. These Fz2C granules did not appear to colocalize with NPCs as determined both by light and electron microscopy (EM). However, the morphological criteria utilized in this study do not preclude the function of specific NPC components in forming the granules at the NE. The above studies also demonstrated that Fz2C granules were localized at the perinuclear space, between the INM and the ONM, were surrounded by membrane and a coat of lamC. They were also enriched in poly-adenylated RNAs, and sometimes contained Poly(A)-binding protein 2 (PABP2) [51]. Formation of these perinuclear RNPs required the phosphorylation of LamC by atypical-PKC (aPKC), similar to the requirement for cellular and viral Lamin-targeted kinases during HV nuclear egress [51]. The presence of Fz2C granules at the perinuclear space was also demonstrated by the lack of accessibility of fluorescent dextran of molecular mass above the NPC size cut-off injected into the muscle cytoplasm [51]. Thus, Fz2C granules appear to transiently reside at the perinuclear space, similar to HV nucleocapsids during their nuclear egress (Fig. 3).
Figure 3. Similarities between nuclear HV egress and cellular RNP NE-budding.
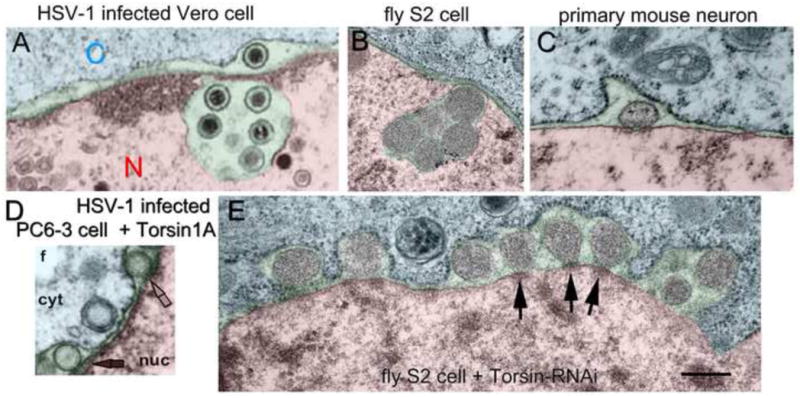
An HSV-1-infected Vero cell, displaying nucleocapsids in the perinuclear space (pseudo-colored in green) is shown (A). The cytoplasmic region and the nucleoplasm are pseudo-colored in blue and red, respectively. Note that both a cluster of nucleocapsids and a single nucleocapsid are seen in the perinuclear space. Nucleocapsids and large RNP clusters are often seen in convoluted enlargements of the perinuclear space. An RNP cluster localized at the perinuclear space of a S2 cell (B), resembling the nucleocapsids cluster is shown in (A). A single RNP observed in the perinuclear space of a mouse neuron (C). Nucleocapsids at the perinuclear space of an HSV-1-infected PC6-3 cell in which TorA is overexpressed (D). RNPs accumulating at the perinuclear space of an S2 cell treated with Torsin-dsRNA (E). Note the electron dense structures observed at the “necks” of RNPs which tether the RNPs to the INM resembling the nucleocapsids shown in (D). Also note the widely expanded perinuclear space. The calibration bar shown in (E) is 250 nm for all panels. Panel A is from Fig. 3E of [55], Panel D is from Fig. 2F of [28], Panel 3 is from Figure 2B from [52] and all are used with permission. Panels B and C are VB’s unpublished data.
What might be the function of large RNPs that exit the nucleus through NE-budding? Some of the first indications were derived from the observation that Fz2C RNPs contained transcripts encoding the scaffolding proteins Par6, MAGI as well as 4 other proteins that are normally enriched at the postsynaptic region of the NMJ, and thus likely locally translated at synaptic sites. Blocking NE-budding by downregulating or eliminating LamC as well as by mutations in Torsin (see below) eliminated the synaptic enrichment of par6 and/or magi RNA, suggesting that NE-budding serves as a pathway for the nuclear export of mRNAs that are localized at postsynaptic sites. Whether the large RNPs contain multiple RNA species or multiple copies of a single species remains to be determined.
Similar to its possible involvement in HV nuclear egress, NE-budding of large RNPs required the TorA ortholog Torsin, the sole Torsin member in Drosophila [52]. Treatment of Drosophila cultured S2 cells, which recapitulate many, if not all, aspects of the FNI/NE-budding pathway, with dsRNA targeting Torsin, or expressing mutant Torsin forms modelled after dystonia patients, resulted in perinuclear space accumulation of RNP granules. These granules appeared surrounded by INM, but were still tethered to the INM by a dense structure forming a neck (Fig. 3E). Notably, a very similar structure was observed at the perinuclear space of HV-infected cells when overexpressing TorA [28]. Expressing a Torsin substrate trap mutant (TorsinE→Q) led to an accumulation of the mutant protein at the neck of the budding granule, suggesting that the neck is the site of Torsin action. This led to the proposal that Torsin is likely involved in some aspect of the scission of the INM during primary envelopment of large RNPs. Finally, confirming and extending the previously mentioned observation that there are at least some localized transcripts within Fz2C-RNPs, postsynaptic levels of par6 and magi, were significantly reduced when Torsin action was blocked [52].
Together, these studies point to the existence of an endogenous cellular pathway that transports large RNPs to the cytoplasm via NE-budding. The similarity between NE-budding of RNPs and HV nuclear egress is striking and likely indicates that HVs have, once again, taken advantage of a host cell pathway to further their propagation. Is NE-budding conserved across species? Does it have tissue-specificity? So far observations in Drosophila indicate the presence of Fz2C RNPs in salivary gland epithelial cell nuclei, S2 cells derived from a macrophage cell lineage [51], somatic muscle cells, glial cells, and developing oocytes (V. Johki, S. Speese and V. B., unpublished), indicating that NE-budding is widely used in different tissue types. Large RNPs present between the INM and ONM have also been observed in neurons of young mice (Fig. 3), as well as in skeletal and cardiac muscle (J. Ashley, B. Ding and V. B., unpublished). As expected from large RNPs that exit the nucleus via NE-budding, these perinuclear granules are enriched in poly(A) RNA and often contain a mammalian Fz2 homolog, Fz8. NE-budding, however, appears to be transiently utilized during specific stages of cell differentiation, making it difficult to study. For example, only 10–30% of the cells of a given type that utilize NE-budding appear to contain large RNPs at the perinuclear space at any one time. Nevertheless, blocking NE-budding at specific stages (e.g., through mutations in Torsin) can result in the accumulation of RNPs between the two nuclear membranes in esssentially 100% of the cells. Together with the reports that similar structures are observed at the perinuclear space in both plants and animals [11–15], it seems probable that NE-budding of large RNPs is a widely-used eukaryotic strategy.
Given the key role played by A-type Lamin during NE-budding, we anticipate that at least some of the laminopathies resulting from mutations in LMNA might be the consequence of defective NE-budding. This is supported by unpublished studies modelling LMNA mutations in LamC after progeroid syndrome-causing LMNA mutations in the fly also result in premature aging in the fly (Y. Li and V. B., in preparation).
Finally, it has been observed that perinuclear vesicles in the conditional Torsin-deficient mouse, resembling those that contain RNA granules in the perinuclear space, contain polyubiquitinated proteins [53]. This has led to the suggestion that NE-budding might also serve in the disposal of large, obsolete, nuclear material [53, 54]. Although polyubiquitin aggregates have not been observed in perinuclear granules at the NMJ (S. Speese and V. B., unpublished), a multifunctional role for NE-budding to export both large RNPs and large protein aggregates is conceivable.
Future directions
While much remains to be understood about the endogenous NE-budding pathway employed by eukaryotic cells and likely co-opted by HVs, here we highlight several fundamental questions which, in our opinion, are current priorities to be addressed. First and foremost, what are the cellular orthologs of the virally-encoded NEC proteins? Proteomic study of isolated budding RNPs and screens for genes whose mutation block the early stages of nuclear egress should prove useful in answering this question. Secondly, what mechanisms control nuclear RNP formation and composition? Thirdly, how are post-budding RNPs targeted to their destinations? Finally, what additional cellular processed utilize NE-budding as a strategy to export cargo out of the nucleus?
More generally, further comparative studies of the common steps during HV vs RNP nuclear egress might be critical to our knowledge about a) which aspects of viral nuclear budding can be therapeutically targeted for the treatment of HV infections?, b) how localized translation of RNPs might be controlled, at least in part, by regulation of RNP nuclear budding rates or differential cargo loading? and c) What are the precise cargoes whose diminished export possibly underlies normal aging and/or laminopathies in general?
Acknowledgments
We apologize to our colleagues who we could not cite due to the strict number of references allowed in this review. We gratefully acknowledge Drs. Thomas Mettenleiter and Richard Roller for allowing us to use their published images and Drs. Thomas Mettenleiter and Barbara Klupp for their comments on the manuscript. This work is supported by NIH NS063228 to VB and the “Nederlandse Organisatie voor Wetenschappelijk Onderzoek” ZonMw TOPGrant (40-00812-98-10058) to LGF and Jasprina N. Noordermeer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
References of particular interest published in the period of review have been highlighted below.
* Papers of special interest
** papers of outstanding interest
- 1.Kite GL. Studies on the physical properties of protoplasm. American J Physiol. 1913:32. [Google Scholar]
- 2.Devos DP, Graf R, Field MC. Evolution of the nucleus. Curr Opin Cell Biol. 2014;28:8–15. doi: 10.1016/j.ceb.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–64. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber KH, Kennedy BK. When lamins go bad: nuclear structure and disease. Cell. 2013;152(6):1365–75. doi: 10.1016/j.cell.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callan HG, Tomlin SG. Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc R Soc Lond B Biol Sci. 1950;137(888):367–78. doi: 10.1098/rspb.1950.0047. [DOI] [PubMed] [Google Scholar]
- 6.Kabachinski G, Schwartz TU. The nuclear pore complex--structure and function at a glance. J Cell Sci. 2015;128(3):423–9. doi: 10.1242/jcs.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Biol. 2012;13(11):687–99. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- 8.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13(2):425–34. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solmaz SR, Blobel G, Melcak I. Ring cycle for dilating and constricting the nuclear pore. Proc Natl Acad Sci U S A. 2013;110(15):5858–63. doi: 10.1073/pnas.1302655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solmaz SR, Chauhan R, Blobel G, Melcak I. Molecular architecture of the transport channel of the nuclear pore complex. Cell. 2011;147(3):590–602. doi: 10.1016/j.cell.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson HG. Nucleo-Cytoplasmic Interaction following Meiosis in the Young Microspores of Lilium Longiflorum; Events at the Nuclear Envelope. Grana. 1971;11:2, 117–127. [Google Scholar]
- 12.Gay H. Nucleocytoplasmic relations in Drosophila. Cold Spring Harb Symp Quant Biol. 1956;21:257–69. doi: 10.1101/sqb.1956.021.01.021. [DOI] [PubMed] [Google Scholar]
- 13.Hadek R, Swift H. Nuclear extrusion and intracisternal inclusions in the rabbit blastocyst. J Cell Biol. 1962;13:445–51. doi: 10.1083/jcb.13.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochstrasser M, Sedat JW. Three-dimensional organization of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organization and gene regulation. J Cell Biol. 1987;104(6):1471–83. doi: 10.1083/jcb.104.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szollosi D. Extrusion of nucleoli from pronuclei of the rat. J Cell Biol. 1965;25(3):545–62. doi: 10.1083/jcb.25.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mehlin H, Skoglund U, Daneholt B. Transport of Balbiani ring granules through nuclear pores in Chironomus tentans. Exp Cell Res. 1991;193(1):72–7. doi: 10.1016/0014-4827(91)90539-7. [DOI] [PubMed] [Google Scholar]
- 17.Brangwynne CP. Phase transitions and size scaling of membrane-less organelles. J Cell Biol. 2013;203(6):875–81. doi: 10.1083/jcb.201308087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bigalke JM, Heldwein EE. The Great (Nuclear) Escape: New Insights into the Role of the Nuclear Egress Complex of Herpesviruses. J Virol. 2015;89(18):9150–3. doi: 10.1128/JVI.02530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol. 2011;9(5):382–94. doi: 10.1038/nrmicro2559. [DOI] [PubMed] [Google Scholar]
- 20.Klupp BG, Granzow H, Mettenleiter TC. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J Virol. 2000;74(21):10063–73. doi: 10.1128/jvi.74.21.10063-10073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mettenleiter TC, Muller F, Granzow H, Klupp BG. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol. 2013;15(2):170–8. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- 22.Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science. 2002;297(5582):854–7. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J Virol. 2001;75(18):8803–17. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J Virol. 2000;74(1):117–29. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang YE, Van Sant C, Krug PW, Sears AE, Roizman B. The null mutant of the U(L)31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J Virol. 1997;71(11):8307–15. doi: 10.1128/jvi.71.11.8307-8315.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marschall M, Feichtinger S, Milbradt J. Regulatory roles of protein kinases in cytomegalovirus replication. Adv Virus Res. 2011;80:69–101. doi: 10.1016/B978-0-12-385987-7.00004-X. [DOI] [PubMed] [Google Scholar]
- 27.Leach NR, Roller RJ. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology. 2010;406(1):127–37. doi: 10.1016/j.virol.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Maric M, Shao J, Ryan RJ, Wong CS, Gonzalez-Alegre P, Roller RJ. A functional role for TorsinA in herpes simplex virus 1 nuclear egress. J Virol. 2011;85(19):9667–79. doi: 10.1128/JVI.05314-11. This paper and reference 52 describe apparently similar roles for TorsinA in HSV and cellular RNP egress, respectively. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodchild RE, Kim CE, Dauer WT. Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron. 2005;48(6):923–32. doi: 10.1016/j.neuron.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Gerace L. TorsinA and torsion dystonia: Unraveling the architecture of the nuclear envelope. Proc Natl Acad Sci U S A. 2004;101(24):8839–40. doi: 10.1073/pnas.0402441101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Granata A, Warner TT. The role of torsinA in dystonia. Eur J Neurol. 2010;17(Suppl 1):81–7. doi: 10.1111/j.1468-1331.2010.03057.x. [DOI] [PubMed] [Google Scholar]
- 32.Laudermilch E, Schlieker C. Torsin ATPases: structural insights and functional perspectives. Curr Opin Cell Biol. 2016;40:1–7. doi: 10.1016/j.ceb.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *33.Turner EM, Brown RS, Laudermilch E, Tsai PL, Schlieker C. The Torsin Activator LULL1 Is Required for Efficient Growth of Herpes Simplex Virus 1. J Virol. 2015;89(16):8444–52. doi: 10.1128/JVI.01143-15. This paper reports that TorA/B double knockout Hela cells show almost wild type levels of HV production which contrasts the findings in TorA-overexpressing cells described in reference 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bigalke JM, Heldwein EE. Structural basis of membrane budding by the nuclear egress complex of herpesviruses. EMBO J. 2015;34(23):2921–36. doi: 10.15252/embj.201592359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bigalke JM, Heuser T, Nicastro D, Heldwein EE. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun. 2014;5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **36.Hagen C, Dent KC, Zeev-Ben-Mordehai T, Grange M, Bosse JB, Whittle C, Klupp BG, Siebert CA, Vasishtan D, Bauerlein FJ, et al. Structural Basis of Vesicle Formation at the Inner Nuclear Membrane. Cell. 2015;163(7):1692–701. doi: 10.1016/j.cell.2015.11.029. This tour de force structural characterization of the NEC, building on the authors’ and others’ previous work, employs multimodal imaging and x-ray scattering data to detail the roles of oligomerized viral protein dimers in forming the NEC coat which induces curvature of the INM prior to capsid budding into the nuclear cleft. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leigh KE, Sharma M, Mansueto MS, Boeszoermenyi A, Filman DJ, Hogle JM, Wagner G, Coen DM, Arthanari H. Structure of a herpesvirus nuclear egress complex subunit reveals an interaction groove that is essential for viral replication. Proc Natl Acad Sci U S A. 2015;112(29):9010–5. doi: 10.1073/pnas.1511140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lye MF, Sharma M, El Omari K, Filman DJ, Schuermann JP, Hogle JM, Coen DM. Unexpected features and mechanism of heterodimer formation of a herpesvirus nuclear egress complex. EMBO J. 2015;34(23):2937–52. doi: 10.15252/embj.201592651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Passvogel L, Janke U, Klupp BG, Granzow H, Mettenleiter TC. Identification of conserved amino acids in pUL34 which are critical for function of the pseudorabies virus nuclear egress complex. J Virol. 2014;88(11):6224–31. doi: 10.1128/JVI.00595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Passvogel L, Trube P, Schuster F, Klupp BG, Mettenleiter TC. Mapping of sequences in Pseudorabies virus pUL34 that are required for formation and function of the nuclear egress complex. J Virol. 2013;87(8):4475–85. doi: 10.1128/JVI.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walzer SA, Egerer-Sieber C, Sticht H, Sevvana M, Hohl K, Milbradt J, Muller YA, Marschall M. Crystal Structure of the Human Cytomegalovirus pUL50-pUL53 Core Nuclear Egress Complex Provides Insight into a Unique Assembly Scaffold for Virus-Host Protein Interactions. J Biol Chem. 2015;290(46):27452–8. doi: 10.1074/jbc.C115.686527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeev-Ben-Mordehai T, Weberruss M, Lorenz M, Cheleski J, Hellberg T, Whittle C, El Omari K, Vasishtan D, Dent KC, Harlos K, et al. Crystal Structure of the Herpesvirus Nuclear Egress Complex Provides Insights into Inner Nuclear Membrane Remodeling. Cell Rep. 2015;13(12):2645–52. doi: 10.1016/j.celrep.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai PJ, Pryce EN, Henson BW, Luitweiler EM, Cothran J. Reconstitution of the Kaposi’s sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J Virol. 2012;86(1):594–8. doi: 10.1128/JVI.05988-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci U S A. 2007;104(17):7241–6. doi: 10.1073/pnas.0701757104. This study established that expression of two virally-encoded NEC proteins is sufficient to bud out empty envelopes into the nuclear cleft in absence of viral infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.Lorenz M, Vollmer B, Unsay JD, Klupp BG, Garcia-Saez AJ, Mettenleiter TC, Antonin W. A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J Biol Chem. 2015;290(11):6962–74. doi: 10.1074/jbc.M114.627521. This study reported that the membrane-anchored pUL34 protein serves to recruit pUL31 which itself has the activity to cause membrane budding and abscission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 47.Mathew D, Ataman B, Chen J, Zhang Y, Cumberledge S, Budnik V. Wingless signaling at synapses is through cleavage and nuclear import of receptor DFrizzled2. Science. 2005;310(5752):1344–7. doi: 10.1126/science.1117051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Packard M, Koo ES, Gorczyca M, Sharpe J, Cumberledge S, Budnik V. The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell. 2002;111(3):319–30. doi: 10.1016/s0092-8674(02)01047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ataman B, Ashley J, Gorczyca D, Gorczyca M, Mathew D, Wichmann C, Sigrist SJ, Budnik V. Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP. Proc Natl Acad Sci U S A. 2006;103(20):7841–6. doi: 10.1073/pnas.0600387103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mosca TJ, Schwarz TL. The nuclear import of Frizzled2-C by Importins-beta11 and alpha2 promotes postsynaptic development. Nat Neurosci. 2010;13(8):935–43. doi: 10.1038/nn.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **51.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149(4):832–46. doi: 10.1016/j.cell.2012.03.032. The first report of large RNP budding from the nucleus and its roles in localizing synaptic mRNAs to the Drosophila neuromuscular junction which are required for proper synaptic development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52.Jokhi V, Ashley J, Nunnari J, Noma A, Ito N, Wakabayashi-Ito N, Moore MJ, Budnik V. Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 2013;3(4):988–95. doi: 10.1016/j.celrep.2013.03.015. See comment for reference 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang CC, Tanabe LM, Jou S, Chi F, Dauer WT. TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J Clin Invest. 2014;124(7):3080–92. doi: 10.1172/JCI72830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rose A, Schlieker C. Alternative nuclear transport for cellular protein quality control. Trends Cell Biol. 2012;22(10):509–14. doi: 10.1016/j.tcb.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Granzow H, Klupp BG, Fuchs W, Veits J, Osterrieder N, Mettenleiter TC. Egress of alphaherpesviruses: comparative ultrastructural study. J Virol. 2001;75(8):3675–84. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


